Abstract
Key to Pseudomonas aeruginosa’s ability to thrive in a diversity of niches is the presence of numerous genomic islands that confer adaptive traits upon individual strains. We reasoned that P. aeruginosa strains capable of surviving in the harsh environments of multiple hosts would therefore represent rich sources of genomic islands. To this end, we identified a strain, PSE9, that was highly virulent in both animals and plants. Subtractive hybridization was used to compare the genome of PSE9 to the less virulent strain PAO1. Nine genomic islands were identified in PSE9 that were absent in PAO1; seven of these had not been previously described. One these seven islands, designated PAGI-5, has already been shown to carry numerous interesting ORFs, including several required for virulence in mammals. Here we describe the remaining six genomic islands, PAGI-6, -7, -8, -9, -10, and -11, which include a prophage element and two Rhs elements.
Introduction
Pseudomonas aeruginosa is a ubiquitous environmental gram-negative bacterium that can be found in lakes, streams, soil, and plant matter (Green, et al., 1974, Hoadley, 1977, Rhame, 1979). In addition to thriving in multiple environmental niches, P. aeruginosa can infect many different organisms, including yeast (Hogan & Kolter, 2002), the nematode Caenorhabditis elegans (Mahajan-Miklos, et al., 1999), insects (Jander, et al., 2000), plants (Elrod & Braun, 1942, Rahme, et al., 1995), and mammals (Glazebrook, et al., 1978, Hammer, et al., 2003). In humans, it is considered an opportunistic pathogen and is a significant cause of both acute infections (e.g. hospital-acquired pneumonia, urinary tract infections, and wound infections) and chronic infections (e. g. respiratory infections in individuals with cystic fibrosis) (Stryjewski & Sexton, 2003).
Two aspects of P. aeruginosa’s genome evidently allow it to exploit differing environmental niches and infect a broad range of host organisms. First, it has an approximately 6.3 Mb genome (Stover, et al., 2000), one of the largest among bacteria. Thus it harbors a large amount of genetic material necessary for environmental versatility. Consistent with its ability to inhabit diverse niches, P. aeruginosa’s large genome has one of the highest proportions of predicted regulatory genes observed among bacterial genomes—8.4% of all predicted genes (Stover, et al., 2000). Second, the P. aeruginosa genome contains a large number of genomic islands. About 90% of the P. aeruginosa chromosome is conserved (Wolfgang, et al., 2003), but inserted within this core genome are genomic islands, which are found in some strains but not others (Schmidt, et al., 1996). Genomic islands are segments of DNA acquired by horizontal transfer (Dobrindt, et al., 2004, Lawrence, 2005). They are frequently integrated adjacent to tRNA genes, have a G+C content distinct from that of the host core chromosome, and contain components of mobile genetic elements (Reiter, et al., 1989, Cheetham & Katz, 1995). (Although the term “genomic island” usually implies a large region of DNA, we will here use it to refer to both large and small segments of integrated DNA.) In P. aeruginosa, genomic islands constitute an accessory genome that may account for 10% of an individual isolate’s genetic material (Spencer, et al., 2003, Shen, et al., 2006) and are thought to contribute to the ability of some P. aeruginosa strains to inhabit extreme environments.
Although the conserved core genome of P. aeruginosa has now been characterized by the sequencing of several strains (Stover, et al., 2000, Lee, et al., 2006, Mathee, et al., 2008), the wealth of genetic material present in genomic islands remains relatively unexplored. Studies performed to date have identified and characterized several islands. For example, a 49 kb island called P. aeruginosa Genomic Island 1 (PAGI-1) was identified in a urinary tract infection isolate and was found to be present in 85% of clinical strains tested (Liang, et al., 2001). The large genomic islands PAGI-2 and PAGI-3 were identified by sequencing a hypervariable region in two different strains: a cystic fibrosis lung isolate and an environmental aquatic isolate (Larbig, et al., 2002). PAPI-1 is representative of a large family of genomic islands derived from an ancestral pKLC102-like plasmid. pKLC102 is an 103.5 kb plasmid initially found in P. aeruginosa clone C strains that can exist as a plasmid or integrate into the chromosome, and can excise from the chromosome at a rate of up to 10% (He, et al., 2004, Klockgether, et al., 2004, Klockgether, et al., 2007). A recent study comparing the genomes of five sequenced P. aeruginosa strains identified 62 genomic locations where at least one strain differed from the others by at least 4 open reading frames (ORFs) (Mathee, et al., 2008). These loci were designated “regions of genomic plasticity (RGPs)” and represent hot spots for the presence of genomic islands. Therefore characterized genomic islands represent a small fraction of the genomic diversity present in P. aeruginosa (Wolfgang, et al., 2003).
Virulence is a complex trait requiring multiple steps, including entry into the host, adherence to and spread through host tissues, subversion of host defense systems, and induction of tissue damage (Finlay & Falkow, 1997). In P. aeruginosa, distinct strains appear to use a varying combination of factors to progress through these steps, and some of these factors appear to be encoded by genomic islands (Lee, et al., 2006). We therefore reasoned that unusually virulent strains of P. aeruginosa are likely to harbor a larger number of novel and interesting genomic islands. Thus in a previous report, we described the measurement of the virulence of a large collection of P. aeruginosa isolates to identify a candidate strain for studies aimed at identifying novel genomic islands (Battle, et al.). For this purpose, a set of 35 previously characterized P. aeruginosa clinical isolates (designated PSE isolates), all of which were originally cultured from patients with ventilator-associated pneumonia, was used (Hauser, et al., 2002). Each isolate was screened for virulence in a mouse model of acute pneumonia and a lettuce leaf model of virulence in plants (Schulert, et al., 2003, Battle, et al.). One isolate, PSE9, was noted to be highly virulent in both models and was chosen for further analysis. Subtractive hybridization was used to compare the genome of PSE9 to that of the less virulent but fully sequenced strain PAO1. This yielded 35 non-redundant sequences found in PSE9 but not PAO1. Of these, 13 sequences corresponded to previously identified P. aeruginosa genetic elements. Three of the remaining 22 novel sequences were used to identify a 99 kb pKLC102/PAPI-1 related genomic island designated PAGI-5, as described elsewhere (Battle, et al.). Here we describe the identification and characterization of an additional six PSE9 islands by this approach.
Materials and Methods
Construction and screening of a PSE9 fosmid library
Construction of the fosmid library of PSE9 genomic DNA has been previously described (Battle, et al.). The complete library was stored in ten 96-well plates. These plates were screened for the presence of subtractive hybridization sequences by PCR amplification using primers corresponding to the sequences (Table S2 of Supplemental Data). A three-tiered screening method was used, as previously described (Battle, et al.).
Sequencing of fosmids
Inserts in fosmids identified as containing subtractive hybridization products were sequenced using the EZ::TN <KAN-2> transposon-mediated sequencing approach (Epicentre) as previously described (Battle, et al.).
Sequence assembly, annotation, and analysis
Vector NTI Contig Express (InforMax, Inc, Frederick, MD) was used to assemble contiguous sequences. ORFs were identified using GenDB (Meyer, et al., 2003) and GeneMark (Lukashin & Borodovsky, 1998). G+C content was calculated by Vector NTI BioPlot (InforMax, Inc) from a sliding 100 bp window. BLASTN and BLASTP were used to identify nucleotide and amino acid sequence similarity, respectively (Altschul, et al., 1990). Vector NTI AlignX (Informax, Inc) was used to align sequences. The Dense Alignment Surface Transmembrane prediction method was used to identify potential transmembrane domains (Cserzo, et al., 1997).
Nucleotide sequence accession number
The sequences of PAGI-6, -7, -8, -9, -10, and -11 have been submitted to the National Center for Biotechnology Information (NCBI) gene bank under the accession numbers EF611302, EF611303, EF611304, EF611305, EF611306, and EF611307, respectively.
Results and Discussion
Identification of fosmids containing PSE9 genomic islands
As mentioned, subtractive hybridization of PSE9 with PAO1 had yielded 22 PSE9 sequences that did not correspond to characterized P. aeruginosa genomic islands (Battle, et al.). Three of these sequences were used to identify PAGI-5 (Battle, et al.). The remaining 19 sequences were used to screen a fosmid library of PSE9 genomic DNA to identify fosmids that contained these sequences. The library was screened as pools using primers designed to amplify subtractive hybridization sequences by PCR. One of the sequences was not present in the library, but the remaining 18 sequences were all found between one and five times. A number of different subtractive hybridization sequences were found together on each of several different fosmid clones, suggesting that they were contained within the same genomic island. Overall, 23 fosmid clones that contained at least one of the 18 subtractive hybridization sequences were identified. A subset of seven fosmid clones cumulatively contained all 18 of the subtractive hybridization sequences. This subset of clones was used in subsequent analyses.
Location of novel genomic islands
The locations of the identified genomic islands within the P. aeruginosa chromosome were determined. Primers were designed to hybridize to the fosmid backbone sequence flanking the insert cloning site to allow sequencing of the ends of each PSE9 genomic insert. Sequencing analysis was then performed. In five of seven fosmids, PAO1 sequence was found at both ends of the fosmid insert, and the remaining two had PAO1 sequence at one end, allowing placement of the insert in the P. aeruginosa core genome. The proximity of the flanking PAO1 sequence found in the latter two fosmids indicated that they represented opposite ends of a single genomic island. Overall, this analysis suggested that the set of analyzed fosmid inserts represented six distinct genomic islands located at different sites in the P. aeruginosa genome (Table 1 and Fig. S1 of the Supplemental Data).
Table 1.
Characteristics of PSE9 genomic islands.
| Genomic island | Size (bp) | Location* | Number of predicted ORFs | G+C% | Insertion site |
|---|---|---|---|---|---|
| PAGI-5# | 99,276 | 1,061,197 | 121 | 59.6 | tRNALys |
| PAGI-6 | 44,302 | 5,810,047 | 47 | 60.8 | tRNAThr |
| PAGI-7 | 22,479 | 4,439,857 | 20 | 55.8 | |
| PAGI-8 | 16,195 | 5,798,636 | 11 | 54.1 | tRNAPhe |
| PAGI-9 | 7,192 | 4,294,706 | 1 | 61.6 | |
| PAGI-10 | 2,194 | 2,759,146 | 1 | 52.7 | |
| PAGI-11 | 2,003 | 2,116,708 | 0 | 50.5 |
Genomic locations are given with respect to the PAO1 genome nomenclature (Stover, et al., 2000).
The identification and characterization of PAGI-5 is presented elsewhere (Battle, et al.). It is included here for completeness.
Sequencing of PSE9 genomic islands
To further characterize the PSE9 genomic islands, the complete nucleotide sequence of the subset of fosmids containing all 18 subtractive hybridization products was obtained. In cases in which the PSE9 genomic island extended beyond the end of the fosmid insert, PCR primers were designed to amplify a sequence at the border of the insert, and the fosmid library was rescreened for the presence of this sequence. In this way, the complete sequence of each PSE9 genomic island was obtained.
Altogether, six distinct genomic islands varying in size from 44 kb to 2 kb were identified (Table 1). Using the nomenclature system of Liang et al., Larbig et al., Klockgether et al., and Battle et al., who identified PAGIs -1, -2, -3, -4, and -5 (Liang, et al., 2001, Larbig, et al., 2002, Klockgether, et al., 2004, Battle, et al.), we named these six novel genomic islands PAGI-6, -7, -8, -9, -10, and -11 (Table 1). Each island was in turn annotated.
PAGI-6
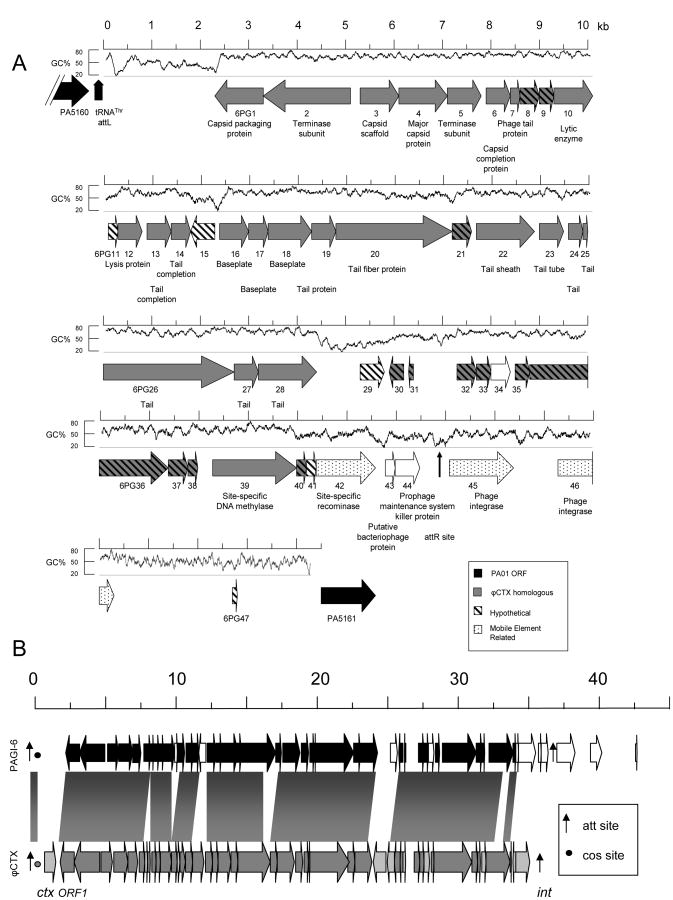
The first genomic island, PAGI-6, is 44,302 bp in size and has a G+C content of 60.6%, 6% less than that of the overall genome of P. aeruginosa (Fig. 1A). It is integrated into a site immediately flanking a tRNAThr gene (PA5160.1) between genes annotated to encode a drug efflux transporter (PA5160) and a dTDP-D-glucose 4,6-dehydratase (PA5161). (Unless otherwise noted, PAO1 gene designations will be used throughout this discussion (Stover, et al., 2000).) PAGI-6 has many large regions highly similar to ϕCTX, a cytotoxin-converting phage isolated from P. aeruginosa strain PA158 (Nakayama, et al., 1999). ϕCTX is a member of the P. aeruginosa R-pyocin related family of phages (Hayashi, et al., 1994). As their name implies, these phages have similarities to R-pyocin type bacteriocins (Shinomiya & Ina, 1989), bacterially derived proteins with antimicrobial properties (Riley & Wertz, 2002). It has been proposed that R-pyocins are defective phages that have been evolutionarily selected to function as bacteriocins (Nakayama, et al., 2000). Thus PAGI-6 appears to be or to have evolved from a prophage. Interestingly, PAGI-6 and ϕCTX have different chromosomal locations; in PSE9 PAGI-6 is integrated into the tRNAThr gene PA5160.1, but in P. aeruginosa strain PA158 the ϕCTX phage genome is integrated in tRNASer gene PA2603.1 (Hayashi, et al., 1993), neither of which were previously identified as RGPs (Mathee, et al., 2008).
Figure 1.
The PAGI-6 genomic island. (A) Map of PAGI-6. Arrows represent ORFs and are oriented in the direction of transcription. Arrows with gray backgrounds represent ORFs with similarity to ϕCTX sequences, and arrows with white backgrounds represent ORFs that lack ϕCTX similarity. Black arrows represent PAO1 ORFs that flank PAGI-6. ORFs without similarity to characterized ORFs are indicated with diagonal stripes, and ORFs expected to play a role in DNA mobility are speckled. tRNA attL and attR sites are represented by vertical arrows. G+C content is shown above the ORFs, calculated from a sliding 100 bp window. PAGI-6 ORFs are referred to as “6PG#”, where “#” is the sequential number of the ORF within the genomic island. (B) PAGI-6 alignment with the ϕCTX genome. Dark bands and ORFs represent conserved nucleotide sequences whereas light gray and white ORFs indicate unrelated sequences. att sites are represented by vertical arrows, and cos sites by circles.
PAGI-6 is somewhat larger than the genome of ϕCTX, which is 35,538 bp in size, but both elements are relatively conserved over the majority of their sequences (Fig. 1B). Notable differences include the absence of ϕCTX cytotoxin and ϕCTX integrase genes (ctx and int) from PAGI-6, and the presence of 7,403 bp segment of DNA that follows the attR site of PAGI-6 containing two predicted integrase genes and a stretch of DNA devoid of ORFs except for a small 99 bp predicted ORF. A more detailed description can be found in the PAGI-6 section of the Supplemental Data.
PAGI-7
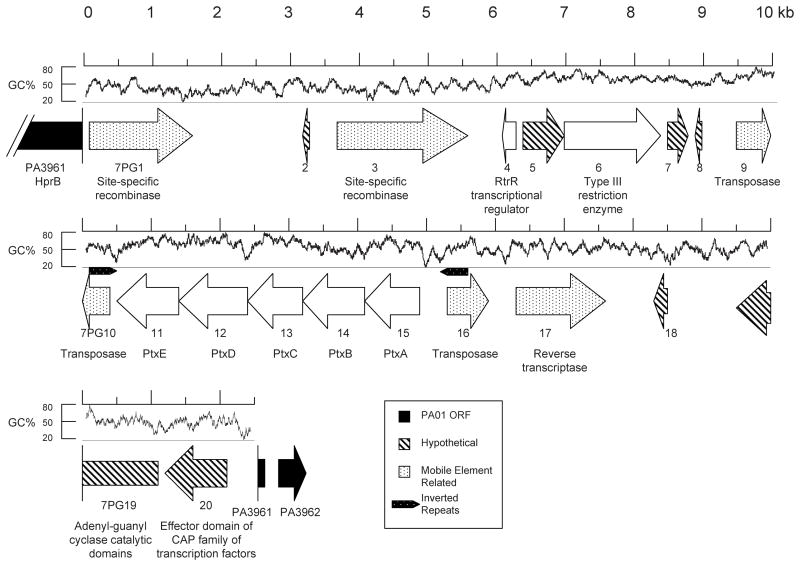
The next largest identified island was PAGI-7 (Fig. 2). This island is 22,479 bp in size and has a G+C content of 55.8%. PAGI-7 is not found within a tRNA gene, but instead is integrated within PAO1 ORF PA3961, which is predicted to encode HprB, a probable ATP-dependent helicase is also not a previously identified RGP. Although the island interrupts PA3961, no portion of this ORF is deleted or repeated. PAGI-7 contains 20 ORFs (Fig. 2, Table S1), including multiple mobility associated ORFs, predicted transcriptional regulators, and a predicted ptxABCDE operon. The latter was first identified in Pseudomonas stutzeri, where it is required for the oxidation of phosphite to phosphate (Metcalf & Wolfe, 1998, Costas, et al., 2001). A more detailed description can be found in the PAGI-7 section of the Supplemental Data.
Figure 2.
Map of PAGI-7. Arrows represent ORFs and are oriented in the direction of transcription. Arrows with white backgrounds represent PAGI-7 ORFs, and black arrows represent PAO1 ORFs that flank PAGI-7. ORFs without similarity to characterized ORFs are indicated with diagonal stripes, and ORFs expected to play a role in DNA mobility are speckled. The locations of inverted repeat sequences are indicated. G+C content is shown above the ORFs, calculated from a sliding 100 bp window. PAGI-7 ORFs are referred to as “7PG#”, where “#” is the sequential number of the ORF within the genomic island.
PAGI-8
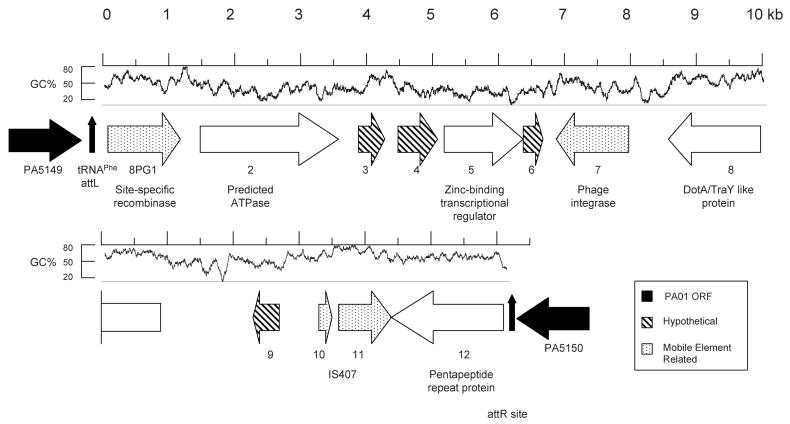
The next largest identified island was PAGI-8 (Fig. 3). This island, which is 16,195 bp in size and has a G+C content of 54.1%, is inserted into the genome immediately flanking a tRNAPhe gene (PA5149.1) at a site designated as RGP60 by Mathee and colleagues (Mathee, et al., 2008). A 44 bp duplication of the tRNA gene (representing an attR site) is at the end of the island. PA5149.1 is located between PA5149 and PA5150, which encode a hypothetical protein and a probable short-chain dehydrogenase, respectively. PAGI-8 contains 12 ORFs, including one predicted to encode a protein with 69% identity and 78% similarity to the TraY/DotA-like type IV secretion system protein of Cupriavidus metallidurans strain CH34 (formerly Ralstonia metallidurans), and 19% and 21% identity and 32% and 37% similarity to DotA of Legionella pneumophila and TraY of E. coli, respectively. Also in PAGI-8 are ORFs similar to an ATPase, and a zinc-binding transcriptional regulator, but no additional ORFs with similarity to other type IV secretion system genes. A more detailed description can be found in the PAGI-8 section of the Supplemental Data.
Figure 3.
Map of PAGI-8. Arrows represent ORFs and are oriented in the direction of transcription. Arrows with white backgrounds represent PAGI-8 ORFs, and black arrows represent PAO1 ORFs that flank PAGI-8. ORFs without similarity to characterized ORFs are indicated with diagonal stripes, and ORFs expected to play a role in DNA mobility are speckled. tRNA attL and attR sites are represented by vertical arrows. G+C content is shown above the ORFs, calculated from a sliding 100 bp window. PAGI-8 ORFs are referred to as “8PG#”, where “#” is the sequential number of the ORF within the genomic island.
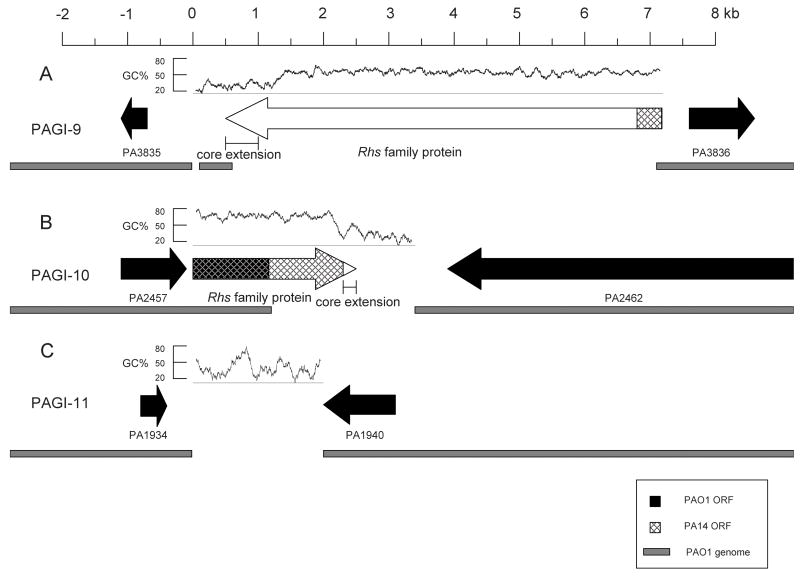
PAGI-9 and PAGI-10
The PAGI-9 island is 6,581 bp in size and is located in an intergenic region between PA3835 and PA3836, both of which encode hypothetical proteins (Fig. 4A). This region was not identified as an RGP by Mathee and colleagues (Mathee, et al., 2008). It has a G+C content of 63.4%. This island consists of a single very large ORF of 6,672 bp that is similar to the rearrangement hot spot (Rhs) family of genetic elements (Fig. 4A and Table S1 in Supplemental Data) (Hill, 1999). PAGI-10 is 2,194 bp in size and has a G+C content of 56.6%. It is located in RGP25 between PA2457 and PA2462 of PAO1, which encode a hypothetical protein with partial similarity to an Rhs core protein and an extremely large hypothetical protein (5,628 amino acids) with a low level of similarity to hemagglutinin, respectively (Fig. 4B). PAGI-10 replaces the PAO1 ORFs PA2458-61. PA2458 has partial similarity to an Rhs element, and PA2459-61 do not have similarity to known genes. Similar to PAGI-9, PAGI-10 contains a single 2,457 bp ORF with similarity to an Rhs core ORF. For additional information on PAGI-9 and -10 and Rhs elements, see the PAGI-9 and PAGI-10 section of the Supplemental Data.
Figure 4.
Maps of PAGI-9 (A), PAGI-10 (B), and PAGI-11 (C). Arrows with white backgrounds represent genomic island ORFs, and black arrows represent flanking PAO1 ORFs. The underlying gray bars indicate the extent of PAO1 conserved sequence. Cross-hatching represents conserved PA14 sequence. The predicted locations of the Rhs element core extensions are indicated. G+C content is shown above the ORFs, calculated from a sliding 100 bp window.
PAGI-11
PAGI-11 is the smallest of the identified genomic islands, consisting of 2,003 bp (Fig. 4C). As such, it verifies the power of the subtractive hybridization approach to detect genetic elements as small as 2 kb. PAGI-11 has a G+C content of 50.5% and does not contain any ORFs or repeated sequences. It is located in RGP52 between PA1934 and PA1940 of the PAO1 genome, which encode hypothetical proteins, although a region of PA1940 is similar to a catalase domain. PAGI-11 replaces a 5870 bp segment of the PAO1 genome containing PA1935-1939. PA1935 and PA1936 are predicted to encode proteins of unknown function, and PA1939 is similar to a gene encoding an ATP-dependant endonuclease. PA1937 and PA1938 are similar to two different IS911 ORFs predicted to encode transposases, and are almost completely identical to PA0979 and PA0978, which are part of the 8.9 kb tRNALys-associated genomic island of strain PAO1. In addition to PAO1, unique genomic insertions containing between one and five ORFs are found at this locus in strains PA14, PACS2, 2192, and C3719 (Mathee, et al., 2008). Thus it appears that these strains contain mobile genetic elements at the locus where PAGI-11 resides in PSE9.
Distribution of genomic islands in clinical isolates
To determine the frequency and distribution of PAGI-6, -7, -8, -9, and -10 in P. aeruginosa strains, we screened the collection of 35 clinical isolates for sequences found within these genomic islands (Table 2). PCR was used to amplify a sequence from each island in each isolate (Table S3). PAGI-6 was found in 2 (6%) of the 35 isolates. PAGI-7 and PAGI-9 where both present in the same 16 (46%) isolates. PAGI-8 and PAGI-10 where both found only in strain PSE9 (3%).
Table 2.
Distribution of PAGI sequences throughout collection of clinical isolates.
| Isolate | PAGI-6 | PAGI-7 | PAGI-8 | PAGI-9 | PAGI-10 |
|---|---|---|---|---|---|
| PSE1 | + | ||||
| PSE2 | + | + | |||
| PSE3 | + | + | |||
| PSE4 | |||||
| PSE5 | + | + | |||
| PSE6 | + | + | |||
| PSE7 | |||||
| PSE8 | + | + | |||
| PSE9 | + | + | + | + | + |
| PSE10 | |||||
| PSE11 | |||||
| PSE12 | + | + | |||
| PSE13 | |||||
| PSE14 | |||||
| PSE15 | |||||
| PSE16 | + | + | |||
| PSE17 | |||||
| PSE18 | |||||
| PSE19 | + | + | |||
| PSE20 | |||||
| PSE21 | |||||
| PSE22 | |||||
| PSE23 | + | + | |||
| PSE24 | |||||
| PSE25 | + | + | |||
| PSE26 | + | + | |||
| PSE27 | + | + | |||
| PSE28 | |||||
| PSE29 | |||||
| PSE30 | |||||
| PSE33 | + | + | |||
| PSE35 | |||||
| PSE37 | + | + | |||
| PSE39 | |||||
| PSE41 | + | + |
Conclusion
In conclusion, the information presented here along with that previously reported (Battle, et al.) demonstrates the utility of targeting a hypervirulent strain of P. aeruginosa as a source of genetic information found in the accessory genome. Applying this approach to a panel of clinical isolates has led to the identification of seven novel genomic islands varying in size from 99 kb to 2 kb and together containing 201 ORFs. Several are related to known pathogenicity islands, phages, or Rhs elements while others are quite novel. Many of these islands appear to be chimeric in nature, further demonstrating that composite genomic islands occur commonly in the evolution P. aeruginosa. While three of the seven islands are located in or adjacent to tRNA genes, the remaining four are not, indicating that alternative sites are also capable of being targeted for integration in P. aeruginosa. Together, these results shed additional light on the evolution of genomic islands in P. aeruginosa and attest to the vast amount of genetic information carried by these elements.
Supplementary Material
Acknowledgments
We wish to thank Peter Agron for advice and technical assistance with the subtractive hybridization technique, Laurence Rahme for assistance with the lettuce model of infection, and Kathryn Bieging for experimental assistance. This work was supported by an American Heart Association Predoctoral Fellowship (SEB) and NIH (K02 AI065615 and RO1 AI053674, ARH).
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Battle SE, Meyer F, Rello J, Kung VL, Hauser AR. The hybrid pathogenicity island PAGI-5 contributes to the highly virulent phenotype of a Pseudomonas aeruginosa isolate in mammals. J Bacteriol. 2008 doi: 10.1128/JB.00785-08. [ePub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheetham BF, Katz ME. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 4.Costas AM, White AK, Metcalf WW. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J Biol Chem. 2001;276:17429–17436. doi: 10.1074/jbc.M011764200. [DOI] [PubMed] [Google Scholar]
- 5.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 6.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 7.Elrod R, Braun A. Pseudomonas aeruginosa: Its role as a plant pathogen. J Bacteriol. 1942;44:633–645. doi: 10.1128/jb.44.6.633-645.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlay BB, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glazebrook JS, Campbell RS, Hutchinson GW, Stallman ND. Rodent zoonoses in North Queensland: the occurrence and distribution of zoonotic infections in North Queensland rodents. Aust J Exp Biol Med Sci. 1978;56:147–156. [PubMed] [Google Scholar]
- 10.Green SK, Schroth MN, Cho JJ, Kominos SK, Vitanza-jack VB. Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl Microbiol. 1974;28:987–991. doi: 10.1128/am.28.6.987-991.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammer AS, Pedersen K, Andersen TH, Jorgensen JC, Dietz HH. Comparison of Pseudomonas aeruginosa isolates from mink by serotyping and pulsed-field gel electrophoresis. Vet Microbiol. 2003;94:237–243. doi: 10.1016/s0378-1135(03)00103-2. [DOI] [PubMed] [Google Scholar]
- 12.Hauser AR, Cobb E, Bodí M, Mariscal D, Vallés J, Engel JN, Rello J. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002;30:521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi T, Matsumoto H, Ohnishi M, Terawaki Y. Molecular analysis of a cytotoxin-converting phage, phi CTX, of Pseudomonas aeruginosa: structure of the attP-cos-ctx region and integration into the serine tRNA gene. Mol Microbiol. 1993;7:657–667. doi: 10.1111/j.1365-2958.1993.tb01157.x. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi T, Matsumoto H, Ohnishi M, Yokota S, Shinomiya T, Kageyama M, Terawaki Y. Cytotoxin-converting phages, phi CTX and PS21, are R pyocin-related phages. FEMS Microbiol Lett. 1994;122:239–244. doi: 10.1111/j.1574-6968.1994.tb07174.x. [DOI] [PubMed] [Google Scholar]
- 15.He J, Baldini RL, Deziel E, et al. The broad host range pathogen Pseudomonas aeruginosa strain PA14 carries two pathogenicity islands harboring plant and animal virulence genes. Proc Natl Acad Sci U S A. 2004;101:2530–2535. doi: 10.1073/pnas.0304622101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill CW. Large genomic sequence repetitions in bacteria: lessons from rRNA operons and Rhs elements. Res Microbiol. 1999;150:665–674. doi: 10.1016/s0923-2508(99)00125-4. [DOI] [PubMed] [Google Scholar]
- 17.Hoadley AW. Pseudomonas aeruginosa in surface waters. In: Young VM, editor. Pseudomonas aeruginosa: Ecological aspects and patient colonization. Raven Press; New York: 1977. pp. 31–57. [Google Scholar]
- 18.Hogan DA, Kolter R. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 2002;296:2229–2232. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 19.Jander G, Rahme LG, Ausubel FM. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klockgether J, Reva O, Larbig K, Tummler B. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol. 2004;186:518–534. doi: 10.1128/JB.186.2.518-534.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klockgether J, Wurdemann D, Reva O, Wiehlmann L, Tummler B. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J Bacteriol. 2007;189:2443–2459. doi: 10.1128/JB.01688-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larbig KD, Christmann A, Johann A, et al. Gene islands integrated into tRNA(Gly) genes confer genome diversity on a Pseudomonas aeruginosa clone. J Bacteriol. 2002;184:6665–6680. doi: 10.1128/JB.184.23.6665-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence JG. Horizontal and vertical gene transfer: the life history of pathogens. Contrib Microbiol. 2005;12:255–271. doi: 10.1159/000081699. [DOI] [PubMed] [Google Scholar]
- 24.Lee DG, Urbach JM, Wu G, et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang X, Pham XQ, Olson MV, Lory S. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J Bacteriol. 2001;183:843–853. doi: 10.1128/JB.183.3.843-853.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukashin AV, Borodovsky M. GeneMark. hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahajan-Miklos S, Tan M-W, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 28.Mathee K, Narasimhan G, Valdes C, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metcalf WW, Wolfe RS. Molecular genetic analysis of phosphite and hypophosphite oxidation by Pseudomonas stutzeri WM88. J Bacteriol. 1998;180:5547–5558. doi: 10.1128/jb.180.21.5547-5558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer F, Goesmann A, McHardy AC, et al. GenDB--an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama K, Kanaya S, Ohnishi M, Terawaki Y, Hayashi T. The complete nucleotide sequence of phi CTX, a cytotoxin-converting phage of Pseudomonas aeruginosa: implications for phage evolution and horizontal gene transfer via bacteriophages. Mol Microbiol. 1999;31:399–419. doi: 10.1046/j.1365-2958.1999.01158.x. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama K, Takashima K, Ishihara H, et al. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol. 2000;38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 33.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 34.Reiter WD, Palm P, Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989;17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhame FS. The ecology and epidemiology of Pseudomonas aeruginosa. In: Sabath LD, editor. Pseudomonas aeruginosa: The organism, diseases it causes, and their treatment. Hans Huber Publishers; Bern: 1979. pp. 31–51. [Google Scholar]
- 36.Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol. 2002;56:117–137. doi: 10.1146/annurev.micro.56.012302.161024. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt KD, Tummler B, Romling U. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J Bacteriol. 1996;178:85–93. doi: 10.1128/jb.178.1.85-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulert GS, Feltman H, Rabin SDP, Martin CG, Battle SE, Rello J, Hauser AR. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J Infect Dis. 2003;188:1695–1706. doi: 10.1086/379372. [DOI] [PubMed] [Google Scholar]
- 39.Shen K, Sayeed S, Antalis P, et al. Extensive genomic plasticity in Pseudomonas aeruginosa revealed by identification and distribution studies of novel genes among clinical isolates. Infect Immun. 2006;74:5272–5283. doi: 10.1128/IAI.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinomiya T, Ina S. Genetic comparison of bacteriophage PS17 and Pseudomonas aeruginosa R-type pyocin. J Bacteriol. 1989;171:2287–2292. doi: 10.1128/jb.171.5.2287-2292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spencer DH, Kas A, Smith EE, et al. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J Bacteriol. 2003;185:1316–1325. doi: 10.1128/JB.185.4.1316-1325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 43.Stryjewski ME, Sexton DJ. Pseudomonas aeruginosa infections in specific types of patients and clinical settings. In: Hauser AR, Rello J, editors. Severe infections caused by Pseudomonas aeruginosa. Vol. 7. Kluwer Academic Publishers; Boston: 2003. pp. 1–15. [Google Scholar]
- 44.Wolfgang MC, Kulasekara BR, Liang X, et al. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2003;100:8484–8489. doi: 10.1073/pnas.0832438100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.