Abstract
Efficient gene delivery is a fundamental goal of biotechnology and has numerous applications in both basic and applied science. Substrate-mediated delivery and reverse transfection enhance gene transfer by increasing the concentration of DNA in the cellular microenvironment through immobilizing a plasmid to a cell culture substrate prior to cell seeding. In this report, we examine gene delivery of plasmids that were complexed with cationic polymers (polyplexes) or lipids (lipoplexes) and subsequently immobilized to cell culture or biomaterial substrates by adsorption. Polyplexes and lipoplexes were adsorbed to either tissue culture polystyrene or serum-adsorbed tissue culture polystyrene. The quantity of DNA immobilized increased with time of exposure, and the deposition rate and final amount deposited depended upon the properties of the substrate and complex. For polyplexes, serum modification enhanced reporter gene expression up to 1500-fold relative to unmodified substrates and yielded equivalent or greater expression compared to bolus delivery. For lipoplexes, serum modification significantly increased the number of transfected cells relative to unmodified substrates yet provided similar levels of expression. Immobilized complexes transfect primary cells with improved cellular viability relative to bolus delivery. Finally, this substrate-mediated delivery approach was extended to a widely used biomaterial, poly(lactide-co-glycolide). Immobilization of DNA complexes to tissue culture polystyrene substrates can be a useful tool for enhancing gene delivery for in vitro studies. Additionally, adapting this system to biomaterials may facilitate application to fields such as tissue engineering.
Keywords: gene delivery, reverse transfection, solid-phase delivery, lipofectamine 2000, polyethylenimine, biomaterial, substrate-mediated
INTRODUCTION
Controlled and efficient gene delivery and transgene expression is a fundamental goal of biotechnology, with numerous applications in both basic and applied science. Nonviral vectors provide a versatile, simple to implement, and cost-effective approach for gene delivery and are continually being developed to increase transfection efficiency while reducing cellular toxicity. Traditionally, nonviral vectors are formed by complexation of plasmid DNA with cationic lipids (e.g., Lipofectamine) or cationic polymers (e.g., polyethylenimine, PEI), which reduces the negative surface charge, protects against degradation, and facilitates cellular internalization and intracellular trafficking of the plasmid. In vitro studies typically employ bolus addition of these complexes to the media, resulting in internalization of approximately 20–50% of this plasmid (Tseng et al., 1997; Varga et al., 2001). Alternative delivery strategies have shown that increasing the concentration of DNA in the cellular microenvironment can increase transfection (Luo and Saltzman, 2000).
Immobilizing DNA to the cell culture substrate prior to cell seeding has been proposed as a mechanism to increase the concentration of DNA in the cellular microenvironment. Gene delivery from the culture surface, termed substrate-mediated delivery (Segura and Shea, 2002; Segura et al., 2003), reverse transfection (Ziauddin and Sabatini, 2001), or solid-phase delivery (Bielinska et al., 2000), is being developed to enhance gene transfer and to create transfected cell arrays (Yoshikawa et al., 2004; Ziauddin and Sabatini, 2001). Several strategies have been developed to associate DNA complexes with the substrate including specific binding of complexes to substrate through the biotin–avidin interaction (Segura and Shea, 2002; Segura et al., 2003), gelatin entrapment (Ziauddin and Sabatini, 2001), or nonspecific adsorption (Yoshikawa et al., 2004). These systems have shown the capacity to transfect cells cultured on the substrate, with deposition by adsorption transfecting the greatest number of cell types. The continued development and application of this approach will benefit from an understanding of vector and substrate properties that mediate gene delivery.
This report characterizes substrate binding and transfection of DNA polyplexes and lipoplexes that are nonspecifically adsorbed to either tissue culture polystyrene (PS) or fetal bovine serum-coated polystyrene (FBS-PS). Maximal transfection from this approach requires that the affinity of the DNA complex for the substrate be sufficient to support immobilization yet not so excessive as to limit cellular internalization. These studies have focused on manipulating the properties of the complex and substrate to modulate this binding affinity. Polyplexes and lipoplexes were examined based on their differences in chemical composition, size, and zeta potential, which may affect their interaction with the substrate. Additionally, serum adsorption was used as a simple approach to modulate the interaction of these complexes with the culture substrate. Substrate binding and cellular transfection (i.e., transfected cells, transgene expression) were measured for polyplexes and lipoplexes formed with varying amounts of cationic polymer or lipid, respectively. The mechanism of gene transfer was examined through the distribution and stability of complexes on the substrate. Furthermore, we demonstrate that substrate-mediated delivery can transfect primary human-derived cells and that this approach can be extended to widely used biomaterials.
MATERIALS AND METHODS
Materials
Plasmid DNA encoding for luciferase or enhanced green fluorescent protein (EGFP) with a CMV promoter was purified from bacteria culture using Qiagen (Santa Clara, CA) reagents and stored in Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 7.4). Branched polyethylenimine (PEI, 25 kDa) was purchased from Aldrich (St. Louis, MO). Lipofectamine 2000 was purchased from Life Technologies (Gaithersburg, MD). All other reagents were obtained from Fisher Scientific (Fairlawn, NJ) unless otherwise noted.
Complex Formation and Immobilization
Addition of cationic polymers or lipids to plasmid DNA results in self-assembly to form colloidal polyplexes and lipoplexes, respectively. The complexing agents used were the branched cationic polymer polyethylenimine (PEI) and Lipofectamine 2000. Complexes with PEI, termed polyplexes, were formed at the desired N/P ratio (range 5–25), while DNA complexed with Lipofectamine 2000, termed lipoplexes, were formed at several ratios of DNA/lipid (μg/ μL, range 0.5–5). For polyplexes, polymer solution (final volume of 40 μL in Tris-buffered saline, TBS) was added dropwise to DNA (final volume of 60 μL in TBS), vortexed, and incubated for 15 min at room temperature. For lipoplexes, lipid (final volume 50 μL in serum-free cell growth media, DMEM) was added dropwise to DNA (final volume 50 μL in serum-free cell growth media, DMEM), mixed by gentle pipetting, and incubated for 20 min. Lipoplexes were formed in serum-free cell growth media. Zeta potential and z-average diameter were measured with a Zetasizer Nano ZS (Malvern, Worcestershire, United Kingdom).
Complexes were immobilized by incubation on either untreated or serum-coated tissue culture polystyrene. Tissue culture polystyrene (PS, 96-well strip plate; Corning, Corning, NY) was serum-coated (FBS-PS) by incubation with heat-inactivated fetal bovine serum (FBS, 200 μL, 10% in PBS, pH 7.4) for 2 h, followed by two wash steps with PBS. Complexes were immobilized immediately following complex formation by incubation of DNA complexes (100 μL) with the substrate for 1–24 h and were then washed twice with TBS for polyplexes and serum free cell growth media for lipoplexes.
Surface Characterization
Tissue culture polystyrene (PS) was used as provided or after modification with fetal bovine serum (FBS-PS). Modification was characterized using X-ray photoelectron spectroscopy (XPS), which excites a surface by X-ray irradiation, allowing determination of the binding energy of ejected electrons. These binding energies can be correlated to the atomic species present on the surface as well as the chemical of the atoms. XPS spectra were recorded using an Omicron ESCAProbe system with an Al/Mf anode X-ray source. The wettability of the surfaces was determined through measurement of the contact angle of water droplets on the surface in air. Contact angles were determined at room temperature with a goniometer (Ramé-Hart, Mountain Lakes, NJ), which is equipped with a camera system and DROPimage Standard software. A water droplet was dispensed, and angle measurements were recorded as the angle between the point of contact of the droplet with the solid surface and a tangent with the droplet profile.
Quantification of Immobilized DNA and DNA Release
The binding and release of DNA from the substrates was monitored using plasmid DNA radiolabeled with α-32PdATP. Briefly, a nick translation kit (Amersham Pharmacia Biotech, Piscataway, NJ) was used following the manufacturer's protocol with minor modifications (Segura et al., 2003). Following immobilization and washing, the surface density of the complex was determined by immersing individual wells in scintillation cocktail (5 mL, ScintiVerse II) for measurement with a scintillation counter. The counts were correlated to DNA mass using a standard curve. To determine the release profile, substrates with immobilized DNA were incubated with PBS or conditioned media (cell growth media that had been in culture with cells for at least 24 h; 200 μL) at 37°C in a humid chamber. At predetermined time points, half of the solution (100 μL) was removed and replaced with fresh solution. The activity of the collected sample was measured in a scintillation counter. At the final time point, the counts remaining on the substrate were also determined. The percentage of DNA released was calculated as the ratio of the cumulative counts released through a given time divided by the total counts initially on the substrate.
Cell Culture and Transfection
Transfection studies were performed with NIH/3T3 (ATCC, Manassas, VA) cells cultured at 37°C and 5% CO2 in DMEM (Life Technologies) supplemented with 1% sodium pyruvate, 1% penicillin–streptomycin, 1.5 g/L NaHCO3, and 10% fetal bovine serum (FBS). Human dermal fibroblasts (HDF) were obtained per protocol of the IRB and Office for the Protection of Research Subjects at Northwestern University and cultured as previously described (Mogford et al., 2002). Cells were seeded at a density of 5,000 cells per well in 96-well plates. For substrate-mediated delivery, cells were seeded immediately following complex immobilization and wash. Control studies were performed with bolus delivery to cells plated in a 48-well dish at a density of 15,000 cells/well and cultured overnight before complexes were added. The quantity of DNA delivered as a bolus was equivalent to the amount of DNA immobilized to the substrate, with normalization to the larger well. Complexes were formed as described above, but with a final volume of 30 μL and polyplexes and 75 μL for lipoplexes. Transfection was analyzed following a 48-h culture.
Transfection was characterized through the extent of transgene expression (luciferase levels) and the number of transfected cells (GFP expression). The extent of transgene expression was quantified by measuring the luciferase activity using the Luciferase Assay System (Promega, Madison, WI). Cells were lysed and assayed for enzymatic activity after 48 h. The luminometer was set for a 3-s delay with signal integration for 10 s. Luciferase activity was normalized to the number of cells seeded. Transfected cells were visualized and manually counted using an epifluorescence microscope (Leica, Bannockburn, IL) with a FITC filter and equipped with a digital camera. The percentage of transfected cells was calculated as the ratio of the number of transfected cells divided by total cell number, which was determined by manual counting of phase images. Viability of the cell population was analyzed using the (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS) assay (Promega, Madison, WI), which reports on the metabolic activity (Barltrop et al., 1991). The manufacturer's protocol was followed. Briefly, MTS reagent was added (20% of culture volume) to the cultures, and after a 1.5-h incubation, the absorbance at 490 nm was measured with a spectrophotometer. The relative cell viability is reported as the absorbance for the experimental condition divided by the absorbance for cells grown on PS substrates (no serum, DNA, or transfection reagent).
Confocal Microscopy
DNA complexes were fluorescently labeled with tetramethyl rhodamine (Mirus, Madison, WI). Polyplexes were then immobilized followed by cell seeding as described above. Cells were cultured for 20 h and then labeled with the cytoplasmic stain fluorescein diacetate (Sigma, St. Louis, MO). The distribution of cells and complexes on the substrate was visualized with confocal microscopy (Leica).
PLG Disk
The biomaterial poly(lactide-co-glycolide) (PLG) was fabricated into a disk using a gas foaming process (Jang and Shea, 2003; Nof and Shea, 2002). After fabrication, disks were coated with serum, and complexes were incubated with the disk for 24 h as described above. After being washed, cells were seeded onto the disk. Transgene expression was determined using the luciferase assay as previously described. Transfected cells were visualized by delivering a plasmid encoding for nuclear targeted β-galactosidase and staining with X-gal following culture (Shea et al., 1999).
RESULTS
Complex Formation and Surface Modification
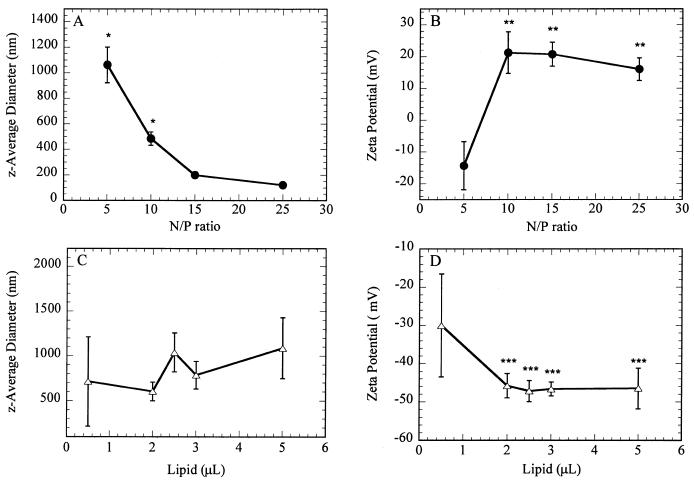
The z-average diameter and zeta potential of DNA complexes, which may impact substrate immobilization and transfection, were measured (Fig. 1) and are consistent with published reports (Ogris et al., 1999; Son et al., 2000; Zhang et al., 2001). The z-average diameter (dz) of the polyplexes decreased with increasing amounts of cationic polymer in the complex, with mean diameters ranging from 1062 ±140 nm to 121 ± 10 nm (P < 0.05) for N/P ratios of 5 and 25, respectively (Fig. 1A). Increasing the amount of cationic polymer led to increases in the zeta potential of polyplexes, which ranged from −14.33 mV to a maximum of 21.21 mV at an N/P of 10 (Fig. 1B). N/P ratios greater than 10 did not significantly affect the zeta potential (P > 0.05). Lipoplexes had a mean dz that ranged from 603 ± 104 nm to 1089 ± 340 nm (P < 0.05) for increasing amounts of lipid (Fig. 1C). For DNA/lipid ratios greater than 1:2, increasing the amount of lipid had no significant effect on the zeta potential (P > 0.05) and averaged −50 mV (Fig. 1D).
Figure 1.
Diameter and zeta-potential of DNA complexes. (A) z-Average diameter and (B) zeta potential of PEI polyplexes with increasing polymer/DNA ratio (N/P). (C) z-Average diameter and (D) zeta potential of Lipofectamine 2000 lipoplexes with increasing volumes of lipid used to complex 1 μg of DNA. Data are presented as average ± standard deviation of the mean. *P < 0.05 relative to remaining data (A); **P < 0.05 relative to N/P equal to 5 (B); ***P < 0.05 relative to 0.5 μL of lipid. For this and all other figures, P values were obtained using Student's t-test with single comparisons.
The altered surface chemistry induced by serum coating was confirmed with X-ray photon spectroscopy (XPS) and contact angle measurements. The XPS spectrum indicates that nitrogen groups are present on FBS-PS but not on PS (Fig. 2). Serum coating also increased the wettability of the surfaces as determined by contact angle measurements (Table I). The FBS treated surfaces had a low contact angle (<15°) relative to the PS surface (48°).
Figure 2.
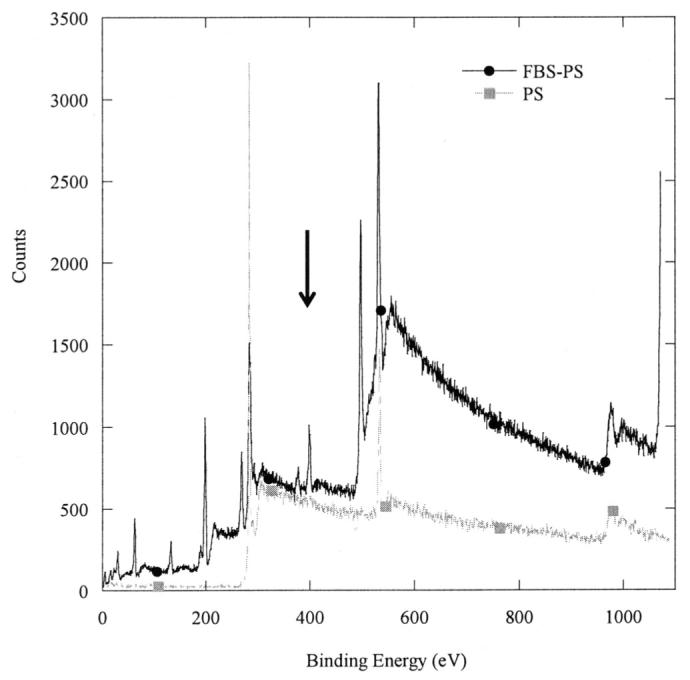
X-ray photoelectron spectrum of polystyrene and serum-modified polystyrene surfaces; peak denoted by the arrow indicates the presence of nitrogen on the substrate.
Table I.
Contact angle measurements for substrates (water).
| Surface | Contact angle (deg) |
|---|---|
| Tissue culture polystyrene (PS) | 47.75 ± 6.64 |
| FBS modified tissue culture polystyrene (FBS-PS) | 12.67 ± 3.13 |
Complex Immobilization and Stability
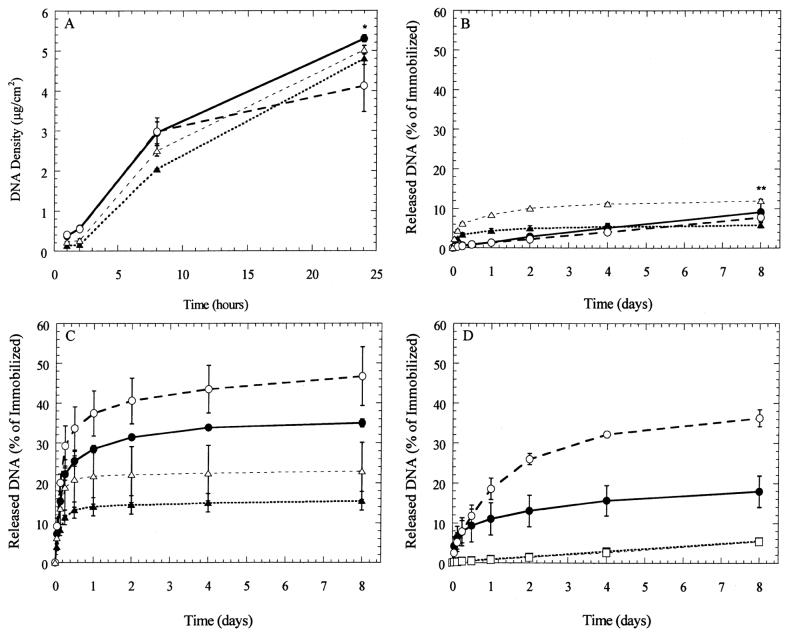
The amount of DNA immobilized to the surfaces increased with time of incubation on the surface and was dependent upon the chemical properties of the surface and DNA complex (Fig. 3A). Incubation of DNA complexes with the substrate for 24 h resulted in surface densities of at least 4 μg/cm2, which corresponds to immobilization of more than 60% of the DNA. Polyplex deposition on PS and FBS-PS occurred at similar rates during the first 8 h of incubation; however, deposition onto FBS-PS slowed relative to PS during the final 16 h, resulting in surface densities of 4.1 and 5.3 μg/cm2 (P < 0.05), respectively. Lipoplex binding to the substrate was not substantially affected by the surface properties during the 24-h incubation (P > 0.05).
Figure 3.
DNA deposition and stability. (A) Polyplexes (circles) and lipoplexes (triangles) were deposited onto polystyrene (●, ▲) and serum-coated polystyrene (○, △) for 24 h. Substrates were incubated with 2 μg of DNA; (B) immobilized polyplexes and lipoplexes were exposed to phosphate-buffered saline (PBS, pH 7.4); (C) conditioned media; (D) polyplexes incubated with cell growth media (●, PS; ○, FBS-PS), and trypsin (■, PS; □, FBS-PS) at 37°C. Data are presented as average ± standard deviation of the mean. *P < 0.05 relative to FBS-PS deposition of polyplexes after 24 h; **P × 0.05 relative to lipoplexes released from PS at 8 days.
The retention of polyplexes, but not lipoplexes, on both PS and FBS-PS substrates during incubation in PBS was unaffected by surface modification. For polyplexes, incubation in PBS resulted in 9% and 8% (P < 0.05) release after 8 days from PS and FBS-PS, respectively (Fig. 3B). For lipoplexes, incubation in PBS resulted in 6% and 12% (P < 0.05) release after 8 days from PS and FBS-PS, respectively (Fig. 3B). The release of polyplexes gradually increased during the 8-day incubation in PBS, whereas lipoplexes reach maximal release after 2–4 days. Transfection experiments typically involve cell culture for 24–96 h, which corresponds to release of 2.75–10% of immobilized DNA.
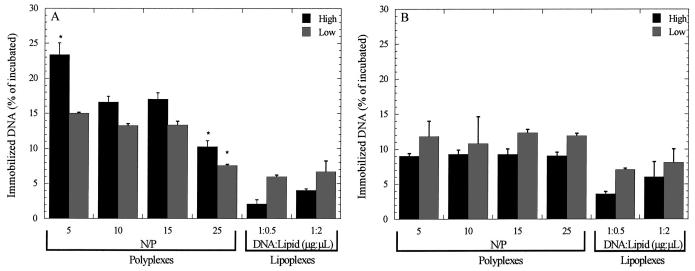
Increasing the quantity of DNA incubated with the substrate resulted in increased quantities of immobilized DNA; however, the percentage of DNA immobilized after a 2-h incubation is dependent upon surface modification and complex properties (Fig. 4). For polyplexes immobilized to PS substrates, increasing the quantity of DNA incubated on the surface led to a greater percentage immobilized for all N/P ratios (P < 0.05) (Fig. 4A). Furthermore, increasing the N/P ratio resulted in decreasing quantities of immobilized DNA. Conversely, polyplexes immobilized to FBS-PS substrates exhibited no dependence on DNA quantities or N/P ratio (P > 0.05). Lipoplex binding to PS substrates exhibited a dependence on DNA concentration, with lower concentrations binding more effectively than higher concentrations. This trend was significant for FBS-PS at the lower lipid content (P < 0.05). The lipid content of the complexes did not affect the percentage bound to the substrate on both PS and FBS-PS substrates.
Figure 4.
DNA adsorption to surface. Polyplexes and lipoplexes were incubated on (A) PS and (B) FBS-PS surfaces for 2 h. Black bars indicate 2 μg, and gray bars indicate 0.5 μg of DNA incubated on the surface. Data are presented as average ± standard deviation of the mean. High doses have P < 0.05 relative to corresponding low doses at the same N/P (A). *P < 0.05 relative to other polyplex conditions.
Cellular Transfection
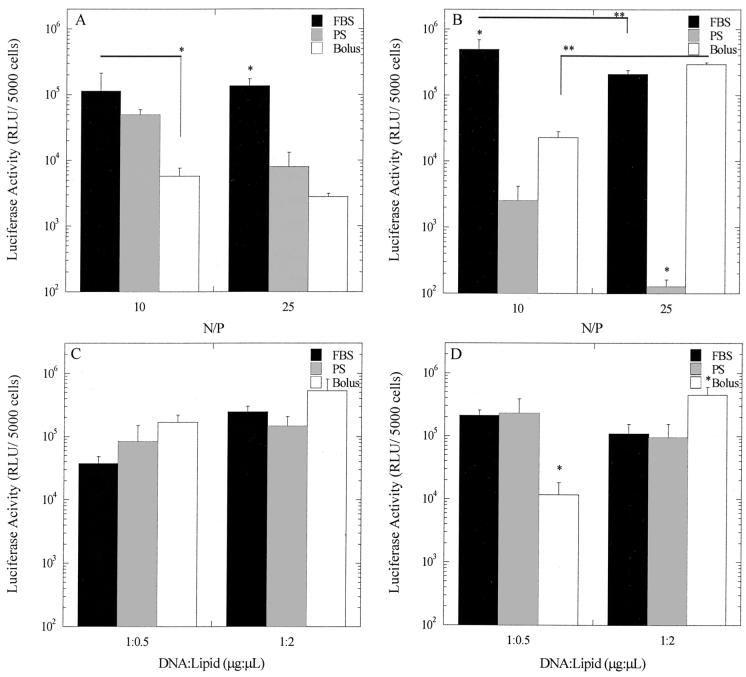
The extent of transgene expression on substrates with immobilized polyplexes and lipoplexes depended upon substrate modification and complex formulation (Fig. 5). Polyplexes and lipoplexes immobilized to FBS-PS substrates yielded equal or greater transgene expression than delivery from PS. For immobilization of small quantities of DNA (0.5 μg), FBS-PS substrates yielded 17-fold greater transgene expression than PS (P < 0.05) at an N/P of 25, and similar levels for an N/P of 10 (Fig. 5A). For immobilization of large quantities of DNA (2 μg), transgene expression on FBS-PS was increased 194- and 1638-fold compared to PS (P < 0.05) surfaces for N/P equal to 10 and 25, respectively (Fig. 5B). Polyplex delivery from FBS-PS surfaces enhanced expression 20- to 49-fold relative to equivalent doses delivered in bolus (P < 0.05), with the exception that the largest dose and N/P ratio provided similar expression levels.
Figure 5.
Transgene expression by substrate-mediated delivery. (A,B) Polyplexes and (C,D) lipoplexes with plasmid encoding for luciferase were incubated on substrates at low (0.5 μg; A, C) and high (2.0 μg; B, D) doses for 2 h. Bolus delivery used equivalent amounts of DNA that were immobilized to the substrate: (A) 0.2 μg/cm2, (B) 0.5 μg/cm2, (C) 0.25 μg/cm2, and (D) 0.1 μg/cm2. Activity was normalized to the number of cells seeded. Data are presented as average ± standard deviation of the mean. *P < 0.05 relative to other delivery methods for that condition; **P < 0.05 at different complex conditions.
Lipoplex delivery from PS and FBS-PS yielded equal levels of luciferase activity under similar dosage and complex conditions. At low-dose conditions, increasing the lipid content of the complexes led to a 6.5-fold enhancement in luciferase activity (Fig. 5C), which was not observed at the larger dose (Fig. 5D). For lipoplexes with the lowest lipid content and at the highest dose, an 18-fold enhancement in activity was observed from serum-modified surfaces when compared to bolus delivery. Otherwise, transgene expression was similar between substrate-mediated and bolus delivery.
The similar levels of transgene expression obtained by lipoplex delivery from both PS and FBS-PS substrates were further examined by quantifying the percentage of transfected cells (Fig. 6). The percentage of transfected cells increased nearly 2-fold on FBS-PS relative to PS, independent of quantity immobilized and lipoplex formulation (Fig. 6A,B; P < 0.05). Substrate-mediated delivery from FBS-PS resulted in similar or greater percentages of transfected cells relative to bolus delivery. Polyplexes delivered from FBS-PS surfaces also had greater percentages of cells transfected compared to PS surfaces; however, transfection efficiencies were low (<10%, data not shown).
Figure 6.
Transfected cell numbers by substrate-mediated delivery. Lipoplexes with plasmid encoding for GFP were incubated on substrates with quantities of (A) 0.5 μg and (B)2 μg for 2 h. Bolus delivery used equivalent amounts of DNA that were immobilized to the substrate: (A) 0.25 μg/cm2 and (B) 0.1 μg/cm2. Data are presented as average ± standard deviation of the mean. *P < 0.05 relative to the other delivery methods for that condition.
The viability of NIH/3T3 cells was similar for transfection by both bolus and substrate-mediated delivery, with the viability affected by the transfection reagent but not the substrate preparation. Substrate mediated delivery showed similar trends to bolus delivery on the different substrates (PS and FBS-PS). Modification of the polystyrene substrate with serum does not influence cell viability as cells yield similar metabolic activities on both substrates (Fig. 7). Polyplex delivery from the substrate or as a bolus did not affect viability (P > 0.05). Delivery of lipoplexes reduced viability relative to polyplex delivery; however, similar levels of viability were observed for bolus and substrate delivery.
Figure 7.
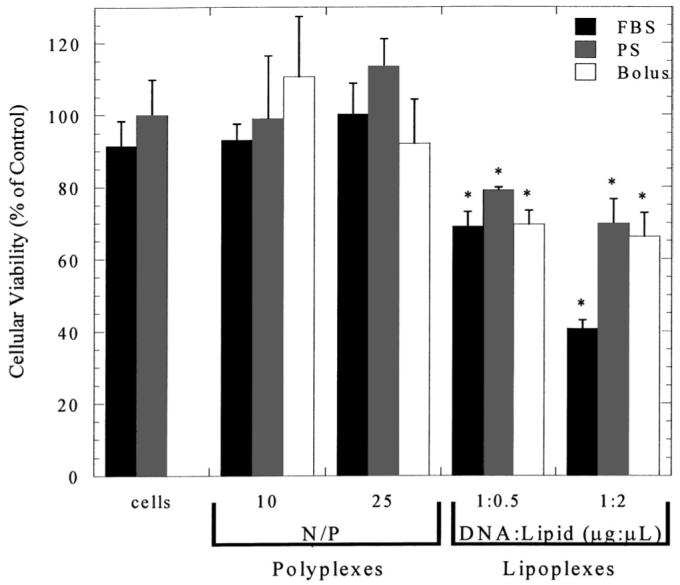
Cellular viability. The MTS assay was used to determine cellular viability on serum-modified substrate. The MTS assay was also used to determine cellular viability after delivery of complexes from FBS-PS and PS substrates and through bolus delivery. Data are presented as average ± standard deviation of the mean. *P < 0.05 relative to cellular viability on PS (control).
Cell–Complex Interaction
The ability of cells to promote release of complexes from the substrate for subsequent internalization was examined by performing release studies with conditioned growth media (Fig. 3). In conditioned media, the release of DNA complexes was greater from FBS-PS compared to PS substrates (Fig. 3C). After 8 days, 35% of polyplexes were released from PS substrates and 47% were released from FBS-PS substrates (P < 0.05) in conditioned media. These numbers are significantly greater than the release in PBS (P < 0.05) (Fig. 3B). Lipoplexes exhibited similar dependence on serum coating, with 23% and 15% released from FBS-PS and PS substrates, respectively. Immobilized polyplexes were also incubated with either serum-containing media (not conditioned) or trypsin in order to identify which components of conditioned media mediate release (Fig. 3D). After 8 days, 18% and 36% of complexes released from PS and FBS-PS substrates in serum-containing media, and 5% released in trypsin from both substrates.
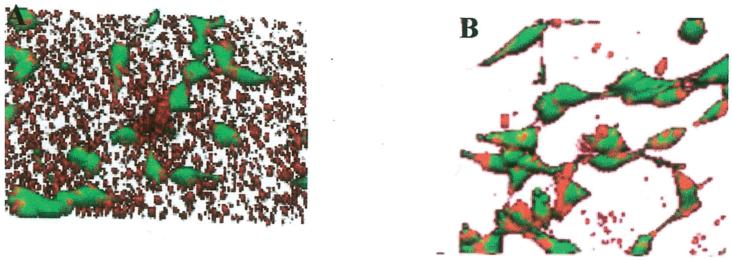
The differences in transfection observed between PS and FBS-PS were subsequently examined using confocal microscopy to determine the relative distribution of polyplexes and cells on the substrate. Conditions with relatively low (PS) and high (FBS-PS) rates of transfection were compared to identify differences in the complex distribution (N/P 25, 2 μg incubated on the surface). Polyplexes immobilized to PS, which had a low transfection rate, were distributed across the surface, and only a fraction of the complexes were associated with cells (Fig. 8A). Seeding cells seeded onto DNA adsorbed FBS-PS substrates that yielded higher transfection led to polyplexes that were not distributed across the substrate but localized to the cells or in their vicinity. Thus, polyplexes had a higher cellular association when delivered from serum-modified surfaces compared to unmodified surfaces.
Figure 8.
Distribution of cells and polyplexes on substrate following cell culture. Confocal microscopic images of polyplexes on (A) PS and (B) FBS-PS 20 h after cell seeding. Polyplexes (2 μg, N/P = 25) were incubated on substrates for 2 h followed by washing and cell seeding. Images were captured 20 h after cell seeding. Plasmid was labeled with tetramethylrhodamine, and cells were stained with fluorescein diacetate.
Primary Cell Transfection
The ability of substrate-mediated delivery to transfect primary cells was assessed using human dermal fibroblasts (HDF), a primary cell type that has been difficult to transfect due to cytotoxicity (Uchida et al., 2002). Lipoplexes delivered as a bolus resulted in large amounts of cellular debris, poor cell morphology, and reduced cellular viability (Fig. 9A). For substrate-mediated delivery, the fibroblasts had a morphology similar to that observed during typical cell culture (Fig. 9B). Transgene expression was greatest for HDFs cultured on FBS-PS, for both immobilized polyplexes and lipoplexes (Fig. 9C,D). Polyplexes delivered from an FBS-PS surface yielded 10.7-fold greater luciferase expression when compared to PS (P < 0.05), with bolus delivery producing luciferase activity similar to FBS-PS. Lower doses of complexes delivered as a bolus resulted in insignificant luciferase activity (data not shown). Consistent with the NIH/3T3 culture, lipoplex delivery by substrate immobilization resulted in similar expression levels independent of substrate modification (FBS-PS, PS) and the lipid content. Additionally, transgene expression for polyplexes and lipoplexes were similar; however, the percentage of transfected cells using lipoplexes was approximately 26%, whereas polyplex immobilization resulted in less than 5% of the cells transfected.
Figure 9.
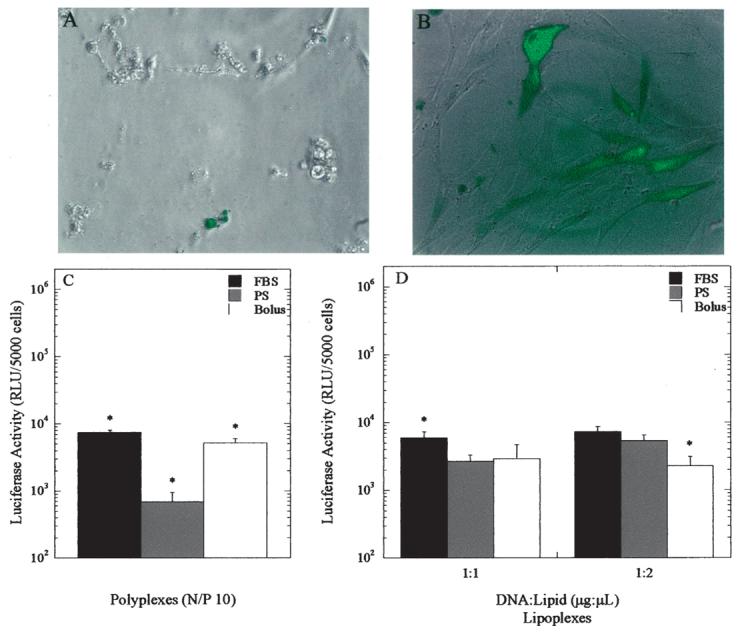
Transfection of primary human dermal fibroblasts. (A) GFP expression from bolus delivery of 0.5 μg lipoplexes. (B) GFP expression from FBS-PS substrate-mediated delivery of 1 μg lipoplexes incubated on substrate. Luciferase activity from substrate and bolus delivery of (C) polyplexes and (D) lipoplexes. Data are presented as average ± standard deviation of the mean. *P < 0.05 relative to other delivery methods for that condition.
Transfection From a Biomaterial Scaffold
The extension of substrate-mediated delivery by complex adsorption to biomaterial scaffolds was subsequently examined, which could facilitate the application of this approach to in vivo gene transfer. Poly(lactide-co-glycolide) (PLG) disks were incubated with polyplexes, resulting in the immobilization of 1.3 μg onto a 5-mm disk. Transfected NIH/3T3 cells were visible across the substrate (Fig. 10), and transgene expression was measured at 1.3 × 105 RLU/5000 cells seeded. These results demonstrate the potential to apply substrate-mediated delivery to alternative substrates other than PS.
Figure 10.
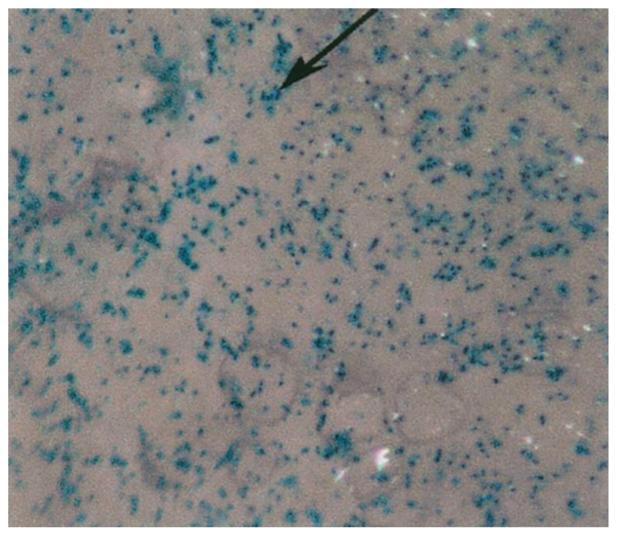
Substrate-mediated transfection of NIH/3T3 fibroblasts from serum-coated PLG substrates. Polyplexes (5 μg) containing plasmid encoding for β-galactosidase were incubated for 24 h. Transfected cells appear blue, as indicated with the arrow.
DISCUSSION
This report characterizes gene transfer by DNA complexes adsorbed to tissue culture polystyrene and extends this approach to a biomaterial substrate. Polyplexes and lipoplexes were adsorbed to either unmodified (PS) or serum-adsorbed (FBS-PS) polystyrene. Complex immobilization to the substrate increased with time of exposure, while deposition depended upon the properties of the substrate and complex. Polyplexes immobilized to FBS-PS provided greater transfection than polyplexes immobilized to PS and equivalent or greater transfection than bolus delivery. Lipoplexes, however, provided similar levels of transgene expression on serum-coated and uncoated substrates. Interestingly, transfection on serum-coated surfaces increased the number of transfected cells relative to unmodified substrates. Immobilized complexes are able to transfect primary cells and resulted in improved cellular viability relative to bolus delivery. Finally, this delivery mechanism was adapted to a biomaterial substrate, PLG, which is commonly used in biomedical applications such as tissue engineering.
Reverse transfection and substrate-mediated delivery are based on the immobilization of DNA complexes to the culture substrate. Substrate-mediated delivery mimics the intrinsic binding of viruses to extracellular matrix. For example, retroviral vectors bind to fibronectin (Hanenberg et al., 1996; Lei et al., 2002) to facilitate internalization. Viral vectors have also been designed that specifically interact with natural and synthetic biomaterials through the use of antibodies or covalent coupling (Abrahams et al., 2002; Klugherz et al., 2002; Stachelek et al., 2004). Reverse transfection was performed by suspending plasmid in gelatin and spotting onto a slide and is followed by lipid addition to the culture media (Ziauddin and Sabatini, 2001). Substantial transfection of HEK 293 cells was observed using this approach, and may require some modification to transfect other cell types (Bailey et al., 2002; Wu et al., 2002). An alternative to the DNA/gelatin immobilization procedure involves the complexation of plasmid with cationic lipids or polymers and then immobilization of these complexes to the substrate. Biotin residues have been attached to cationic polymers, and mixed with plasmid to facilitate specific binding of DNA complexes to an avidin-modified substrate (Segura and Shea, 2002; Segura et al., 2003). Increasing the number of biotin residues enhanced substrate binding, whereas maximal transfection of HEK293T and NIH/3T3 cells was observed with few numbers of biotin residues. Alternatively, complexes can be nonspecifically adsorbed to the substrate, likely through interactions between the cationic lipid or polymer and the substrate. Complexes have been formed in the presence of extracellular matrix components and dried onto the substrate (Yoshikawa et al., 2004). This approach transfected a variety of cell types, including human mesenchymal stem cells. We report on polyplex and lipoplex binding to the substrate from solution and the ability to transfect cell lines and primary human-derived cells. Interestingly, primary cells demonstrated an increased cellular viability, which may result from washing of the substrates to remove unbound lipid or polymer that can be cytotoxic (Godbey et al., 1999). Our studies characterize properties of the substrate and the complexes that mediate substrate immobilization and transgene expression, which serve to elucidate substrate and vector properties that optimize gene transfer.
Incubation of lipoplexes or polyplexes with the tissue culture substrates results in nonspecific adsorption of the complexes. The adsorption of polyplexes was expected based on the use of PEI as a coating material for culture substrates. Although the mechanisms of DNA complex adsorption to substrates have not been well-studied, the mechanisms of protein adsorption may provide insight regarding these interactions. Nonspecific adsorption occurs through electrostatic interactions, van der Waals forces, and hydrophobic interactions. In this report, an increasing polyplex charge did not increase polyplex adsorption suggesting that electrostatic interactions are not the only mechanism of binding. Interactions between proteins and substrates have been characterized by (i) changes in the hydration of the surface and protein molecule, (ii) charge interactions between the protein and the surface, and (iii) structural rearrangement of the adsorbing molecule (Norde and Lyklema, 1991). The affinity for the substrate depends upon the substrate–biomolecule interactions, conformational changes that occur upon binding and the solution properties from which the molecule is adsorbing. Adsorption to hydrophobic substrates is regulated by entropy (Norde, 1986), and the conformation changes induced upon binding may contribute to irreversible binding (Stuart et al., 1991). Hydrophilic substrates generally result in reversible interactions, and the driving force for adsorption is typically enthalpic. The different responses observed on PS and on FBS-PS may reflect changes in the hydrophobicity of the substrate or variations in the molecular interactions between the complex and substrate. Similarly, complexes were expected to adsorb to serum-modified substrates due to their known interaction with serum components (Moret et al., 2001; Zelphati et al., 1998). We propose that the molecular-scale design of the DNA complex and the substrate can be employed to regulate the molecular interactions (e.g., electrostatic, van der Waals, hydrophobic) and will lead to optimal transfection by a substrate-mediated approach.
Maximal transfection from the substrate is hypothesized to represent a balance between complex binding to the substrate and cellular internalization. Transfection was observed with both polyplexes and lipoplexes adsorbed to serum modified and unmodified polystyrene, suggesting that substrate-mediated delivery is applicable to a variety of transfection reagents and substrates. Serum coating of polystyrene can enhance transgene expression (polyplexes) or the percentage of transfected cells (lipoplexes). Serum coating of PS was hypothesized to influence either complex adsorption and release or enhance the cellular viability. For NIH/3T3 cells, serum-modified substrates did not enhance their viability relative to PS (Fig. 7), suggesting that the serum may affect adsorption or release. Addition of serum containing media (pre- and post-conditioning) to the immobilized complexes significantly enhanced release of the complexes relative to incubation with PBS, whereas incubation with trypsin yielded release levels less than PBS (Fig. 3). These observations suggest that complex release from the substrate mediated by competitive binding of serum that displaces the complexes. The internalization of released and substrate-associated complexes, and DNA distribution among the cell populations are the focus of continuing studies.
Substrate-mediated delivery yields comparable levels of transgene expression to bolus delivery but provides a mode of transfection useful in the development of research tools, such as transfected cell arrays, and that may be translated to in vivo gene transfer from biomaterials. Substrate immobilization distributes the DNA complexes across the substrate, which may enhance the cellular distribution and increase the number of transfected cells. Immobilization to the substrate also limits complex aggregation (Segura et al., 2003), and smaller particles may traffic differently than larger complexes (Ogris et al., 2001). Transgene expression levels and the number of cells expressing the transgene may be improved by substrate mediated delivery and this report identifies some of the design considerations. Cellular transfection by the immobilized DNA complexes was not equivalent across substrates and transfection reagents. Measurements of transgene expression characterize the total protein production, whereas the transfected cell number reflects the efficacy of delivery throughout the cell population. These differences in the transfection profile do not reflect substantial differences in the amount of DNA immobilized (Fig. 4) and may result from differences in cellular internalization from the substrate and intracellular trafficking resulting from the different vectors. Conditions with relatively low transfection (PS) had DNA localized to the substrate, whereas conditions with higher transfection (FBS-PS) showed less substrate-associated DNA and more cell-associated DNA (Fig. 8).
Maximal transgene expression for polyplexes and lipoplexes by substrate-mediated delivery is similar, yet the transfection profile and dependence on the substrate properties reflect differences in composition and intrinsic transfection properties. Polyplex delivery from serum-modified substrates can increase transgene expression relative to delivery from PS and to equivalent quantities delivered as a bolus. Serum modification enhanced transgene expression up to 1600-fold relative to unmodified PS, indicating that substrate properties dramatically affect delivery. Lipoplex delivery, however, yielded similar levels of expression across delivery methods, yet serum modification was able to significantly increase the percentage of transfected cells. Increasing the amount of immobilized lipoplexes depressed the expression levels, which is similar to observations with bolus lipoplex delivery. Cellular exposure to excessive quantities of lipoplexes can adversely affect proliferation and viability, thereby reducing transgene expression. These differences in transfection between polyplexes and lipoplexes likely result from variations in the complexation agent chemistry (e.g., lipid, polymer) and the physical properties (e.g., size, zeta potential) of the DNA complex, which would affect molecular interactions between the complex and substrate. Polyplexes and lipoplexes have substantially different z-average diameters, zeta-potential, and chemical composition (Fig. 1). Increasing the N/P ratio for polyplexes increases their zeta potential and reduces their size (Boussif et al., 1995; Choosakoonkriang et al., 2003; Ogris et al., 1999). Conversely, altering the composition of lipoplexes has less marked effects on the complex diameter and zeta-potential. Transfection of primary cells or cell lines can have significant variations due to numerous cell and vector properties, such as proliferation, metabolism, internalization, and intracellular trafficking. Transfection of primary cells can involve further obstacles, such as the differential expression of cell surface proteoglycans or decreased viability caused by the transfection reagent (Uchida et al., 2002). Substrate-mediated delivery may decrease the amount of DNA, or the quantity of transfection reagent, which may enhance either transfection or viability for primary cells.
Immobilization of DNA complexes by adsorption can be extended to biomaterials for applications such as tissue engineering. Expression levels achieved were comparable to transfection on polystyrene and suggest that substrate-mediated delivery is a generalizable phenomenon; the optimal design of biomaterial substrates may also facilitate the translation of this approach to in vivo applications. Porous PLG or collagen scaffolds have been fabricated with encapsulated polyplexes or lipoplexes and have achieved substantial transfection in vitro (Cohen-Sacks et al., 2004; Huang et al., 2003; Scherer et al., 2002) and in vivo (Scherer et al., 2002). The presence of the cationic lipid or polymer can significantly alter the release profile compared to plasmid DNA (Huang et al., 2003; Shea et al., 1999). The biotin–avidin interaction has been used to bind both viral and nonviral vectors to a variety of materials (Levy et al., 2001; Segura et al., in press). Alternatively, complexes formed with polyamidoamine (PAMAM) dendrimers have been dried onto PLG and collagen membranes coated with phosphatidyl glycerol (1–5%) (Bielinska et al., 2000). In vitro transfection was comparable to bolus controls, while in vivo delivery enhanced expression 6- to 8-fold relative to plasmid delivery. Alternatively, plasmid DNA has also been incorporated into inorganic calcium phosphate co-precipitates that are adsorbed onto PLG matrices, and are mostly released by 48 h (Kofron and Laurencin, 2004). This report demonstrates the ability to adsorb complexes to PLG to obtain significant transfection on the substrate and suggests that this strategy may be useful for tissue-engineering applications.
Acknowledgments
Support for this research was provided in part by grants from National Institutes of Health, the Specialized Program of Research Excellence (SPORE) in Breast Cancer of the National Cancer Institute, and the Christopher Reeve Paralysis Foundation. A.K.P. is supported by a NSF Graduate Research Fellowship. XPS measurements were performed at Keck-II/NUANCE, and confocal images were obtained in the Biological Imaging Facility at Northwestern University (Evanston, IL). We thank Kinjal Shah and Biliana Marcheva (Northwestern University) for technical assistance.
Contract grant sponsors: NIH; NCI; Christopher Reeve Paralysis Foundation; NSF
Contract grant numbers: RO1 GM066830 (to L.D.S.) and RO1 GM 41303 to (T.A.M.); P50-CA89018 (to L.D.S.)
References
- Abrahams JM, Song C, DeFelice S, Grady MS, Diamond SL, Levy RJ. Endovascular microcoil gene delivery using immobilized anti-adenovirus antibody for vector tethering. Stroke. 2002;33(5):1376–1382. doi: 10.1161/01.str.0000014327.03964.c0. [DOI] [PubMed] [Google Scholar]
- Bailey SN, Wu RZ, Sabatini DM. Applications of transfected cell microarrays in high-throughput drug discovery. Drug Discovery Today. 2002;7(18 Suppl):S113–118. doi: 10.1016/s1359-6446(02)02386-3. [DOI] [PubMed] [Google Scholar]
- Barltrop JA, Owen TC, Cory AH, Cory JG. 5-(3-Carboxymethoxyphenyl)-2-(4,5-dimethylthiazolyl)-3-(4-sulfophenyl)tetrazolium, inner salt (MTS) and related analogs of 3-(4,5-dimethylthiazolyl)-2, 5-diphenyltetrazolium bromide (MTT) reducing to purple water-soluble formazans as cell-viability indicators. Bioorg Med Chem Lett. 1991;1(11):611–614. [Google Scholar]
- Bielinska AU, Yen A, Wu HL, Zahos KM, Sun R, Weiner ND, Baker JR, Jr, Roessler BJ. Application of membrane-based dendrimer/DNA complexes for solid phase transfection in vitro and in vivo. Biomaterials. 2000;21(9):877–887. doi: 10.1016/s0142-9612(99)00229-x. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci USA. 1995;92(16):7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choosakoonkriang S, Lobo BA, Koe GS, Koe JG, Middaugh CR. Biophysical characterization of PEI/DNA complexes. J Pharm Sci. 2003;92(8):1710–1722. doi: 10.1002/jps.10437. [DOI] [PubMed] [Google Scholar]
- Cohen-Sacks H, Elazar V, Gao J, Golomb A, Adwan H, Korchov N, Levy RJ, Berger MR, Golomb G. Delivery and expression of pDNA embedded in collagen matrices. J Control Release. 2004;95(2):309–320. doi: 10.1016/j.jconrel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Godbey WT, Wu KK, Hirasaki GJ, Mikos AG. Improved packing of poly(ethylenimine)/DNA complexes increases transfection efficiency. Gene Ther. 1999;6(8):1380–1388. doi: 10.1038/sj.gt.3300976. [DOI] [PubMed] [Google Scholar]
- Hanenberg H, Xiao XL, Dilloo D, Hashino K, Kato I, Williams DA. Colocalization of retrovirus and target cells on specific fibronectin fragments increases genetic transduction of mammalian cells. Nat Med. 1996;2(8):876–882. doi: 10.1038/nm0896-876. [DOI] [PubMed] [Google Scholar]
- Huang YC, Connell M, Park Y, Mooney DJ, Rice KG. Fabrication and in vitro testing of polymeric delivery system for condensed DNA. J Biomed Mater Res. 2003;67A(4):1384–1392. doi: 10.1002/jbm.a.20036. [DOI] [PubMed] [Google Scholar]
- Jang JH, Shea LD. Controllable delivery of non-viral DNA from porous scaffolds. J Control Release. 2003;86(1):157–168. doi: 10.1016/s0168-3659(02)00369-3. [DOI] [PubMed] [Google Scholar]
- Klugherz BD, Song C, DeFelice S, Cui X, Lu Z, Connolly J, Hinson JT, Wilensky RL, Levy RJ. Gene delivery to pig coronary arteries from stents carrying antibody-tethered adenovirus. Hum Gene Ther. 2002;13(3):443–454. doi: 10.1089/10430340252792576. [DOI] [PubMed] [Google Scholar]
- Kofron MD, Laurencin CT. Development of a calcium phosphate co-precipitate/poly(lactide-co-glycolide) DNA delivery system: release kinetics and cellular transfection studies. Biomaterials. 2004;25(13):2637–2643. doi: 10.1016/j.biomaterials.2003.09.042. [DOI] [PubMed] [Google Scholar]
- Lei P, Bajaj B, Andreadis ST. Retrovirus-associated heparan sulfate mediates immobilization and gene transfer on recombinant fibronectin. J Virol. 2002;76(17):8722–8728. doi: 10.1128/JVI.76.17.8722-8728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy RJ, Song C, Tallapragada S, DeFelice S, Hinson JT, Vyavahare N, Connolly J, Ryan K, Li Q. Localized adenovirus gene delivery using antiviral IgG complexation. Gene Ther. 2001;8(9):659–667. doi: 10.1038/sj.gt.3301452. [DOI] [PubMed] [Google Scholar]
- Luo D, Saltzman WM. Enhancement of transfection by physical concentration of DNA at the cell surface. Nat Biotechnol. 2000;18(8):893–895. doi: 10.1038/78523. [DOI] [PubMed] [Google Scholar]
- Mogford JE, Tawil N, Chen A, Gies D, Xia Y, Mustoe TA. Effect of age and hypoxia on TGFbeta1 receptor expression and signal transduction in human dermal fibroblasts: impact on cell migration. J Cell Physiol. 2002;190(2):259–265. doi: 10.1002/jcp.10060. [DOI] [PubMed] [Google Scholar]
- Moret I, Esteban Peris J, Guillem VM, Benet M, Revert F, Dasi F, Crespo A, Alino SF. Stability of PEI–DNA and DOTAP–DNA complexes: effect of alkaline pH, heparin and serum. J Control Release. 2001;76(12):169–181. doi: 10.1016/s0168-3659(01)00415-1. [DOI] [PubMed] [Google Scholar]
- Nof M, Shea LD. Drug-releasing scaffolds fabricated from drug-loaded microspheres. J Biomed Mater Res. 2002;59(2):349–356. doi: 10.1002/jbm.1251. [DOI] [PubMed] [Google Scholar]
- Norde W. Adsorption of proteins from solution at the solid–liquid interface. Adv Colloid Interface Sci. 1986;25(4):267–340. doi: 10.1016/0001-8686(86)80012-4. [DOI] [PubMed] [Google Scholar]
- Norde W, Lyklema J. Why proteins prefer interfaces. J Biomater Sci Polym Ed. 1991;2(3):183–202. doi: 10.1080/09205063.1991.9756659. [DOI] [PubMed] [Google Scholar]
- Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin–PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6(4):595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- Ogris M, Steinlein P, Carotta S, Brunner S, Wagner E. DNA/polyethylenimine transfection particles: influence of ligands, polymer size, and PEGylation on internalization and gene expression. AAPS PharmSci. 2001;3(3):E21. doi: 10.1208/ps030321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer F, Schillinger U, Putz U, Stemberger A, Plank C. Nonviral vector-loaded collagen sponges for sustained gene delivery in vitro and in vivo. J Gene Med. 2002;4(6):634–643. doi: 10.1002/jgm.298. [DOI] [PubMed] [Google Scholar]
- Segura T, Chung PH, Shea LD. DNA delivery from hyaluronic acid-collagen hydrogels via a substrate-mediated approach. Biomaterials. doi: 10.1016/j.biomaterials.2004.05.007. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura T, Shea LD. Surface-tethered DNA complexes for enhanced gene delivery. Bioconjugate Chem. 2002;13(3):621–629. doi: 10.1021/bc015575f. [DOI] [PubMed] [Google Scholar]
- Segura T, Volk MJ, Shea LD. Substrate-mediated DNA delivery: role of the cationic polymer structure and extent of modification. J Control Release. 2003;93(1):69–84. doi: 10.1016/j.jconrel.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Shea LD, Smiley E, Bonadio J, Mooney DJ. DNA delivery from polymer matrices for tissue engineering. Nat Biotechnol. 1999;17(6):551–554. doi: 10.1038/9853. [DOI] [PubMed] [Google Scholar]
- Son KK, Patel DH, Tkach D, Park A. Cationic liposome and plasmid DNA complexes formed in serum-free medium under optimum transfection condition are negatively charged. Biochim Biophys Acta. 2000;1466(12):11–15. doi: 10.1016/s0005-2736(00)00176-0. [DOI] [PubMed] [Google Scholar]
- Stachelek SJ, Song C, Alferiev I, Defelice S, Cui X, Connolly JM, Bianco RW, Levy RJ. Localized gene delivery using antibody tethered adenovirus from polyurethane heart valve cusps and intra-aortic implants. Gene Ther. 2004;11(1):15–24. doi: 10.1038/sj.gt.3302129. [DOI] [PubMed] [Google Scholar]
- Stuart MAC, Fleer GJ, Lyklema J, Norde W, Scheutjens JMHM. Adsorption of ions, polyelectrolytes and proteins. Adv Colloid Interface Sci. 1991;34:477–535. doi: 10.1016/0001-8686(91)80056-p. [DOI] [PubMed] [Google Scholar]
- Tseng WC, Haselton FR, Giorgio TD. Transfection by cationic liposomes using simultaneous single cell measurements of plasmid delivery and transgene expression. J Biol Chem. 1997;272(41):25641–25647. doi: 10.1074/jbc.272.41.25641. [DOI] [PubMed] [Google Scholar]
- Uchida E, Mizuguchi H, Ishii-Watabe A, Hayakawa T. Comparison of the efficiency and safety of non-viral vector-mediated gene transfer into a wide range of human cells. Biol Pharm Bull. 2002;25(7):891–897. doi: 10.1248/bpb.25.891. [DOI] [PubMed] [Google Scholar]
- Varga CM, Hong K, Lauffenburger DA. Quantitative analysis of synthetic gene delivery vector design properties. Mol Ther. 2001;4(5):438–446. doi: 10.1006/mthe.2001.0475. [DOI] [PubMed] [Google Scholar]
- Wu RZ, Bailey SN, Sabatini DM. Cell-biological applications of transfected-cell microarrays. Trends Cell Biol. 2002;12(10):485–488. doi: 10.1016/s0962-8924(02)02354-1. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Uchimura E, Kishi M, Funeriu DP, Miyake M, Miyake J. Transfection microarray of human mesenchymal stem cells and on-chip siRNA gene knockdown. J Control Release. 2004;96(2):227–232. doi: 10.1016/j.jconrel.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Zelphati O, Uyechi LS, Barron LG, Szoka FC., Jr Effect of serum components on the physico-chemical properties of cationic lipid/oligonucleotide complexes and on their interactions with cells. Biochim Biophys Acta. 1998;1390(2):119–133. doi: 10.1016/s0005-2760(97)00169-0. [DOI] [PubMed] [Google Scholar]
- Zhang X, Collins L, Fabre JW. A powerful cooperative interaction between a fusogenic peptide and lipofectamine for the enhancement of receptor-targeted, non-viral gene delivery via integrin receptors. J Gene Med. 2001;3(6):560–568. doi: 10.1002/jgm.224. [DOI] [PubMed] [Google Scholar]
- Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]