Abstract
The genome of the opportunistic pathogen Pseudomonas aeruginosa encodes over 60 two-component sensor kinases and uses several (including RetS and GacS) to reciprocally regulate the production of virulence factors involved in the development of acute or chronic infections. We demonstrate that RetS modulates the phosphorylation state of GacS by a direct and specific interaction between these two membrane-bound sensors. The RetS–GacS interaction can be observed in vitro, in heterologous systems in vivo, and in P. aeruginosa. This function does not require the predicted RetS phosphorelay residues and provides a mechanism for integrating multiple signals without cross-phosphorylation from sensors to noncognate response regulators. These results suggest that multiple two-component systems found in a single bacterium can form multisensor signaling networks while maintaining specific phosphorelay pathways that remain insulated from detrimental cross-talk.
Keywords: Molecular switch, two-component system, signal transduction, histidine kinase, cystic fibrosis, biofilm
Two-component system (TCS) signaling pathways are a major signaling mechanism in bacteria and archaea, and are also found in simple eukaryota and higher plants (Wolanin et al. 2002). These diverse organisms capitalize on TCS pathways to monitor critical external and internal stimuli (including levels of nutrients, concentration of ions and gases, temperature, redox states, and cell density) and translate these signals into adaptive responses. Classical TCS pathways share a conserved core architecture: a homodimerizing histidine kinase protein domain (the “sensor”) and a cognate receiver domain (the “response regulator”), coupled mechanistically through a histidine-aspartic acid phosphorelay (Stock et al. 2000). Most cognate sensor response regulator pairs are also linked genetically, encoded by adjacent loci in the chromosome (Alm et al. 2006). Although a single bacterial species can encode up to hundreds of genes specifying TCS pathways, it appears that these systems are insulated against detrimental cross-phosphorylation between sensors and noncognate response regulators (Bijlsma and Groisman 2003; Baker and Stock 2007; Laub and Goulian 2007). The identification of multistep phosphorelays (with intermediary proteins between sensor and regulator) and branched pathways (phosphotransfer between one sensor and multiple response regulators and vice versa) (Laub and Goulian 2007) suggests that TCS pathways have the capacity to form sensitive and complex signaling networks.
In contrast to microorganisms with restricted habitats, the genomes of bacteria capable of occupying a number of diverse environments typically contain a disproportionately large number of genes encoding signal transduction and regulatory systems, including TCSs that allow them to sense and respond to a wide range of environmental signals. For opportunistic bacterial pathogens, a number of these systems regulate the expression of genes necessary for transitioning from the environmental reservoir to the host, overcoming innate defense mechanisms and initiating the disease process. The bacterial pathogen Pseudomonas aeruginosa dedicates a large portion (∼8%) of its genome toward transcriptional regulation that includes coding sequences for the components of ∼70 TCS pathways (Stover et al. 2000). This organism draws on a broad arsenal of virulence factors to shift from its primary reservoir in the environment to a habitat inside its human host, where it causes acute infections in sites compromised by injury or lacking adequate immune surveillance. Animal models have identified specific virulence mechanisms, including flagellar motility and the Type III toxin secretion system (TTSS) that are required for acute infection (Fleiszig et al. 1997; Feldman et al. 1998). In addition to acute infections, changes in lung pathology resulting from cystic fibrosis (CF) lead to chronic P. aeruginosa infections that are the primary cause of mortality for patients with this common hereditary disease. Isolates of P. aeruginosa from CF patients, in contrast to those from acutely infected patients, are characterized by acquisition of stable mutations leading to derepression of alginate production (mucoidy) auxotrophy, loss of motility, O-antigen production, and TTSS function (Hancock et al. 1983; Mahenthiralingam et al. 1994; Jain et al. 2004; Li et al. 2005). Comparative genomic analyses of clonal isolates from acute and chronic infections suggest a high frequency of mutations in regulatory genes (Smith et al. 2006). These observations suggest that some virulence factors associated with acute infections may be dispensable in chronic disease.
Recent studies suggest that P. aeruginosa coordinates the activities of multiple orphan TCS sensor kinases as a genetic “switch” that reciprocally regulates genes involved in acute and chronic infections. The unusual hybrid sensor kinase RetS (Fig. 1A) is required for TTSS activation, concomitant repression of biofilm formation, and colonization/dissemination in murine acute infection models (Goodman et al. 2004; Laskowski et al. 2004; Zolfaghar et al. 2005). Genes under RetS control are inversely regulated by two other sensor kinases: GacS and LadS. Microarray studies have shown that the regulatory activity of RetS appears to be channeled through a GacS and GacA TCS (Brencic and S. Lory, in prep.), found in a number of different microorganisms. Genetic studies in P. aeruginosa suggest that in analogy with the homologous BarA/UvrY system in Escherichia coli, the sensor kinase GacS may phosphorylate the response regulator GacA to initiate downstream signaling (Lapouge et al. 2008). In turn, phosphorylated GacA activates the transcription of the two small RNAs, RsmZ and RsmY, which are antagonists of the translational regulator RsmA. RsmA binds to specific mRNA targets, stabilizing some and inducing the degradation of others. In addition to GacS, another sensor kinase, LadS, functions as an activator of expression of RsmZ and RsmY (Ventre et al. 2006). It therefore appears that the signaling network consisting of RetS, LadS, and GacS/GacA controls the expression of a significant number of P. aeruginosa virulence genes primarily (and perhaps exclusively) at the level of mRNA translation and/or stability. Taken together, these genetic approaches identify multiple components in a “switch” between acute and chronic infection phenotypes and suggest that these TCS proteins may function outside of the classical sensor kinase response regulator signaling paradigm.
Figure 1.
RetS suppresses rmsZ expression via the TCS GacS/GacA. (A) Domain organization of the Pseudomonas aeruginosa GacS and RetS sensor kinases and the map of the deletion constructs used for protein purification and two-hybrid analysis (GacSc, GacSHK, RetSc, and RetSHK). The locations of conserved phosphorelay residues are marked above each protein (amino acid positions); Pfam domain designations and construct boundaries are indicated (domain sizes not to scale). (B) RetS function is dependent on GacS and GacA. Wild-type P. aeruginosa and various deletion strains carrying chromosomal rsmZ-lacZ fusions were grown in LB for 8 h and assayed for β-galactosidase activity. Error bars represent one standard deviation from the mean.
Here we report that RetS modulates the phosphorylation state of GacS through a direct and specific protein interaction. This interaction determines the rate of phosphotransfer between GacS and its cognate response regulator GacA. The observation that two sensor kinases can modulate downstream signaling by direct interaction suggests that higher-order TCS signaling networks can be achieved without cross-phosphorylation between sensors and noncognate response regulators.
Results
GacS and GacA are required for RetS function
Previous work suggests that three physically unlinked sensor kinases (gacS, retS, and ladS) and the response regulator gacA regulate virulence factor expression via the small RNA rsmZ (Supplemental Fig. S1). The observation that rsmZ promoter activity is similarly derepressed in retS and retS ladS strain backgrounds indicates that LadS is not directly required for RetS function but instead functions independently (Ventre et al. 2006). In order to establish the epistatic relationships between RetS, GacS, and GacA in regulating rsmZ expression, we measured the activity of an rsmZ-lacZ fusion in varying genetic backgrounds. In agreement with previous reports, transcription from the rsmZ promoter is activated by GacS or GacA and repressed by RetS (Fig. 1B; data not shown). Introduction of the retS gene tagged with the coding sequence for the VSV-G epitope into the retS mutant strain returns rsmZ-lacZ activity to wild-type levels (Supplemental Fig. S2) with a concomitant restoration of other phenotypes disregulated in a retS mutant, including abolishment of hyperbiofilm formation and of Type III secretion (data not shown). Derepression of rsmZ promoter activity in the retS mutant is abolished by in-frame deletion of gacS or gacA, indicating that RetS function requires the presence of GacS and GacA (Fig. 1B).
One possible explanation for the observation that the activation of the rsmZ promoter activity in the retS mutant requires gacS and gacA is RetS-dependent repression of transcription of one or both of these genes. This explanation for RetS function, however, seems very unlikely. Genomewide transcriptional profiling indicates that gacS and gacA transcript levels are not altered in a retS mutant, suggesting that RetS does not influence the expression of either one of these genes (Goodman et al. 2004). To assess whether other regulatory factors are required for RetS-mediated rsmZ regulation, we screened an ∼100,000-colony transposon insertion library for mutants that induce or repress rsmZ-lacZ transcription. Repeated identification of retS as the strongest negative regulator (approximately fivfold to 10-fold effects; eight independent insertions) and gacS, gacA, and rsmA as the strongest positive regulators (∼50- to 500-fold effects; three, five, and four independent insertions, respectively) of rsmZ transcription is consistent with the hypothesis that proteins encoded by these genes are the central components of the regulatory network and could interact at some level other than transcription.
GacS interacts with RetS in P. aeruginosa
The observation that RetS function requires GacS, the initial signaling element in the Gac/Rsm pathway, suggests that these proteins may function through direct interaction. To test this hypothesis, we assessed whether RetS forms a complex with GacS in vivo and they can be copurifed from detergent solubilized P. aeruginosa membrane preparations. We coexpressed GacS-His6 with RetS-VSV-G in a P. aeruginosa gacS gacA retS mutant and used Ni-NTA affinity chromatography to capture complexes with GacS-His6 (Fig. 2). Pull-down of a complex of RetS-VSV-G and GacS-His6 could be clearly demonstrated, while two inner membrane proteins XcpT or XcpY were not detected in a complex with GacS-His6, even after sevenfold concentration. To further test the avidity of the GacS:RetS interaction, we treated the cells with the chemical cross-linker dithiobis[succinimidylpropionate] (DSP) prior to solubilization of membranes and Ni-NTA affinity chromatography. No enhancement in the recovery of RetS bound to GacS was noted, suggesting that the two proteins form highly stable, detergent resistant complexes.
Figure 2.
Specific pull-down of GacS:RetS multiprotein complexes in vivo. A gacS retS gacA mutant of P. aeruginosa was transformed with plasmids expressing gacSHis6 and/or retSVSV-G genes and grown to OD ∼3.0. Cultures were split into two aliquots of 500 mL, washed with PBS, and treated with 1 mM of the membrane permeable cross-linker DSP (“DSP+”) or DMSO (“DSP−“) for 30 min. Cells were harvested, lysed, divided into two aliquots for total membrane purification (“M”) or for pull-down of GacSHis6 and GacS-associated proteins by Ni(II)-NTA metal affinity chromatography (“NiNTA-E”) in the presence of CHAPS as a membrane-solubilizing detergent. Total membranes (M) and Ni(II)-NTA eluate (“NiNTA-E”) fractions were analyzed by SDS-PAGE and probed by Western immunoblotting with antibodies against the VSV-G and His6 tags, XcpT, and XcpY. NiNTA-E lanes were loaded at a sevenfold higher cell equivalent per lane.
The cytoplasmic domains of RetS and GacS interact directly in vitro
In order to assess whether RetS and GacS interaction is direct, we determined the binding affinity of the purified cytoplasmic (soluble) domains of these proteins by isothermal titration calorimetry. We cloned, expressed, and purified the cytoplasmic portions of RetS and GacS (and of the unrelated sensor kinase PilS) as N-terminal His6 fusions (Fig. 1A). The resulting proteins (RetSc, GacSc, PilSc) were soluble, expressed at high levels, and were purified to >95% purity by a combination of metal affinity chromatography and gel filtration (Supplemental Fig. S3). Interaction of GacSc or PilSc with RetSc was assessed by isothermal titration microcalorimetry. We used an ∼100-fold molar excess of RetSc relative to GacSc or PilSc over a maximum of 30 microinjections. These calorimetric measurements and subsequent modeling of the binding isotherm using a single-binding-site model provide a characterization of GacSc–RetSc protein–protein-binding thermodynamics (Fig. 3A), revealing a large entropic loss counteracted by a large binding enthalpy (−855 kcal/mol). The estimated dissociation constant (K d) was 33 nM. These results imply significant and extensive structural rearrangements upon protein–protein interaction. In contrast, similar experiments using the unrelated sensor kinase PilSc as the titrated protein did not show any measurable interaction with RetSc above background (Fig. 3B), further supporting the hypothesis that GacSc–RetSc complex formation is direct and specific, and does not require the transmembrane domains. Isothermal titration calorimetry was also performed with GacSc as the titrant and PilSc as the receptor, and the results showed also no signal above the background dilution heat (Fig. 3C).
Figure 3.
Isothermal titration calorimetric analysis of RetSc binding to GacSc. Titration of RetSc (0.1 μM) with 10 μM GacSc (A) or PilSc (B) or titration of PilSc (0.1 μM) with 10 μM GacSc (C). Heat of binding was measured by isothermal titration calorimetry in a VP-ITC microcalorimeter. The top panels show the baseline corrected titration data, and the bottom panels show the binding isotherm fitted using nonlinear binding models.
The kinase core domains of GacS and RetS are sufficient for in vivo interaction
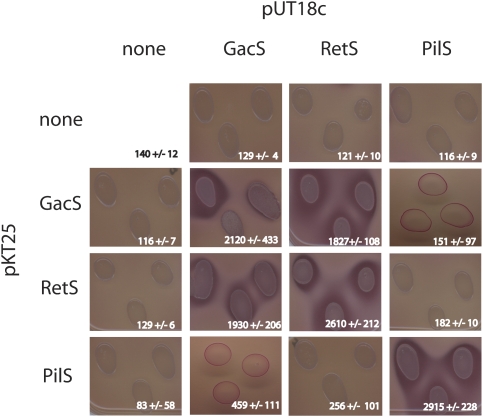
TCS sensor kinases have been shown to form homodimers via their kinase core HisKA-H+ATPase domains of each monomer (Tomomori et al. 1999). We used a bacterial two-hybrid system to determine whether these domains also mediate the GacS–RetS interaction in vivo. The kinase core domains of RetS, GacS, and PilS were cloned in bacterial two-hybrid expression vectors to create C-terminal fusions to two complementary fragments of the Bordetella pertussis adenylate cyclase (CyaA). The GacS fusion also included most of the HAMP domain except for the first 18 amino acids. When introduced into a reporter strain of E. coli, interacting protein fusions reconstitute a functional adenylate cyclase, increasing the cellular pool of cAMP (Karimova et al. 1998). As expected for this family of proteins, strong signals indicative of interactions were detected between homodimers of RetS, GacS, and PilS (Fig. 4). Strains of E. coli coexpressing the HisKA/H + ATPase domain of RetS and the corresponding domain of GacS, as CyaA fusions, also resulted in a significant increase in cAMP levels, suggesting that these domains mediate heterodimer formation between RetS and GacS monomers in vivo. In contrast, the fusions containing the HisKA/H + ATPase domains of RetS and PilS or PilS and GacS were unable to reconstitute significant levels of active CyaA when coexpressed in E. coli. Reciprocal exchange of the fusion domains resulted in the detection of both the homo- and heterodimeric interactions.
Figure 4.
Cytoplasmic domains of RetS and GacS interact in vivo. DNA fragments encoding the HisKA/H+ATPase domains of GacS, RetS, and PilS (as a control) were expressed as fusions with one of two fragments of Bordetella adenylate cyclase from vectors pKT25 and pUT18c. Reconstitution of adenylate cyclase in the E. coli strain DHM1 was detected by the ability of the cells to ferment maltose when grown on MacConkey agar plates. The interactions were also quantified by β-galactosidase assays using liquid cultures of the same cells (average Miller units and standard deviations are shown in each panel).
In vitro reconstitution of the GacS–GacA phosphorelay
The observations that GacS and GacA are required for RetS function in P. aeruginosa and that RetS and GacS interact directly in vivo and in vitro raised the possibility that RetS could function by acting as a phosphatase of GacS∼P or by interfering with GacS autophosphorylation. Since sensor kinases posses an intrinsic ATPase activity, we examined the abilities of RetS and GacS to hydrolyze ATP. Purified GacSc and RetSc proteins were incubated with γ[32P]ATP or α[32P]ATP followed by an analysis of the reaction products by PEI-cellulose thin-layer chromatography. Depending on the location of the label in the substrate ATP, [32P]Pi and α[32P]ADP were detected in the presence of GacSc but not with RetSc (Fig. 5A), implying a lack of autophosphorylation/ATP-phosphatase activity of the latter protein. For some hybrid sensor kinases, fast rates of phosphotransfer to the conserved aspartic acid residues in the receiver domain and subsequent loss of the phosphate group prevent the accumulation of phosphorylated protein; this autophosphatase activity can be blocked by mutation of these aspartic acid residues (Rasmussen et al. 2006). We therefore mutated each of the conserved residues—H424, D713, and D858—of RetSc (Fig. 1A). No autophosphorylation or phosphatase activity was observed in any of these proteins (Fig. 5A), suggesting that RetSc very likely lacks this intrinsic enzymatic activity.
Figure 5.
GacS autophosphorylation and phosphotransfer to GacA in vitro. (A) ATPase activity of GacSc. GacSc, and allelic variants of RetSc were incubated with [α32P ] or [γ32P]ATP for 60 min and reactions were spotted on PEI-cellulose TLC plates for nucleotide analysis. In the presence of [α32P]ATP, GacSc produced labeled ADP, consistent with γ-phosphatase activity. Incubation of GacSc with [γ32P]ATP released [32P]Pi and produced a distinct radioactive signal at the origin, implying the formation of GacSc∼P. (B) Autophosphorylation of GacS and phosphotransfer to GacA. GacSc was incubated with [γ32P]ATP for up to 60 min in the presence or absence of the response regulator GacA (purified as an MBP fusion) and reaction products were analyzed by SDS-PAGE. Phosphorylation was determined by storage phosphorimaging followed by staining with Coomassie Brilliant Blue. (C) GacSc∼P half-life. GacSc was incubated with [γ32P]ATP for 30 min and unincorporated nucleotides were removed by G-25 spin chromatography. At different time intervals, the phosphoprotein preparation was treated with SDS-sample buffer and analyzed by SDS-PAGE. Phosphorylation was determined by storage phosphorimaging and data were fitted into an exponential decay model.
In order to define the consequences of RetS/GacS interaction on GacA phosphorylation, we first developed an in vitro phosphotransfer system between GacS and GacA. GacSc was incubated in the presence of γ[32P]ATP and the labeled reaction products were visualized following SDS-PAGE and autoradiography (Fig. 5B). GacSc autophosphorylation reached saturation between 45 and 60 min and was Mg2+-dependent, as is common for proteins of this class. The half-life of GacSc∼P was determined to be ∼120 min after separation of unincorporated ATP (Fig. 5C). Mn2+ could also substitute for Mg2+ as cofactor (data not shown). When GacSc was incubated in the presence of GacA (purified as a maltose-binding protein [MBP] fusion protein), phosphotransfer to the response regulator was observed after as early as 5 min (Fig. 5B). No accumulation of phosphorylated RetSc was observed over a 120-min time course, in the presence of either Mg2+ or Mn2+ (data not shown).
RetS suppresses GacS autophosphorylation in vitro
In order to determine whether the purified RetSc protein affects the autophosphorylation reaction of GacSc, we incubated these two proteins in the presence of γ[32P]ATP. RetSc was also incubated with γ[32P]ATP and PilS (Fig. 6). Incubation of RetSc and GacSc at increasing molar ratios (from 1:1 to 20:1) lead to a significant decrease in autophosphorylation of GacSc. In contrast, RetSc had no effect on the ability of PilSc to autophosphorylate (Fig. 6A).
Figure 6.
RetS suppresses GacS autotophosphorylation in a phosphorelay-independent fashion. (A) Dose-dependent inhibition of GacSc phosphorylation by RetSc. Purified GacSc and the unrelated sensor kinase PilSc were incubated in the presence of increasing amounts (0–32 μM) of RetSc for 5 min, followed by addition of 500 μM [γ32P]ATP at 0.6 Ci/mmol for 15 min. Reaction products were analyzed by SDS-PAGE and storage phosphorimaging. Signal intensities are quantified in the bottom panel. (B) The conserved phosphorelay residues of RetSc are not required for its ability to inhibit GacSc phosphorylation. Purified GacSc (or PilSc) were incubated with 500 μM [γ32P]ATP for 15 min in the presence of wild-type RetSc and variants with alanine or asparagine substitutions for the canonical histidine and aspartate residues, respectively. The extent of phosphorylation was determined following SDS-PAGE for storage phosphorimaging. (C) RetSc reduces the phosphorylation level of prephosphorylated GacSc. GacSc was preincubated with [γ32P]ATP and subjected to size-exclusion chromatography on a G-25 column to remove unincorporated [γ32P]. Phospho-GacSc was incubated with wild-type RetSc or its allelic variants for 30 min.
We next determined whether the RetSc-mediated inhibition of GacSc autophosphorylation is dependent on the canonical residues involved in TCS phosphorelay. We expressed and purified three RetSc variants with substitutions in the conserved histidine and/or aspartic acid residues (RetSc [H], RetSc [DDD], and RetSc [HDDD]). As observed for RetSc, each mutant protein was able to reduce phosphorylation of GacSc but not PilSc, suggesting that the putative phosphorelay residues in RetS are not required for its interaction with GacS (Fig. 6B). To determine whether the phosphate on GacS can be removed by RetS, we first purified GacSc∼P by gel filtration on a G-25 column followed by incubation with RetSc. RetSc was able to reduce the phosphorylation level of GacSc∼P under these conditions, suggesting that RetS can modulate the steady-state phosphorylation level of GacS homodimers in the absence of free ATP (Fig. 6C). To test whether the RetS core kinase domain (shown to be sufficient for in vivo homo- and heterodimer formation in the two-hybrid system) is also sufficient for the suppression of GacS autophosphorylation, we purified the HisKA-H + ATPase kinase core fragment of RetS (RetSHK) and its derivative with a null mutation in the H424 residue (Fig. 1A). Both of these proteins were also able to reduce GacS∼P levels (Fig. 6C). In none of these experiments we were able to detect accumulation of RetSc∼P.
RetS does not require its classical phosphorelay residues for regulation of rsmZ in vivo
The observation that the mutant alleles of RetSc still function to prevent GacSc phosphorylation in vitro suggests that these mutant proteins should also be functional in regulating the transcription of rsmZ in P. aeruginosa. To test this hypothesis, we introduced these mutations into the native chromosomal copy of the retS gene in an rsmZ-lacZ parental strain and assessed their activity by their ability to repress transcription from the rsmZ promoter (Fig. 7). Unlike the retS deletion strain, each mutant showed wild-type levels of rsmZ transcription, suggesting that the canonical phosphorelay residues are not required for RetS function in vivo. Low-copy plasmids carrying these alleles were similarly able to restore RetS activity to a retS mutant strain (Supplemental Fig. S4). Analogous results were obtained when using the inversely regulated exoS-lacZ fusion (data not shown).
Figure 7.
RetS-dependent regulation of rsmZ is independent of conserved phosphorelay residues in vivo. A P. aeruginosa carrying a chromosomal rsmZ-lacZ fusion and various chromosomal alleles of retS (expressing wild-type RetS and variants with single or multiple point mutations inactivating predicted phosphorelay residues) were grown in LB for 8 h and assayed for β-galactosidase activity. The conserved residues tested are all dispensable for RetS function in this assay.
Discussion
Bacteria use TCS as the primary mechanism for environmental adaptation. Pathogenic microorganisms make extensive use of TCS phosphorelays to assimilate signals in infected hosts and coordinate the expression of virulence determinants. Genomes of most bacteria contain multiple copies of genes encoding TCS proteins ranging from a few to 278 in Myxococus xanthus (Whitworth and Cock 2008). Branched pathways, where multiple sensors target the same response regulator or a single sensor phosphorylates several response regulators, capitalize on the conserved domain organization in these proteins. However, despite similarities between the components of distinct pathways, phosphorelays are well-insulated from cross-talk between systems, and cells have evolved mechanisms to avoid potentially deleterious interference between signals transmitted through specific pathways (Laub and Goulian 2007).
Previous genetic studies of the opportunistic pathogen P. aeruginosa implicated at least three orphan sensor kinases (GacS, RetS, and LadS) in coordinating the expression of virulence factors associated with the transition between acute and chronic infections (Goodman et al. 2004; Laskowski and Kazmierczak 2006; Ventre et al. 2006). A comparison of transcriptomes of wild-type P. aeruginosa with those of a retS and a retS gacS double mutant shows a significant overlap. In total, 84% of the 397 genes of the retS regulon were also reciprocally controlled by GacS, including type III secretion, type VI secretion, pili and biofilm-related genes (Brencic and S. Lory, in prep.).Based on genetic data in various Pseudomonas species, and on biochemical data in E. coli and Salmonella, GacS is the likely cognate sensor kinase for the response regulator GacA, which is the transcription factor controlling the expression of the rsm family of small RNAs. Additionally, neither RetS nor LadS are encoded by genes linked to specific response regulators; however, they also regulate the transcription of the P. aeruginosa small RNA RsmZ (Ventre et al. 2006). The findings described in this study indicate that Pseudomonas GacS can transphosphorylate GacA, and suggest that the sensor kinase RetS exerts its regulatory activity through a direct and specific interaction with GacS, interfering with its phosphorylation and presumably, limiting phosphorylation of GacA. In this way, the signals from two sensor kinases are integrated not at the level of cross-phosphorylation of a shared response regulator, but instead, by the formation of a heteromeric complex in the bacterial membrane.
Several lines of evidence suggest that the interaction between GacS and RetS is direct. GacS bound to RetS can be isolated from P. aeruginosa membranes as a detergent-stable complex. Fusion proteins of putative interactive domains of the proteins expressed in E. coli strongly activate a two-hybrid reporter system that detects heterodimer formation. In vitro, the purified soluble portion of RetS directly binds the corresponding domain of GacS with high affinity. Reconstituted phospho-transfer reactions, consisting of purified components, further suggest that the RetS/GacS interaction modulates the phosphorylation state of GacS. Interestingly, the conserved histidine residue in the histidine phosphotransferase domain (H424) and the aspartate resides in the adjacent two receiver domains (D713 and D858) are not required for RetS activity in regulating rsmZ expression in vivo or for its ability to interfere with GacS phosphorylation in vitro. Recent studies in the P. aeruginosa strain PA103 have shown that a mutant RetS allele missing the conserved residue H424 is partially able to complement a retS deletion for TTSS expression, although this phenotype is reported to be temperature-dependent (Laskowski and Kazmierczak 2006). This group also reports that the residues D713 and D858 are dispensable and required, respectively, for RetS function. The basis of these discrepancies is unclear. Additionally, work by Hsu et al. (2008) has shown that RetS, and three additional P. aeruginosa sensor histidine kinases, can phosphorylate the histidine phosphotransfer protein HptB in vitro. The same four kinases were shown to interact with HptB in a bacterial two-hybrid assay. Hsu et al. (2008) further show that although RetS lacks autophosphorylation activity as we have observed, it can receive phosphate from phosphorylated HptB. Whether this reaction occurs in vivo, requires the conserved RetS phosphorelay residues, or has a role in the downstream rsmZ-dependent signaling pathway examined here is not known. Nevertheless, this work raises the intriguing possibility that in addition to interfering with GacS autophosphorylation, RetS may participate in additional phosphotransfer reactions, which may explain the evolutionary conservation of essential residues in its histidine kinase and receiver domains.
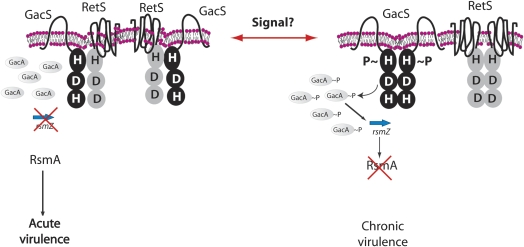
Our findings reported here provide evidence for a model in which the unorthodox sensor kinase RetS regulates the expression of its downstream targets by a novel mechanism, involving interference with another sensor kinase through the formation of heteromeric complexes. This activity is consistent with the dispensability of its conserved phosphorelay residues. Sensor kinases involved in multistep phosphorelay reactions, such as GacS, share a conserved modular architecture and a common mechanism of phosphate transfer. They function as homodimers in which the γ-phosphate from ATP bound by one monomer is transferred intermolecularly to the conserved histidine on the histidine kinase domain of the opposing monomer. The experiments described here are consistent with a model in which RetS blocks GacS phosphorylation at this very early step of the signal transduction pathway, by disrupting the formation of GacS homodimers and instead forming a nonproductive GacS:RetS heterodimer or a higher-order structure of GacS and RetS homodimer (Fig. 8). Several biophysical approaches from structure determination, to dynamic light scattering following size-exclusion chromatography may be used in future studies to test these two hypotheses in detail.
Figure 8.
Model for regulation of GacS/GacA signal transduction by RetS. P. aeruginosa reciprocally regulates genes involved in acute and chronic infection through the interaction between the two membrane-bound sensors RetS and GacS. During the acute virulence phase, RetS forms heterodimers with GacS, blocking GacS autophosphorylation (and subsequent phosphotransfer to the response regulator GacA) leading to reduction in rsmZ expression. RsmA lacking this regulatory RNA promotes the translation of genes for acute virulence factors, while destabilizing transcripts of factors important in chronic infections. Upon perception of unknown signals, GacS and RetS each form homodimers, allowing autophosphorylation of GacS and phosphorylation of GacA. The Gac-dependent small RNA rsmZ sequesters the mRNA-binding protein RsmA, resulting in the expression of genes involved in chronic infection.
Analysis of global gene expression in P. aeruginosa has shown that RetS and GacS are constitutively coexpressed under most conditions. What signals trigger the activation of RetS is unclear. RetS (and LadS) contain a putative periplasm-facing signal receiver domain, referred to by PFAM designation 7TMR-DISMED2, located between a single N-terminal transmembrane segment on one side and seven transmembrane segments on the other. It is conceivable that RetS forms a homodimer where it remains sequestered from GacS. Binding of an external signal to the 7TMR-DISMED2 could dissociate the homodimer and make RetS monomers available for inhibitory interactions with GacS. Identification of the ligands that activate RetS would provide a critical piece of information toward testing this and perhaps other models of RetS activity.
The observation that heterologous sensor kinases can interact directly adds an additional level of signaling potential to this very broadly conserved protein family. By modulating signal transduction at the level of sensor kinase phosphorylation, multiple inputs can be integrated without the need for cross-phosphorylation between sensors and noncognate response regulators. For orphan senor kinases, which are quite abundant in a number of microorganisms, this mechanism could provide a simple means for controlling the expression of genes that respond to multiple signals.
Materials and methods
Bacterial strains growth conditions
Bacterial strains and plasmids are described in Supplemental Table S1. Unless otherwise stated, bacteria were grown at 37°C in LB medium or ECPM1 (Coligan et al. 2007). McConkey-maltose media was prepared as described (Ausubel et al. 1987). Antibiotics were added at the following concentrations: carbenicillin 100 μg mL−1 (for E. coli) or 150 μg mL−1 (for P. aeruginosa); gentamicin 15 μg mL−1 (E. coli) or 75 μg mL−1 (for P. aeruginosa); chloramphenicol 34 μg mL−1; kanamycin 50 μg mL−1. For Pseudomonas selection media, Irgasan at 25 μg mL−1 was used.
Molecular and genetic techniques
DNA purification, molecular cloning, and PCR were performed following standard procedures as described (Ausubel et al. 1987). Deletion strains were constructed by SOE-PCR of deletion alleles, ligation into plasmid pEXG2 (Rietsch et al. 2005), and allelic exchange by sucrose selection on LB agar containing 6% sucrose as described (Horton et al. 1989). Plasmids were introduced into recipient cells by electroporation or conjugation by triparental mating using pRK2013 as the helper plasmid (Ditta et al. 1980). All mutations were verified by PCR and sequencing using a Big Dye fluorescent terminator and an ABI3770 capillary sequencer at the Dana Farber Cancer Center Core Laboratory. Transposon mutagenesis was performed as previously described with a mariner element derivative of Mar2xT7 (Liberati et al. 2006).
Plasmid construction
Genes and deletion alleles were amplified from P. aeruginosa PAK genomic DNA using the primers described in Supplemental Table S1. P. aeruginosa expression constructs were appended with the C-terminal epitopes VSV-G (YTDIEMNRLGK) or His6 by PCR and cloned into expression vectors described in Supplemental Table S1. Plasmid pET15b (Novagen) was used to engineer N-terminally tagged RetS and GacS proteins for expression in E. coli. For expression of GacA fused to the MBP, the gene was amplified from genomic DNA, and ligated into the expression vector pMalc2X.
Site-directed mutagenesis
Oligonucleotide-directed mutagenesis of the pPSV35 and pET15b plasmids carrying a retS allele was performed using the primer pairs described in Supplemental Table S1 and the QuickChange protocol (Clontech) modified by a two-step procedure (Wang and Malcolm 2002). Mutant clones were screened by PCR followed by digestion at restriction sites engineered with silent mutations next to the substituted codons or by direct sequencing.
Bacterial two-hybrid assay
Bacterial two-hybrid experiments were conducted as described (Karimova et al. 1998). PCR fragments corresponding to gacS, retS, and pilS were cloned into the pUT18c and pKT25 vectors to create chimeric proteins with 18-kDa C-terminal adenylate cyclase fragment (amino acids 225–399) or 25-kDa N-terminal adenylate cyclase (CyaA) fragment (amino acids 1–224) fusions, respectively. All fusion proteins were confirmed by DNA sequencing. cAMP production by reconstituted CyaA was measured indirectly by maltose or lactose metabolism in an E. coli cya mutant (DHM1) transformed with the pKT25- and pUT18-derived plasmids. Maltose metabolism was assayed on McConkey plates after incubation 2–4 d at 30°C. Red colonies appeared when reconstitution of active CyaA was obtained. The strength of complementation was quantified by measuring β-galactosidase assays in cells grown overnight at 30°C in LB medium supplemented with 1.0 mM IPTG (Miller 1992).
Protein expression and purification
For His-tagged protein expression and purification, E. coli BL21 carrying pET15b plasmids described in Supplemental Table S1 were grown in 1 L LB or ECPM1 until the culture reached an OD (at 600 nm) of 0.6 (LB) or 5.0 (ECPM1). IPTG was added to a final concentration of 0.3 mM and the cultures were grown at 15°C for 15 h or at 30°C for 5 h. Cells were harvested by centrifugation and resuspended in 40 mL of ice-cold lysis buffer (5 mM imidazole, 500 mM NaCl, 10% glycerol, 0.1% Triton X-100, 1 mM PMSF, 20 mM HEPES at pH 8.0 at 4°C), containing 500 mL of EDTA-free protease inhibitor cocktail (Sigma). The cell suspension was sonicated with a Branson Sonic disruptor at 50% duty, 10 sec on/30 sec off for 25 cycles. The lysate was first centrifuged at 6000g for 10 min, then at 45,000g for 60 min at 4°C. Column chromatography was performed at 4°C. The supernatant was loaded with a Pharmacia P-1 peristaltic pump onto a 1-mL His-Trap FF (fast flow Ni2+-Sepharose 6B; Amershan/GE) column equilibrated with 20 column volumes (CV) of buffer A (5 mM imidazole, 500 mM NaCl, 10% glycerol, 0.1% reduced Triton X-100, 20 mM HEPES at pH 8.0 at 4°C). Chromatography was performed in an AKTA-Prime FPLC apparatus (Amershan/GE). The column was washed with 20 CV of buffer A or until the UV absorption readings stabilized, then weakly bound proteins were washed with a 0%–12% gradient of buffer B (500 mM imidazole, 500 mM NaCl, 10% glycerol, reduced 0.1% Triton X-100, 20 mM HEPES at pH 8.0 at 4°C) until UV absorbance returned to background. His6-tagged proteins were eluted with a 12%–100% gradient of buffer B in 20 CV. Fractions containing the UV-absorbing peak were analyzed by SDS-PAGE, pooled and buffer-exchanged in a G-25 HiLoad column (Amersham/GE) equilibrated with storage buffer (200 mM KCl, 2 mM DTT, 0.2 mM EDTA, 20 mM HEPES at pH 8.0). Proteins were concentrated in Vivaspin 15R (Viva Science) ultrafiltration devices (10 kDa molecular weight cutoff) and stored at −80°C after addition of sterile glycerol to 50% final concentration. His6-GacSc was further purified by size exclusion chromatography using a Sephacryl S-200 HR column (Amersham/GE) equilibrated with storage buffer.
For purification of MBP-tagged GacA, the protein was expressed from the pMalc2X in E. coli T7Express LacIq (New England Biolabs), a BL21(DE3) derivative. Cultures were grown in 1 L of EPCM1 medium with induction at 15°C for 14 h with 0.5 mM IPTG when OD600 reached 1.0. Cleared cell lysates were obtained as described above. Low-pressure chromatography using a high-flow amylose resin (New England Biolabs) packed on a 25-mL column was performed with buffer C (20 mM HEPES at pH 8.0, 250 mM NaCl, 0.2% NP-40, 1 mM EDTA) as the eluant. The bound protein was eluted with 10 mM maltose in buffer C, desalted on a HiLoad G-25 column, and further purified by anion exchange chromatography on a mono-Q HiTrap column. Purified fractions were concentrated by ultrafiltration as described above. Protein concentration was determined by UV spectroscopy using extinction coefficients calculated as described (Gill and von Hippel 1989). The purity of proteins was evaluated by SDS-PAGE followed by Coomassie Brilliant Blue staining (Laemmli 1970).
Isothermal titration calorimetry
Isothermal titration calorimetry experiments were performed using a VP-ITC microcalorimeter (MicroCal). Recombinant proteins (GacSc, RetSc, and PilSc) were buffer-exchanged into 20 mM HEPES (pH 8.0), 200 mM KCl, 0.2 mM EDTA by G-25 spin-column chromatography; concentration was determined by amino acid analysis using a Beckman Model 7300 instrument. A solution containing 5 μM GacSc or PilSc was used as titrant, whereas solutions containing 0.1 μM RetSc were used in the calorimetry cell. The heat of reaction per injection (microcalories per second) was determined by integration of the peak areas using the Origin version 5.0 software (OriginLab). Heat of binding (ΔH°), the stoichiometry of binding (n), and the dissociation constant (K d) were calculated from plots of the heat produced per mole of ligand injected versus the molar ratio of ligand to receptor (Ladbury and Chowdhry 1996; Ladbury 2004).
In vitro phosphorylation assay
For autophosphorylation and phosphotransfer assays, 2 μM GacSc or PilSc was autophosphorylated with 500 μM [γ32P]ATP (0.08 Ci/mmol). GacSc experiments were conducted in presence or absence of the response regulator GacA. All phosphorylation assays were conduced in 10 mM HEPES (pH 8.0), 5 mM MgCl2, 50 mM KCl2, 1 mM DTT, and 0.1 mM EDTA in a final volume of 20 μL. Reactions were stopped by adding Laemli sample buffer (Laemmli 1970) and they were analyzed by SDS-PAGE. The extent of phosphorylation was determined by storage phosphorimaging. When GacSc∼P was required, the autophosphorylation reaction was passed over a G-25 column to separate unincorporated nucleotides from the phosphoprotein. The half life of GacSc∼P was determined by fitting the experimental data into an exponential decay model using GraphPad Prism (version 3.02).
Thin-layer chromatography
For the analysis of the nucleotides, 0.5- to 1.0-μL products from the phosphorylation reactions were spotted on PEI-cellulose plates (Sigma) predeveloped with water and methanol. The samples were run with 1.5 M KH2PO4 (pH 3.65) as the mobile phase and air-dried, and plates were analyzed by storage phosphorimaging.
In vivo cross-linking and purification of RetS:GacS complexes
For in vivo cross-linking experiments, bacteria were grown overnight and inoculated 1:1,000 in 1 L LB with 1 mM IPTG and appropriate antibiotics until cultures reached OD ∼1.0. Cells were pelleted at 5000g at 20°C and resuspended in 100 mL 1× PBS. Where indicated, the cross-linking agent DSP was added to 1 mM final concentration and cells were incubated for 30 min at 37°C with agitation. The cross-linking reaction was quenched with 50 mM Tris-HCl for 20 min and cells were pelleted. Bacterial pellets were resuspended in 35 mL of buffer D (20 mM Na2HPO4 at pH 8.0, 500 mM NaCl, 5 mM imidazole, 2% CHAPS, 0.5% Triton X-100, 1 mM PMSF, 200 μL of Sigma protease inhibitor cocktail, 5 mg/mL lysozyme) and lysed in a Branson sonicator, and the unlysed cells were removed by centrifugation at 20,000g for 45 min. Supernatants were incubated with 2 mL of Ni-NTA slurry for 2 h at 4°C. The Ni2+-IMAC resin was packed into a Bio-Rad Econo-Column and washed with 25 CV of buffer E (20 mM Na2HPO4 at pH 8.0, 500 mM NaCl, 5 mM imidazole, 0.2% CHAPS, 0.1% Triton X-100), followed by 25 CV of buffer E with 50 mM imidazole. The His6-tagged proteins were eluted with buffer E containing 250 mM imidazole. Protein eluates were concentrated in an Amicon ultrafiltration device (10 kDa MWCO) and stored at −80°C until analysis by Western immunoblotting and mass spectrometry. Gel loading was normalized per number of cells. For total membrane purification, bacteria were grown as above, lysed, and total membrane purified as described previously (Merighi et al. 2007).
Western blot analysis
Proteins were separated by SDS-PAGE and transferred to a PVDF membrane. PVDF membranes were blocked overnight with 20 mM Tris-HCl at pH 7.5, 150 mM NaCl (TBS) with 3% nonfat dry milk at 4°C, rinsed in TBST (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.05% Tween-20) and probed with monoclonal antibodies diluted in TBS–1% nonfat dry milk. Anti-His antibody (H3; Santa Cruz Biotechnologies) was diluted 1:1500, monoclonal anti-VSV-G antibodies (Santa Cruz Biotechnologies) was diluted 1:20,000. Antibodies to XcpT and XcpY were described previously (Nunn and Lory 1993; Michel et al. 1998) and were used at a 1:4,000 dilution. The PVDF membranes were washed and probed with goat anti-mouse IgG(H + L) conjugated with HRP (KPL). Supersignal West Pico chemiluminescent substrate (Pierce) was used for the detection of the conjugates. Blots were reprobed following washings with 62.5 mM Tris-HCl (pH 6.8), 2% SDS, 100 mM β-mercaptoethanol for 30 min at 55°C.
Mass spectrometry
Proteins were separated by SDS-PAGE, fixed, and stained with Coomassie Brilliant Blue. Bands of interest were excised and treated with MS-grade trypsin and peptides were extracted follwing treatment with DTT and iodoacetamide alkylation as described (Coligan et al. 2007). Peptides were analyzed using a cyclotron ion trap (FT-ICR) spectrometer at the Harvard Partners Center for Genetics and Genomics.
β-Galactosidase assay
β-Galactosidase assays were carried out using a spectrophotometric method with ortho-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate (Miller 1992). Assays were performed in triplicate. Specific enzyme activities are reported in Miller units (ONPG assays).
Acknowledgments
This work was supported by the NIH grant AI021451. M.H. was supported by a fellowship from the Japan Society for the Promotion of Science.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1739009
Supplemental material is available at http://www.genesdev.org.
References
- Alm E., Huang K., Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput. Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. Current protocols in molecular biology. John Wiley and Sons; New York: 1987. [Google Scholar]
- Baker M.D., Stock J.B. Signal transduction: Networks and integrated circuits in bacterial cognition. Curr. Biol. 2007;17:R1021–R1024. doi: 10.1016/j.cub.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Bijlsma J.J., Groisman E.A. Making informed decisions: Regulatory interactions between two-component systems. Trends Microbiol. 2003;11:359–366. doi: 10.1016/s0966-842x(03)00176-8. [DOI] [PubMed] [Google Scholar]
- Boyer H.W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli . J. Mol. Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Coligan J.E., Dunn B.M., Speicher D.W., Wingfield P.T., Ploegh H.L. Current protocols in protein science. John Wiley and Sons; New York: 2007. [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D.R. Broad host range DNA cloning system for Gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti . Proc. Natl. Acad. Sci. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Bryan R., Rajan S., Scheffler L., Brunnert S., Tang H., Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect. Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiszig S.M., Wiener-Kronish J.P., Miyazaki H., Vallas V., Mostov K.E., Kanada D., Sawa T., Yen T.S., Frank D.W. Pseudomonas aeruginosa-mediated cytotoxicity and invasion correlate with distinct genotypes at the loci encoding exoenzyme S. Infect. Immun. 1997;65:579–586. doi: 10.1128/iai.65.2.579-586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furste J.P., Pansegrau W., Frank R., Blocker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Gill S.C., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Goodman A.L., Kulasekara B., Rietsch A., Boyd D., Smith R.S., Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa . Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Hancock R.E., Mutharia L.M., Chan L., Darveau R.P., Speert D.P., Pier G.B. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: A class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T.T., Kutchma A.J., Becher A., Schweizer H.P. Integration-proficient plasmids for Pseudomonas aeruginosa: Site-specific integration and use for engineering of reporter and expression strains. Plasmid. 2000;43:59–72. doi: 10.1006/plas.1999.1441. [DOI] [PubMed] [Google Scholar]
- Horton R.M., Hunt H.D., Ho S.N., Pullen J.K., Pease L.R. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Hsu J.L., Chen H.C., Peng H.L., Chang H.Y. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Ramirez D., Seshadri R., Cullina J.F., Powers C.A., Schulert G.S., Bar-Meir M., Sullivan C.L., McColley S.A., Hauser A.R. Type III secretion phenotypes of Pseudomonas aeruginosa strains change during infection of individuals with cystic fibrosis. J. Clin. Microbiol. 2004;42:5229–5237. doi: 10.1128/JCM.42.11.5229-5237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladbury J.E. Application of isothermal titration calorimetry in the biological sciences: Things are heating up! Biotechniques. 2004;37:885–887. doi: 10.2144/04376TE01. [DOI] [PubMed] [Google Scholar]
- Ladbury J.E., Chowdhry B.Z. Sensing the heat: The application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem. Biol. 1996;3:791–801. doi: 10.1016/s1074-5521(96)90063-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lapouge K., Schubert M., Allain F.H., Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Laskowski M.A., Kazmierczak B.I. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 2006;74:4462–4473. doi: 10.1128/IAI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M.A., Osborn E., Kazmierczak B.I. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa . Mol. Microbiol. 2004;54:1090–1103. doi: 10.1111/j.1365-2958.2004.04331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub M.T., Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- Li Z., Kosorok M.R., Farrell P.M., Laxova A., West S.E., Green C.G., Collins J., Rock M.J., Splaingard M.L. Longitudinal development of mucoid Pseudomonas aeruginosa infection and lung disease progression in children with cystic fibrosis. JAMA. 2005;293:581–588. doi: 10.1001/jama.293.5.581. [DOI] [PubMed] [Google Scholar]
- Liberati N.T., Urbach J.M., Miyata S., Lee D.G., Drenkard E., Wu G., Villanueva J., Wei T., Ausubel F.M. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahenthiralingam E., Campbell M.E., Speert D.P. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect. Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi M., Lee V.T., Hyodo M., Hayakawa Y., Lory S. The second messenger bis-(3′–5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa . Mol. Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Michel G., Bleves S., Ball G., Lazdunski A., Filloux A. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa . Microbiology. 1998;144:3379–3386. doi: 10.1099/00221287-144-12-3379. [DOI] [PubMed] [Google Scholar]
- Miller J.H. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1992. A short course in bacterial genetics: A laboratory manual and handbook for Escherichia coli and related bacteria . [Google Scholar]
- Nunn D.N., Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J. Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.A., Wegener-Feldbrugge S., Porter S.L., Armitage J.P., Sogaard-Andersen L. Four signalling domains in the hybrid histidine protein kinase RodK of Myxococcus xanthus are required for activity. Mol. Microbiol. 2006;60:525–534. doi: 10.1111/j.1365-2958.2006.05118.x. [DOI] [PubMed] [Google Scholar]
- Rietsch A., Vallet-Gely I., Dove S.L., Mekalanos J.J. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa . Proc. Natl. Acad. Sci. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.E., Buckley D.G., Wu Z., Saenphimmachak C., Hoffman L.R., D'Argenio D.A., Miller S.I., Ramsey B.W., Speert D.P., Moskowitz S.M., et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Taylor R.K., Manoil C., Mekalanos J.J. Broad-host-range vectors for delivery of TnphoA: Use in genetic analysis of secreted virulence determinants of Vibrio cholerae . J. Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomomori C., Tanaka T., Dutta R., Park H., Saha S.K., Zhu Y., Ishima R., Liu D., Tong K.I., Kurokawa H., et al. Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat. Struct. Biol. 1999;6:729–734. doi: 10.1038/11495. [DOI] [PubMed] [Google Scholar]
- Ventre I., Goodman A.L., Vallet-Gely I., Vasseur P., Soscia C., Molin S., Bleves S., Lazdunski A., Lory S., Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Malcolm B.A. Two-stage polymerase chain reaction protocol allowing introduction of multiple mutations, deletions, and insertions, using QuikChange site-directed mutagenesis. Methods Mol. Biol. 2002;182:37–43. doi: 10.1385/1-59259-194-9:037. [DOI] [PubMed] [Google Scholar]
- Whitworth D.E., Cock P.J. Two-component systems of the myxobacteria: Structure, diversity and evolutionary relationships. Microbiology. 2008;154:360–372. doi: 10.1099/mic.0.2007/013672-0. [DOI] [PubMed] [Google Scholar]
- Wolanin P.M., Thomason P.A., Stock J.B. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 2002;3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfaghar I., Angus A.A., Kang P.J., To A., Evans D.J., Fleiszig S.M. Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes Infect. 2005;7:1305–1316. doi: 10.1016/j.micinf.2005.04.017. [DOI] [PubMed] [Google Scholar]