Abstract
Oct1 and Oct4 are homologous transcription factors with similar DNA-binding specificities. Here we show that Oct1 is dynamically phosphorylated in vivo following exposure of cells to oxidative and genotoxic stress. We further show that stress regulates the selectivity of both proteins for specific DNA sequences. Mutation of conserved phosphorylation target DNA-binding domain residues in Oct1, and Oct4 confirms their role in regulating binding selectivity. Using chromatin immunoprecipitation, we show that association of Oct4 and Oct1 with a distinct group of in vivo targets is inducible by stress, and that Oct1 is essential for a normal post-stress transcriptional response. Finally, using an unbiased Oct1 target screen we identify a large number of genes targeted by Oct1 specifically under conditions of stress, and show that several of these inducible Oct1 targets are also inducibly bound by Oct4 in embryonic stem cells following stress exposure.
Keywords: Oct1, Oct4, stress response
Oct1 and Oct4 (products of the Pou2f1 and Pou5f1 genes) are members of the POU (Pit-1, Oct1/2, Unc-86) domain transcription factor family (Herr et al. 1988; Ryan and Rosenfeld 1997). This family is defined by the presence of a bipartite DNA-binding domain in which two subdomains, covalently connected by a flexible linker, typically recognize DNA through major groove interactions on opposite sides of the helix (Klemm et al. 1994). The classical DNA recognition sequence is known as an octamer motif (5′-ATGCAAAT-3′, hereafter called a “simple” octamer). However, we demonstrated recently that native binding sites for Oct4 frequently exist in complex paired, overlapping, and nonconsensus configurations (Tantin et al. 2008).
Oct4 is a master regulator of the stem cell state and has recently been shown to be one of three proteins sufficient to reprogram differentiated adult mouse and human cells to the embryonic stem (ES) cell lineage (Okita et al. 2007; Takahashi et al. 2007; Nakagawa et al. 2008). The biological function of Oct1 is more enigmatic. Oct1 is known to interact with regulatory sites in interleukin, immunoglobulin, and histone genes (Garrity et al. 1994; Zheng et al. 2003; Ushmorov et al. 2004; Murayama et al. 2006). Oct1 also moderately stimulates gene expression reporter constructs linked to target sequences in transient transfection assays (Sive et al. 1986; LeBowitz et al. 1988). However, we showed that Oct1 is nonessential for native H2B, IgH, and Igκ expression (V.E. Wang et al. 2004; V.E.H. Wang et al. 2004). Oct1-deficient cells appear morphologically normal in light microscopy and divide at normal rates. Oct1-deficient mice die in mid–late gestation (embryonic days 12,5–18.5 [E12.5–E18.5]) (V.E.H. Wang et al. 2004).
We determined previously that Oct1−/− mouse embryonic fibroblasts (MEFs) are hypersensitive to oxidative and genotoxic stress (Tantin et al. 2005). One explanation for this result is that constitutive products of Oct1-mediated transcription participate in stress response pathways. Support for an alternative hypothesis, namely that Oct1 directly senses cellular stress, comes from the findings that Oct1 interacts with the Ku70 subunit of DNA-dependent protein kinase (DNA-PK) (Schild-Poulter et al. 2001) and is phosphorylated in vitro by DNA-PK at physiologically important serine and threonine residues (Schild-Poulter et al. 2007). Oct1 also interacts with BRCA1 (Fan et al. 2002; R.H. Wang et al. 2004), and PARP-1 (Nie et al. 1998), known participants in stress response pathways.
Here, using an affinity purification approach we identify in vivo post-translational Oct1 modification events following treatment of HeLa cells with ionizing radiation (IR) or H2O2. Comparison with a second Oct1 modification data set indicated that two DNA-binding domain Oct1 phosphorylation events have the potential to modulate Oct1 association with DNA. Using two model sequences, termed PORE (palindromic octamer-related element) and MORE (more PORE) (Remenyi et al. 2001), we show that DNA-binding domain modifications alter the in vitro affinity of Oct1 and Oct4 specifically at complex binding sites. We demonstrate stress-induced binding of both Oct1 and Oct4 to physiological targets in vivo, show that induced Oct1 binding regulates native gene expression, and expand the repertoire of complex site categories to which Oct1 and Oct4 can bind. Using chromatin immunoprecipitation (ChIP) coupled with deep sequencing (ChIPseq), we identify a large number of constitutive Oct1 targets as well as targets specifically induced in the presence of oxidative stress. These targets frequently contain conserved complex binding sites for Oct1. We demonstrate that Oct4 inducibly binds two of these targets in vivo using mouse ES cells and ChIP. Together, the data show that Oct1 and Oct4 are potent stress response effectors.
Results
Oct1 is phosphorylated in vivo following exposure to genotoxic and oxidative stress
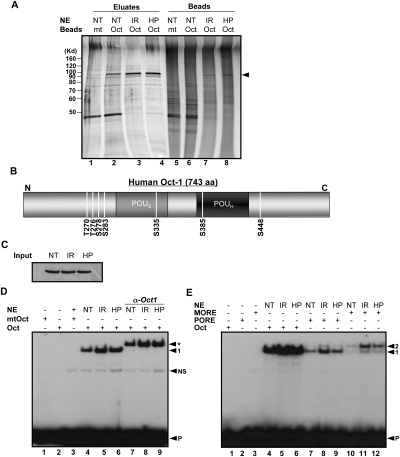
We used a single-step affinity approach to purify Oct1 and other DNA-binding proteins from nuclear extracts (Tantin et al. 2004; V.E.H. Wang et al. 2004). This approach involves ferromagnetic latex nanoparticles (“nanobeads”) coupled to multimerized DNA sequence motifs for rapid and sequence-selective enrichment of DNA-binding proteins. Relative to other supports such as sepharose, nanobeads have lower nonspecific binding (due to their nonporous, moderately hydrophilic surface), higher capacity (due to their smaller size), and will remain in suspension until high-speed centrifugation or magnetic separation (Shimizu et al. 2000; Nishio et al. 2008). We used nanobeads coupled to multimerized consensus or mutant simple octamer sequences to purify Oct1 from HeLa cells following IR or H2O2 treatment. This affinity purification approach will only isolate Oct1 molecules with modifications that do not inhibit DNA binding. As shown in Figure 1A, an ∼100-kDa band was isolated using nanobeads coupled to consensus, but not mutant DNA (Fig. 1A, cf. lane 2 and lane 1). The 100-kDa band was excised, destained, and subjected to tryptic digestion. Mass spectrometry identified peptides consistent with human Oct1. Peptide coverage was good and the sequences were confirmed through sequencing using collision-induced dissociation (CID) in the linear ion trap (data not shown; see the Materials and Methods). The intense band near 40 kDa was identified as β-actin (data not shown).
Figure 1.
Genotoxic and oxidative stress induce Oct1 phosphorylation and dimerization at complex sites. (A) Oct1 was purified from HeLa nuclear extracts using nanoparticles coupled to multimerized octamer or mutant sequences. HeLa cells were treated with IR or H2O2 and incubated for 1 h. The eluted proteins were resolved using SDS-PAGE and silver stained. (NT) No treatment; (HP) H2O2. (Arrow) Oct1 band. (B) Identified Oct1 serine and threonine phosphorylation events superimposed on a schematic of the Oct1 amino acid structure. DNA-binding domain modification events identified separately by the Gygi laboratory are also shown. (C) Oct1 Western blot showing protein levels in the input HeLa nuclear extracts. (D) EMSA using a simple octamer probe and extracts from C. Arrows indicate free probe (P), nonspecific band (NS), monomeric Oct1 (1), and Oct1 antibody supershifted band (*). (E) EMSA using radiolabeled simple octamer, PORE, and MORE probes. Arrows indicate monomeric (1) and dimeric (2) occupancy, and free probe (P).
Using equal amounts of extract from HeLa cells exposed to IR (Fig. 1A, lane 3) or H2O2 (Fig. 1A, lane 4), a small increase in bound protein was identified compared with normal HeLa cells (Fig. 1A, lane 2). This increase is consistent with previous reports (Meighan-Mantha et al. 1999; Zhao et al. 2000; B. Wang et al. 2004; Chen and Currie 2005). We excised these bands and subjected them to mass spectroscopy to identify Oct1 modifications. We unambiguously identified Oct1 phosphopeptides, many of which were identified unphosphoryated in the absence of stress (see Table 1). These data show that Oct1 becomes dynamically modified in vivo shortly following stress treatment. Although our data were consistent with a large-scale screen for protein phosphorylation events in HeLa cells following IR exposure conducted in the laboratory of Dr. Steven Gygi (S.P. Gygi, unpubl.), two Oct1 DNA-binding domain modifications were identified by the Gygi laboratory that were not observed using our affinity purification approach: phospho-S335 and phospho-S385 (Table 1). Because the Gygi laboratory screen did not rely upon DNA binding to isolate phosphopeptides, we focused on these two modifications, hypothesizing that they regulate DNA binding. The complete panel of in vivo phosphorylation events from both studies is shown in Table 1 and in schematic form in Figure 1B.
Table 1.
Oct1 phosphopeptides identified following exposure of HeLa cells to IR (mass error <3 ppm) using LC/MS/MSa
Alteration in Oct1-binding specificity following stress exposure
The above results demonstrated that Oct1 becomes dynamically modified following stress exposure. We therefore attempted to identify dynamic changes in the properties of the protein that could provide a functional stress readout. First, we performed Western blotting using Oct1-specific antibodies and extracts from HeLa cells exposed to IR or H2O2. Little change was observed (Fig. 1C). We also conducted gel mobility shift experiments using wild-type and mutant simple octamer sequences. A strong octamer sequence-dependent band was identified, but again, little change was observed using extracts from IR or H2O2-treated HeLa cells (Fig. 1D, lanes 4–6). There was some increase in DNA-binding activity (Fig. 1D, cf. lanes 5,6 and lane 4), consistent with previous reports (Meighan-Mantha et al. 1999; Zhao et al. 2000; B. Wang et al. 2004; Chen and Currie 2005).
Recently, a PKA-mediated phosphorylation event was identified in the CNS-specific POU domain protein Brn-2/N-Oct-3/Pou3f2 (Nieto et al. 2007). In this case, the modification, at a position homologous to Oct1 S385, was found to alter binding specificity for complex dimeric sites. More recently, we identified native Oct4 genomic binding sites (Tantin et al. 2008). These target sequences were highly enriched for complex multimeric arrangements of half-sites with the capability of binding two or more Oct4 molecules. The potential for these complex arrangements arises from the fact that in the POU domain, the linker connecting the two DNA-binding subdomains is flexible. Because these results showed that native sites for POU transcription factors frequently exist in complex configurations, we tested Oct1 binding to two model complex sites, termed PORE and MORE. We chose these model complex sequences because structural information was available.
PORE and MORE sequences are both capable of binding Oct proteins as a dimer, but in different configurations (Remenyi et al. 2001). We conducted electrophoretic mobility shift assays (EMSA) using these sequences together with extracts from HeLa cells cultured under standard conditions or exposed to stress. Using a PORE, a somewhat stronger induction of binding by stress was observed compared with simple sequences. This was particularly true of extracts from cells exposed to IR (Fig. 1E, cf. lanes 8,9 and lane 7). Using a MORE, an even stronger induction was observed following stress exposure. This induction was again particularly robust using IR and was manifested both at the level of overall binding and increased relative dimerization (Fig. 1E, cf. lane 11 and lanes 10,12). The induced binding could be eliminated by shifting the equilibrium to the bound state, as inclusion of higher amounts of extract in the binding reactions increased dimer occupancy and decreased stress induction (data not shown).
Stress-induced association of Oct1 with endogenous complex sites
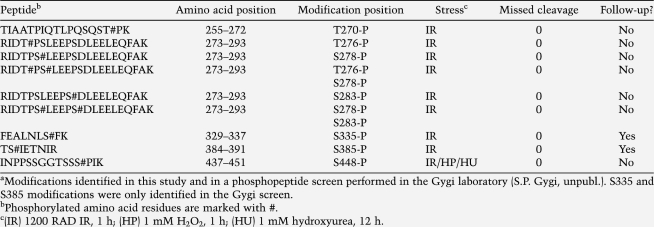
A MORE has previously been identified in the promoter for the human RNA polymerase II (Pol II) large subunit (Polr2a) (Fig. 2A, top panel; Tomilin et al. 2000). We identified a second MORE located immediately upstream. Both MOREs are conserved across multiple mammalian species (Fig. 2A, bottom panel). Using EMSA with HeLa nuclear extract and a probe derived from the Polr2a 2XMORE region, we confirmed that this compound element could interact with four Oct1 molecules, and that Oct1 binding could be strongly induced by stress (Fig. 2B). In Figure 2, B and C, the gel was run further to resolve the multiply bound species, and the labeled free probe was consequently run off the gel. In Figure 2B, increasing amounts of normal and IR-treated HeLa cell extract were incubated with either a simple octamer sequence or a 2XMORE probe of identical size. DNA binding was strongly enhanced by stress (Fig. 2B, cf. lanes 7,12,17). We used an Oct1-specific antibody to confirm that the multiply bound species contained Oct1 (Fig. 2B, lanes 8,13,18). In Figure 2C, point mutations were engineered into the Polr2a 2XMORE probe to verify the stoichiometry of the protein:DNA complex. In comparison with a simple octamer (Fig. 2C, lanes 1–3), the bands produced by the wild type, and mutant Polr2a probes were consistent with four and two bound Oct1 molecules, respectively (Fig. 2C, lanes 4–6,7–12). Complexes of one or three molecules were not observed. Compared with the normal (unstressed) condition (Fig. 2C, lanes 1,4), augmented Oct1 binding was observed using the 2XMORE, but not a simple octamer, following either IR or H2O2 treatment (Fig. 2C, cf. lanes 5,6 and lanes 2,3). Quantification of the band intensities from Figure 2C and two replicate experiments showed that the increase in Oct1-shifted DNA in the presence of either stress was progressively higher with the 1XMORE and 2XMORE probes compared with the simple octamer probe, suggesting that the stress-induced 1XMORE and 2XMORE Oct1:DNA complexes were progressively more cooperative (data not shown).
Figure 2.
An endogenous 2XMORE interacts with four Oct1 molecules in a cooperative stress-induced complex. (A) UCSC Genome Browser (http://genome.ucsc.edu) screenshots of the human Polr2a locus. (Top) The region upstream of the TSS and mammalian conservation. Arrow indicates position of the 2XMORE. (Bottom) Higher-resolution screenshot showing the nucleotide sequence. Boxed sequences contain the conserved MOREs. (B) EMSA using probes designed from the sequence in A. Arrows indicate monomeric (1), dimeric (2), and tetrameric (4) occupancy. (*) Antibody supershift. The free probe was run off the gel. (C) EMSA using probes containing point mutations in one or both MOREs. A simple octamer probe of the same length was used as a control for monomeric binding. Arrows indicate monomeric (1), dimeric (2), and tetrameric (4) occupancy. (NS) Nonspecific band. Probe sequences are shown at bottom. (D) Kinetic dissociation assay of Oct1:DNA complexes assembled using simple octamer, MORE or 2XMORE probes. The top panels show the cropped bands from the imaged gel mobility shift. The plot shows quantification of average band intensities from three experiments together with decay curves calculated using exponential regression. Half-lives were calculated from these curves.
To assess complex stability, we performed kinetic complex dissociation assays using a 200-fold molar excess of cold competitor (Fig. 2D). Complexes were assembled using simple octamer, 1XMORE or 2XMORE probes, and incubated with cold competitor for varying times prior to native gel loading. Band intensities were determined and used to plot exponential decay curves. Complex half-life using a simple octamer was significantly shorter (T1/2 = 10.8 sec) compared with the 2XMORE (80.2 sec). Using a 1XMORE, we obtained an intermediate result (T1/2 = 13.7 sec). These data show that the 2XMORE complex is significantly more stable than complexes formed using a simple octamer sequence or a single MORE.
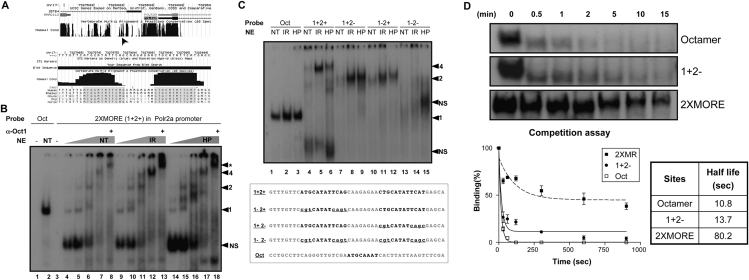
We performed ChIP assays to assess Oct1 binding to the Polr2a promoter in vivo. We observed low-level association of Oct1 to Polr2a in the absence of stress, but a significant augmentation following H2O2 exposure (Fig. 3A). Occupancy was maximal at 60 min and decreased thereafter. In contrast, Oct1 occupancy of the control histone H2B promoter (which contains a single simple octamer) was unaffected by H2O2. Induced Oct1 Polr2a occupancy was also observed following IR treatment; however, in this case the kinetics were different, with Polr2a occupancy peaking at 60 min, but subsequently remaining more stable (Fig. 3B). In contrast to the induced binding at these complex sites, simple sites showed either reduced binding or no change. At the H2B promoter, Oct1 occupancy was significantly decreased at short time points following IR exposure. These results were reproducible, and we conclude that IR stress affects Oct1 binding to Polr2a and H2B, but in opposite ways. We also tested Oct1 binding to Gadd45a, a known Oct1 target (also containing a simple octamer sequence), finding that Oct1 occupancy was stable. Therefore, the Oct1 transcriptional response to stress is not uniform, but varies depending on the spatial context in which Oct1 associates with the DNA of different targets.
Figure 3.
Oct1 functionally regulates Polr2a. (A) Time-course ChIP assay of Polr2a using HeLa cells treated with H2O2. H2B and input DNA are shown as controls. (B) Similar time-course experiment using IR. Oct1 association with a simple octamer site in Gadd45a is additionally shown. (C) Reporter activity of a 2XMORE linked to the SV-40 promoter in a luciferase-based assay. Constructs were transfected into HeLa cells. Experiment was performed in triplicate. Error bars depict standard deviations. (D) Activity of the human Polr2a promoter fragment (−600 to +1 relative to the TSS) linked to luciferase. A 45-bp 2XMORE deletion was made in the context of the full sequence. Empty vector was used as a control. (E, left panel) Real-time RT–PCR using intron-spanning mouse Polr2a primers and either wild-type or Oct1-deficient MEFs. Message levels are shown following treatment with 2 mM H2O2 relative to wild-type fibroblasts under unstressed conditions on a log2 scale. (Right panel) Dose response experiment. Polr2a expression was measured 4 h following exposure to different H2O2 doses. (F) Western blot showing time course of Pol II large subunit expression in immortalized MEFs cells treated with H2O2. β-actin is shown as a loading control.
To determine whether Oct1 binding to the 2XMORE is functional, we linked the 2XMORE to an SV-40 promoter luciferase vector and used it in transfections with HeLa cells together with a cotransfected thymidine kinase promoter-linked Renilla luciferase internal control (Fig. 3C). A small, but significant increase in luciferase activity was observed using a 2XMORE in either orientation, and activity was decreased by MORE point mutation. Because these experiments used the 2XMORE in the context of a heterologous promoter, we also linked 600 base pairs (bp) of native Polr2a promoter sequence to a promoterless luciferase vector. A strong increase was observed relative to the empty vector (Fig. 3D). We engineered a 2XMORE deletion, which significantly decreased activity, although the effect was not complete. We ascribe the residual activity to other bound transcription factors and/or basal promoter activity.
To determine whether Oct1 loss of function effects native Polr2a gene expression under stressed conditions, we used real-time RT–PCR with RNA prepared from early-passage primary MEFs derived from littermate wild-type and Oct1-deficient embryos (V.E.H. Wang et al. 2004). The MEFs were either untreated or exposed to H2O2 (Fig. 3E, left panel). Three biological replicates were performed for each condition and averages are shown. Polr2a is a constitutive gene, and Pol II large subunit levels are thought to be regulated mainly at the protein level in response to stress stimuli. We confirmed this supposition using wild-type cells, in which only small changes in steady-state Polr2a mRNA levels were evident following stress (Fig. 3E, dark line). Over repeated experiments, no difference in baseline Polr2a expression was observed between wild-type and Oct1-deficient cells (data not shown). These results are consistent with our findings that expression of other reported Oct1 targets such as histone H2B, U2 snRNA, and immunoglobulin are unchanged in the Oct1-deficient and unstressed condition (V.E. Wang et al. 2004; V.E.H. Wang et al. 2004). Strikingly, in the absence of Oct1 a significant depletion in Polr2a mRNA levels was observed following stress exposure (gray line). The effect was greatest (approximately fourfold) at 4 h. We also performed a Polr2a dose response experiment using a fixed 4-h time point. Increasing doses of H2O2 resulted in increased Polr2a mRNA repression in the Oct1-deficient condition (Fig. 3E, right panel). These results strongly suggest that Oct1 acts as a functional anti-repressor of Polr2a following stress.
We confirmed an effect at the protein level using Western blotting and immortalized MEFs: In the Oct1-deficient condition, Pol II large subunit levels were not detectable several hours after H2O2 treatment, although, in this case, a transient and Oct1-independent increase in protein levels was also observed (Fig. 3F).
S335 and S385 control MORE Oct1 dimerization
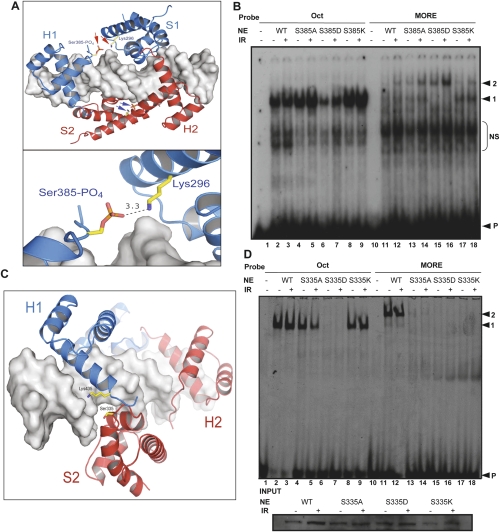
Oct1 bound to MORE DNA adopts a unique structure in which the two subdomains of each molecule no longer span the helix but instead occupy adjacent major grooves on the same face of the DNA (Remenyi et al. 2001). We used molecular modeling to identify a possible role for Oct1 DNA-binding domain modifications in stress-induced MORE dimerization. Figure 4A shows Oct1 occupying MORE DNA, in which a phosphoserine has been modeled at position 385. This phosphoserine lies in the POU-homeodomain and is in close apposition to a lysine residue (K296) in the POU-specific domain of the same molecule (arrows; see inset), suggesting that phosphorylation at this position may structurally preorient Oct1 so as to favor MORE binding.
Figure 4.
Oct1 S385 and S335 mutations modulate DNA selectivity. (A) Molecular models based on the crystal structure of the Oct1 POU domain dimer bound to MORE DNA (Remenyi et al. 2001). Phospho-Ser 385 and Lys 296 are modeled (red and blue arrows). Bottom image shows predicted Å distance. Images generated using PYMOL (http://www.pymol.org). (B) EMSA using nuclear extracts prepared from Oct1-deficient immortalized MEFs infected with retroviruses encoding wild-type, S385A, S385D, or S385K Oct1. Arrows indicate monomeric (1) and dimeric (2) occupancy, and free probe (P). (NS) Nonspecific. (C) MORE DNA-Oct1 dimer structure showing Ser 335 (red). Lys 435 is also shown. (D) EMSA using nuclear extracts prepared from Oct1-deficient immortalized MEFs infected with retroviruses containing either wild-type, S335A, S335D, or S335K Oct1 cDNAs and probes tagged with Cy5. Arrows indicate monomeric (1) and dimeric (2) occupancy, and free probe (P). The gel was scanned using a Typhoon Imaging system (Molecular Dynamics). (Below) Oct1 Western blot of the input extracts.
We used extracts from Oct1-deficient 3T3-immortalized MEFs (V.E.H. Wang et al. 2004) in which Oct1 function was restored using retroviruses encoding human wild-type Oct1, or one of three engineered three point mutations (S385A, S385D, and S385K) to determine the effect on binding in the presence and absence of IR treatment. An alanine substitution at position 385 had little effect (Fig. 4B, lanes 4,5,13,14), suggesting that other modifications may also induce MORE binding (see below). S385D, a potential phosphomimetic mutation, had a deleterious effect on monomer binding (Fig. 4B, lanes 6,7). The same mutation significantly enhanced MORE dimerization (Fig. 4B, lane 16). This mutation also favored MORE binding under unstressed conditions (Fig. 4B, lane 15). In contrast to S385D, the S385K mutation strongly inhibited MORE dimerization, instead favoring the monomeric form (Fig. 4B, lanes 17,18). Band intensity quantification showed that the S385D mutation augmented MORE dimer formation, but only partially decreased dimer stress induction, and that S385K disfavored, but did not completely eliminate MORE dimer stress induction (data not shown), again suggesting that other modification events can also facilitate dimerization (see below). For these experiments, low titers of Oct1 retrovirus were used, because at higher titers, or higher amounts of extract, dimer occupancy was strongly favored and the differences between the various mutants were obviated. These data are consistent with a model in which phospho-S385 makes and contacts with K296. Mutation of K296 (which is also in close apposition to the DNA backbone) (Fig. 4A) to alanine eliminated MORE binding, including when made as a double mutant with S385D (data not shown).
The other modification identified in the Oct1 DNA-binding domain was phospho-S335. This residue is located in the POU-specific domain and is positioned close to the DNA backbone. Figure 2C shows a different view of the same Oct1 dimer/MORE DNA structure with a phosphoserine modeled at position 335. Although there is a lysine (K435) located across the dimer interface in the POU-homeodomain, the intervening distance is too great to support an interaction without changing the modeled coordinates. We hypothesize that in order to form an optimal interation, slight rearrangements of these domains take place. We engineered similar point mutations at this position (S335A, S335D, S335K). The S335 mutants were more robust, such that higher titers of virus could be used to infect cells, or higher amounts of extract could be used in the EMSA and the phenotypes retained (Fig. 3D; see Discussion). Using high viral titers, S335A had minimal effects on simple octamer binding (Fig. 4D, lanes 4,5), but strongly inhibited MORE dimerization (Fig. 4D, lanes 13,14). S335D did not appear to bind DNA under any conditions (Fig. 4D, lanes 7,8,15,16). This serine is closer to the DNA backbone than S385 and it is possible that electrostatic repulsion disfavors binding of the S335D mutant Oct1 to DNA. Interestingly, the homolgous Oct4 mutation binds a MORE in vitro but is not induced by stress (see below). We conclude that either (1) Oct1 S335D is not a true phosphomimetic or (2) phosphorylation of S335 inhibits DNA binding, but not all Oct1 is phosphorylated at this site, and the unphosphorylated fraction has augmented MORE dimerization activity (possibly because of additional modifications). S335K demonstrated the same selectivity effects as the alanine mutation (Fig. 4D, lanes 9,10,17,18). The mutant proteins were expressed at equivalent levels (Fig. 4D, inset).
We generated a double mutation (S335A/S385D) to determine which was dominant. S335A disrupted MORE DNA binding even in the presence of S385D (data not shown). This finding is consistent with the fact that S385 mutation could be compensated by increased amounts of Oct1 protein, whereas the S335 mutation phenotype was protein level-independent.
Control of Oct4 binding to complex sites by homologous residues
Oct1 Ser 385 and Ser 335 are conserved in Oct2 and Oct4 (Fig. 5A), and across mammalian species (data not shown). The conservation at 335 is interesting because alanine and lysine substitutions at this position do not affect DNA binding to simple octamers, suggesting that MORE dimerization is physiologically significant (see below) and is likely regulated by phosphorylation in multiple Oct proteins, including Oct4. We restored Oct protein function in Oct1-deficient fibroblasts using a retroviral construct encoding mouse Oct4 and used this model system to determine whether stress exposure also regulates Oct4 binding. Fibroblasts were exposed to IR and nuclear extracts prepared after a 1-h incubation. Oct4 expression and loading levels were confirmed by Western blot (Fig. 5B, inset). Compared with Oct1-deficient fibroblasts, in which no octamer-directed DNA-binding activity was observed, Oct4 retroviral expression resulted in binding activity (Fig. 5B, lanes 3,5). As with Oct1, augmented MORE binding was observed in IR-treated fibroblasts compared with untreated cells (Fig. 5B, lanes 7,8). We engineered an Oct4 mutation at a position equivalent to Oct1 S335 (Oct4 S186D), and determined the effect on MORE DNA binding using extracts from cells exposed to methyl methanesulfonate (MMS) or cultured under normal conditions. As with Oct1, mutation of this residue resulted in sequence-selective effects. Unlike Oct1, the aspartic acid mutation did not eliminate DNA binding, but instead resulted in failure to generate induced MORE dimerization (Supplemental Fig. 1, lanes 7–10). There may therefore be slight differences between the proteins, such that homologous mutations at specific positions can elicit distinct effects in vitro.
Figure 5.
Oct4 complex site binding is IR-inducible. (A) Alignment of DNA-binding domain amino acid sequences of human Oct1, Oct2, and Oct4. Homologous amino acids are highlighted. Oct1 S335 and S385 and the homologous Oct2/Oct4 serines are boxed. (B) EMSA using nuclear extracts from Oct1-deficient MEFs infected with retroviruses encoding murine Oct4 or uninfected controls. (NS) Nonspecific; (ND) not determined. Probes: (Oct) Simple octamer; (MR) MORE. (Inset) Oct4 Western blot using input extracts. (C) (Top panels) Screenshots of mouse Bmp4 locus intronic region. The MORE is marked with an arrow. Mammalian conservation is also shown. (Below) Higher-resolution image showing the MORE sequence and conservation. (Bottom panel) ChIP assay using mouse ES cells treated with IR and a primer pair spanning the Bmp4 MORE. (D) Microarray output tracks along the human Taf12 locus. Log2 microarray intensities are shown for library oligonucleotides. IR-treated ES extracts were prepared at 0-, 1-, and 3-h time points. (E) (Top panels) Oct1 and Oct4 ChIP assays using mouse ES and human HeLa cells and genomic oligonucleotides encompassing the enriched Taf12 region. (Bottom panels) ChIP time course showing Oct4 and Oct1 association with the Taf12 site following IR exposure of mouse ES cells and HeLa cells.
The above experiments were performed in vitro with Oct4 and a synthetic MORE. To verify inducible in vivo Oct4 binding to sequences that regulate endogenous genes, we used a previously identified MORE (CTGCATATGCAT) from the Bmp4 regulatory region (Fig. 5C, top panel; Tomilin et al. 2000). This sequence is conserved between humans and rodents (Fig. 5C, bottom panel). Bmp4 is an Oct4 target (Sharov et al. 2008). Using ChIP, we verified an interaction between Oct4 and this genomic region and showed that Oct4 occupancy was induced by stress (Fig. 5C). The kinetics of induced Oct4 binding were similar to those observed using Oct1, with occupancy peaking at 1 h
To identify genomic DNA sequences from known Oct4 targets, gene regions that associate with Oct4 in extracts from normal or irradiated mouse ES cells, we generated a pool of tiled oligonucleotides from common human and mouse Oct4 targets (Boyer et al. 2005; Loh et al. 2006) and immunoprecipitation to enrich for Oct4-associated oligonucleotides. Microarray analysis on the purified material identified a number of in vitro targets (W.G. Fairbrother, in prep.), including a site upstream of the Taf12 promoter (Fig. 5D). This sequence, which is highly conserved among mammals (Supplemental Fig. 2A), interacts with Oct4 1 h following IR treatment, but not in the untreated condition or at 3 h. We confirmed the ability of both Oct4 and Oct1, which is also expressed in ES cells, to interact with Taf12 in vivo using mouse ES cells and ChIP. Oct1 also binds this region in HeLa cells (Fig. 5E). Because Oct4 binding was inducible at this sequence in vitro using extracts from irradiated Oct1-deficient fibroblasts complemented with Oct4 retroviruses (Supplemental Fig. 2B), we performed ChIP using mouse ES cells in a time course following IR treatment. Although slight baseline binding was observed, Oct4 binding was potently induced by stress, with maximal binding at 1 h, decreasing thereafter (Fig. 5E). Oct1 also inducibly binds this sequence with similar kinetics in HeLa cells (Fig. 5E). We used EMSA to show that Oct1 also interacts with the same site in vitro, and that multimeric binding could be induced by stress (Supplemental Fig. 2C). The size of the shifted band was consistent with a dimer.
The Taf12 sequence contains half-sites for both the POUH and POUS DNA-binding subdomains, but in a configuration distinct from a MORE or PORE (Supplemental Fig. 2D). We identified highly similar sequences in the mouse Piwil2 (also known as Mili) first intron, and upstream of mouse Foxo4. In both cases the sequence was conserved to human (Supplemental Fig. 2C). We also identified similar sequences in conserved intergenic regions between Zcchc16 and Lhfp11 and between Prr16 and Srfbp1 (data not shown). Using ES cells and ChIP, we confirmed Oct4 binding to Foxo4 and Mili (Supplemental Fig. 2E). We term this sequence a TMFORE (Taf12 Mili Foxo4 octamer-related element) (see Discussion).
High-throughput identification of Oct1 targets confirms stress-responsive binding
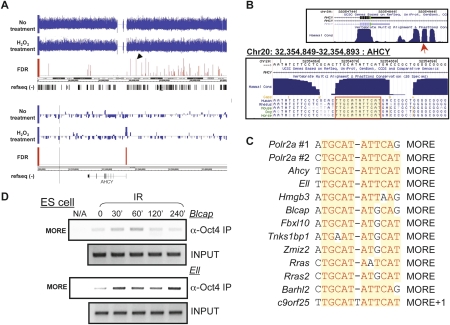
To identify inducible Oct1 targets on a large scale and in an unbiased manner, we used Oct1 ChIP material from normal or H2O2-treated HeLa cells together with deep sequencing (ChIPseq). We used 12,190,661 (input DNA), 4,167,664 (normal cells), and 3,107,208 (H2O2-treated cells) pipeline quality control-filtered reads with an eland alignment score >13 in the analysis. Targets are provided as Supplemental Table 1 (no treatment) and Supplemental Table 2 (H2O2 treatment). In static maps of untreated cells, known Oct1 targets such as genes encoding histones, U snRNAs, and GADD45a were represented with high scores. We also identified differential targets (Supplemental Table 3) using the untreated ChIP material, rather than input DNA, as the control for the H2O2 sample (see the Materials and Methods). The top differential target (Ahcy) is shown in Figure 6A. The target region mapped to the immediate 5′ end of the locus and contained a conserved MORE (Fig. 6B). Additional genes are shown in Supplemental Figure 3 (see Supplemental Table 4 for annotations). Polr2a was identified using this analysis, but not as an induced target. We observed some Oct1 occupancy at this locus in the absence of stress (Fig. 3A,B), suggesting that the ChIPseq analysis represents an underestimate of inducible targets.
Figure 6.
ChIPseq identifies inducible Oct1 targets. (A) Screenshots showing ChIPseq hits covering human chromosome 20. Arrow indicates one significant hit. (Bottom panel) Higher-magnification image showing this hit upstream of Ahcy. (B) Screenshots of murine Ahcy promoter. Sequence conservation is shown. MORE is highlighted. (C) MORE motifs in Oct1 targets identified by ChIPseq. (D) Oct4 ChIP using primer pairs spanning the homologous mouse region of a subset of inducible human Oct1 targets identified by ChIPseq, together with mouse ES cells treated with IR. Input DNA is shown as a control.
A number of the most high-scoring differentially occupied targets were previously identified as Oct4 targets in mouse ES cells (Chen et al. 2008). These included Blcap, Tnks1bp1, Fbxl10, Rras, and c9orf25 (2310028H24Rik in mouse). In many cases, the Oct1- and Oct4-bound regions overlapped perfectly (Supplemental Table 5). These regions also contained highly conserved MOREs that mapped to the immediate 5′ end of their respective targets (see Supplemental Fig. 3 for human and Supplemental Table 5 for mouse positions). A small number of other regions identified using ChIPseq, although not showing strong stress induction, overlapped with known Oct4 target regions. These included Polr2a, Ell, and Zmiz2. Conserved MOREs were also identified within these regions (Supplemental Table 5). An overlapping group of conserved MORE sequences was also identified in the top 500 differential targets that mapped within 250 bp of the nearest TSS (Fig. 6C). We conclude that Oct1 regulates a large number of genes specifically under oxidative stress conditions, a subset of which contain promoter-proximal MOREs.
To both validate the induced Oct1 ChIPseq target results from human HeLa cells, and to determine whether they can be applied to inducible Oct4 target binding in mouse ES cells, we generated primer pairs spanning the conserved equivalent regions of the mouse genome and used them in Oct4 ChIP assays following IR exposure. Oct4 binding was induced by stress at both Blcap and Ell (Fig. 6D).
Discussion
Here we show that Oct1 is dynamically phosphorylated at serine and threonine residues in vivo in response to stress stimuli and that phosphorylation at two conserved DNA-binding domain serine residues regulates Oct1 binding to complex DNA configurations. Mutation of a homologous Oct4 residue also supports a role in binding site selectivity. Oct1 binding to specific complex sites (in Polr2a and Taf12) is inducible by stress in vivo, and Oct1 is critical for maintaining normal gene expression at one such site (Polr2a) following stress exposure. In vivo Oct4 DNA binding to several target loci in ES cells is regulated in the same manner. We also identify a new class of complex site (TMFORE) capable of binding Oct1 and Oct4 as a dimer. We use ChIPseq to confirm inducible target occupancy on a large scale, and to identify new constitutive and inducible Oct1 targets. Finally, using Oct4 ChIP in ES cells, we confirm that Oct4 inducibly interacts with two of these same targets in response to stress stimuli.
We used an affinity purification/mass spectrometry approach to purify Oct1 from HeLa cells under normal and stressed conditions and to identify modifications. Two identified stress-dependent phosphorylation events, T276 and S278, were identified previously as in vitro DNA-PK targets (see the supplemental material in Schild-Poulter et al. 2007). Although we did not identify other modifications described in that report, it is notable that there are few substrates for tryptic digestion in the Oct1 N-terminus, where these events were identified. Comparing our data with an unpublished data set generated by the laboratory of Dr. Steven Gygi, we noted two DNA-binding domain modification events (S335 and S385) that were not identified using our affinity approach. We hypothesized that one or both modifications affect DNA binding. In vitro studies previously identified the phospho-S385 modification, and it was suggested that this modification inhibited DNA-binding activity during M phase (Segil et al. 1991). Our data suggest that the effect of S385 phosphorylation is more nuanced, acting at the level of selectivity for different classes of target sites. Using S385 and S335 mutations, we confirmed a role for both residues in Oct1 target site selectivity.
The Oct1 S385D phosphomimetic mutation partially inhibits binding to simple octamer motifs in EMSA, consistent with the suggestion that S385 modification could inhibit Oct1 DNA binding (Segil et al. 1991). Because simple octamer binding in extracts from irradiated cells is not inhibited, S385D may not fully mimic serine phosphorylation at this position. Alternatively, only a fraction of Oct1 may be phosphorylated at this position and/or other modifications may simultaneously augment simple octamer binding. In contrast, S385D augmented binding to MORE sites. S385K mutation had the opposite effect: Little change in simple octamer binding was observed, but MORE dimerization was strongly inhibited (Fig. 4B). Our interpretation that phosphorylation of S385 results in an intramolecular stabilization of an orientation that favors dimer binding is consistent with the effect of the S385K, which would create an electrostatic clash with K296 in the MORE structure and disfavor dimerization (Fig. 4A). This interpretation is also consistent with our findings that higher protein concentrations reduce the effects of S385 mutation (data not shown).
Oct1 S335 alanine or lysine substitution caused a major change in the DNA-binding spectrum, strongly inhibiting MORE binding, but having little effect on binding to a simple site (Fig. 4D). Unlike S385, increasing the amount of added protein did not alter the mutant phenotypes, arguing for a more direct role in dimer stabilization. An analogous Oct4 mutation also resulted in specific effects on dimer induction following exposure to the DNA-damaging agent MMS (Supplemental Fig. 1).
We identified a new class of complex recognition sequences that includes sites in TAF12, Mili, and Foxo4 (Fig. 5D–F; Supplemental Fig. 2). We term this sequence TMFORE. It is highly likely that there are a number of additional classes of Oct protein-binding sites, each capable of adopting unique protein:DNA configurations, and each capable of responding to different spectrums of Oct protein modification.
Oct protein target genes have functions consistent with a role in the stress response. Examples include Bcl-2 (Heckman et al. 2006), p15INK4b (Hitomi et al. 2007), Cyclin D1 (Magne et al. 2003), GADD45a and GADD45g (Takahashi et al. 2001; Boyer et al. 2005), C-reactive protein (Voleti and Agrawal 2005), iNOS (Kim et al. 1999), PDRG (Luo et al. 2003), and a number of interleukins (Pfeuffer et al. 1994; Duncliffe et al. 1997; Wu et al. 1997; Murayama et al. 2006). Recent studies have also identified a global association between Oct1 recognition sequences and stress response regulation. One study used low pH as a stress agent and identified selective Oct-binding site enrichment in a large group of coordinately up-regulated mRNAs (Duggan et al. 2006). Another study identified sites in >90% of genes, with significant changes in transcription rate following exposure of HeLa cells to camptothecin (Fan et al. 2006). Oct1 also appears to be a mediator of the response to heavy metals (Glahn et al. 2008). These results support our conclusion that Oct1 is a stress response effector. It will now be of importance to re-evaluate previously defined targets to determine whether the sequence composition and Oct protein binding is indicative of simple octamer-, PORE-, MORE-, or TMFORE-type configurations, or less well-defined configurations. It will also be important to define the different kinases and phosphatases that mediate the modification of both proteins and the triggers and signaling cascades involved.
Interestingly, although Oct1 binding to the Polr2a locus is induced following stress exposure rather than modulating gene expression in wild-type cells, Oct1 is necessary for maintenance of normal Polr2a expression: In the Oct1-deficient condition, message levels are inappropriately repressed. Although it is formally possible that there is a difference in message stability specifically in Oct1-deficient cells and specifically at time points following stress, the most likely interpretation is that Oct1 plays an unusual functional role as a stress-dependent Polr2a anti-repressor. In contrast to the effects on expression of the native Polr2a gene, stress exposure had little effect in luciferase-based transient transfection assays (data not shown). It is therefore likely that in response to stress signals, Oct1 relieves repressive activities through a mechanism involving genomic chromatin context not recapitulated in transfections. Our recent results indicate that Oct1 and Oct4 control gene expression, at least in part, through control of a specific negative epigenetic mark, consistent with this hypothesis (J. Kang, A. Shakya, C. Collister, and D. Tartin, unpubl.).
Oct4 also has the capacity to respond to stress signals by selectively altering the affinity for complex binding sites in vitro. Further, Oct4 inducibly associates with a number of in vivo targets, including Taf12, Bmp4, Blcap, and Ell. Oct4 mutation at a serine residue homologous to Oct1 S335 alters binding selectivity for complex sites in vitro, suggesting that the stress responsiveness of Oct4 operates through modification of similar sites. The concept that Oct1 and Oct4 share common modes of regulation is not surprising, as the proteins share high identity over a substantial portion of their amino acid sequence, recognize DNA with similar specificity and affinity in vitro, and regulate common targets (e.g., histones, Gadd45, osteopontin, Taf12, Blcap, and Ell) (Botquin et al. 1998; Wang et al. 2000; Boyer et al. 2005; this study).
The most potent and well-characterized stress response pathways involve transcription factors strongly associated with cancer, such as p53 and NF-κB. Because Oct1 and Oct4 recognize and integrate stress signals to direct changes in gene expression, a logical extension is that deregulation of these proteins may be a causal agent of malignancy. Evidence for this view comes from the findings that malignant T-cell lymphomas arise in transgenic mice overexpressing a fragment of Oct1 under the control of the T-cell-specific Lck promoter (Qin et al. 1994), and that Oct1 is overexpressed in some human breast and intestinal gastric tumors (Jin et al. 1999; Almeida et al. 2005). Oct4 is also strongly associated with hyperplasia and malignancy (Gidekel et al. 2003; Hochedlinger et al. 2005; Covello et al. 2006). One study has demonstrated a significant enrichment in Oct protein target sites in genes overexpressed in a lung cancer model, but not overexpression of Oct1 itself, suggesting that Oct1 activity, rather than levels, may regulate these processes (Reymann and Borlak 2008).
Materials and methods
Oligonucleotides
Sequences for gel-shift probes, site-directed mutagenesis, ChIP, PCR, real-time RT–PCR, and nanoparticle synthesis are presented in Supplemental Table 6.
Plasmids, cloning, and site-directed mutagenesis
pBabe-hOct1 has been described previously (V.E.H. Wang et al. 2004). Mouse Oct4 cDNA was cloned into pBabe by excising the cDNA from pGEX4T2-Oct4 (a gift of Y. Bergman) using EcoRI. The cDNA was inserted into the unique EcoRI site of pBabe. Directionality was confirmed by sequencing. Site-directed mutagenesis was performed using QuickChange (Stratagene).
The 2XMORE from the Polr2a gene was inserted into pGL2 (Promega) by synthesizing an oligonucleotide. pGL2 was digested with SmaI, and the oligo was ligated into the digested backbone creating a dead SmaI site. The ligation mixture was recut with SmaI and transformed into competent cells. Correct clones were confirmed by sequencing.
A 600-bp fragment of Polr2a was amplified using PCR primers with 5′ KpnI and HindIII restriction sites. PCR products were purified, digested with KpnI and HindIII, and ligated into a pGL3 backbone from a plasmid digested with the same enzymes.
Cell culture
Oct1−/− E12.5 MEFs were generated as described (V.E.H. Wang et al. 2004). MEFs and HeLa cells were cultured in Dulbecco's modified Eagle's medium (Hyclone) supplemented with 10% heat-inactivated bovine fetal serum (FBS, Hyclone), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine (Invitrogen), and 50 μM β-mercaptoethanol (Sigma) at 37°C and 5% CO2 in a humidified atmosphere. R1.45 ES cells (a gift from M. Capecchi) used 15% FBS and additionally contained 1 mM sodium pyruvate, 0.1 mM nonessential amino acids (Invitrogen), and 1000 U/mL LIF (Chemicon). For IR, cells were exposed to 1200 RAD using an X RAD 320 X-ray irradiator and a hard beam filter (Precision X-Ray). The flux was ∼100 RAD/min. H2O2 was used at 1 mM except where indicated.
Oct1 purification using latex nanoparticles
HeLa cell spinner cultures were treated with IR or H2O2 and incubated for 1 h at 37°C. Cell pellets were resuspended in 10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, protease inhibitors (PIs), and phosphatase inhibitors (PhIs, Roche). Following dounce homogenization, nuclei were collected by centrifugation at 15,000g. Proteins in the nuclear pellet were extracted on ice for 1 h in 20 mM Hepes (pH 7.9), 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, PIs and PhIs. Nuclear extracts were clarified by centrifugation at 15,000g and dialyzed for 4 h at 4°C in 20 mM Hepes (pH 7.9), 20% glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT, PIs and PhIs. Two milligrams of protein were mixed with 10 μL of pelleted nanobeads in buffer A (50 mM Tris at pH 8.0, 20% glycerol, 0.5 mM EDTA, 0.1% NP-40, 1 mM DTT, PIs and PhIs) plus 100 mM NaCl. The mixture was incubated for 1 h at 4°C. Nanobeads were washed with buffer A containing 200 mM and 300 mM NaCl, respectively. Bound proteins were eluted with buffer A containing 300 mM NaCl and 30 μM octamer oligomers for 1 h on ice.
Liquid chromatography–mass spectrometry–mass spectrometry (LC/MS/MS)
Gel slabs were excised from silver-stained polyacrylamide gels and destained using a 1:1 mixture of 30 mM Potassium Ferricynide/100 mM Sodium Thiosulfate solution and equilibrated in water. Protein gel bands were digested overnight with trypsin (Promega), and reconstituted in 10 μL of 5% acetonitrile (ACN) with 0.1% formic acid (FA), which was then manually injected (5 μL) using a nano injector (Valco) onto a self-packed column (100 μm × 75 mm, 3-μm particle size; Waters). An 80-min solvent gradient of 5%–85% A:B (A: 5% ACN/0.1% FA; B: 80% acetonitrile /0.1% FA) was used at 350 nL/min at 5% B for the first 3 min, followed by a linear increase to 55% B in 50 min, and finally maintained at 85% B for 5 min. LC/MS/MS data were acquired using a LTQ-FT hybrid mass spectrometer (ThermoElectron Corp.) equipped with a nanospray ionization source. Peptide molecular masses were measured by FT-ICR. Peptide sequencing was performed by collision-induced dissociation (CID) in the linear ion trap. Peptides were assigned from MSDB or NCBI protein database searches, using the MASCOT search engine (Matrix Science). All assigned peptides (including phosphopeptides) showed MASCOT scores >20 and had mass errors <3 ppm.
EMSA
EMSA procedures paralleled those described (V.E.H. Wang et al. 2004; Tantin et al. 2008). A monoclonal anti-Oct1 antibody (Upstate Biotechnologies) was used for supershifts. Reactions contained 10 μg (HeLa cell) or 15 μg (mouse fibroblast) of nuclear extract.
ChIP/ChIPseq
ChIP was performed as described in Boyd and Farnham (1999) and Shang et al. (2000), except that the ChIP dilution buffer included 190 mM NaCl and Protein G nanobeads (ActivMotif) were used. HeLa or ES cells (1 × 107) were used per individual sample. Oct1 and Oct4 antibodies were purchased from Chemicon and Santa Cruz Biotechnologies, respectively.
For ChIPseq, ChIP material was subjected to high-volume sequencing using a Solexa GAI sequencer (Illumina). Libraries were prepared from ∼10 ng of chromatin-precipitated DNA using the Illumina ChIPseq Sample Prep Kit. Adaptor-modified fragments ranging in size from 350 to 450 bp were selected by agarose gel electrophoresis, amplified by PCR, and validated using an Agilent Bioanalyzer. library samples (2–6 pM) were hybridized to an activated flowcell and amplified. DNA sequence identification was performed on an Illumina Genome Analyzer I using a 26 Cycle Sequencing Kit. Reads were mapped to chromosomal locations in the March 2006 NCBI Build 36.1 human genome using the pipeline's eland_extended aligner. Further analysis was performed using the USeq analysis package (http://sourceforge.net/projects/useq). Briefly, a sliding window (500 bp) binomial P-value was used to identify significant regions of enrichment relative to the control. P-values were converted to FDRs using Storey's q-value method, adjacent windows that exceeded an FDR of 0.01 were joined and ranked to generate a list of putative binding peaks for each condition. For analysis of differential Oct1 occupancy under stress conditions, the no treatment ChIP data was substituted for the input. Raw data are available at the NCBI Short Read archive.
Real-time RT–PCR
RNA was prepared using TRIzol (Invitrogen). Five micrograms of RNA were converted into cDNA using Superscipt III and random hexamers (Invitrogen). PCR reactions used SYBR Green (Molecular Probes) to detect signals in proportion to the accumulating target amplicons and intron-spanning primer pairs. PCR results were obtained and analyzed using LightCycler 480 Real-Time PCR System (Roche).
Western blotting
Antibodies used for Western blotting were as follows: mouse anti-Oct1 (Upstate Biotechnologies), mouse anti-Oct4 and β-actin (Santa Cruz Biotechnologies), and rabbit anti-Pol II large subunit (Chemicon).
Acknowledgments
We thank S. Gygi for sharing data prior to publication. We thank D. Stillman for critical reading of the manuscript and M. Capecchi for ES cells. We also thank D. Stillman and B. Graves for helpful technical advice and A. Shakya for providing reagents. We thank C. Nelson and K. Parsawar for mass spectrometry. We thank B. Dalley and D. Nix for high-throughput sequencing and analysis. This work was supported by a March of Dimes/Basil O'Connor Starter Scholar award, a grant from the American Cancer Society (GMC-115196), to D.T., and by NCI Cancer Center Support Grant P30CA042014.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1750709.
Supplemental material is available at http://www.genesdev.org.
References
- Almeida R., Almeida J., Shoshkes M., Mendes N., Mesquita P., Silva E., Van Seuningen I., Reis C.A., Santos-Silva F., David L. OCT-1 is over-expressed in intestinal metaplasia and intestinal gastric carcinomas and binds to, but does not transactivate, CDX2 in gastric cells. J. Pathol. 2005;207:396–401. doi: 10.1002/path.1861. [DOI] [PubMed] [Google Scholar]
- Botquin V., Hess H., Fuhrmann G., Anastassiadis C., Gross M.K., Vriend G., Scholer H.R. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes & Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd K.E., Farnham P.J. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Cell. Biol. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Currie R.W. Heat shock treatment suppresses angiotensin II-induced SP-1 and AP-1 and stimulates Oct-1 DNA-binding activity in heart. Inflamm. Res. 2005;54:338–343. doi: 10.1007/s00011-005-1360-y. [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V.B., Wong E., Orlov Y.L., Zhang W., Jiang J., et al. Integration of external signaling pathways with the core transcriptional network in embyonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Covello K.L., Kehler J., Yu H., Gordan J.D., Arsham A.M., Hu C.J., Labosky P.A., Simon M.C., Keith B. HIF-2α regulates Oct-4: Effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes & Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan S.P., Gallagher W.M., Fox E.J., Abdel-Latif M.M., Reynolds J.V., Kelleher D. Low pH results in co-ordinate regulation of gene expression in oesophageal cells. Carcinogenesis. 2006;27:319–327. doi: 10.1093/carcin/bgi211. [DOI] [PubMed] [Google Scholar]
- Duncliffe K.N., Bert A.G., Vadas M.A., Cockerill P.N. A T cell-specific enhancer in the interleukin-3 locus is activated cooperatively by Oct and NFAT elements within a DNase I-hypersensitive site. Immunity. 1997;6:175–185. doi: 10.1016/s1074-7613(00)80424-0. [DOI] [PubMed] [Google Scholar]
- Fan W., Jin S., Tong T., Zhao H., Fan F., Antinore M.J., Rajasekaran B., Wu M., Zhan Q. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J. Biol. Chem. 2002;277:8061–8067. doi: 10.1074/jbc.M110225200. [DOI] [PubMed] [Google Scholar]
- Fan J., Zhan M., Shen J., Martindale J.L., Yang X., Kawai T., Gorospe M. Enmasse nascent transcription analysis to elucidate regulatory transcription factors. Nucleic Acids Res. 2006;34:1492–1500. doi: 10.1093/nar/gkj510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity P.A., Chen D., Rothenberg E.V., Wold B.J. Interleukin-2 transcription is regulated in vivo at the level of coordinated binding of both constitutive and regulated factors. Mol. Cell. Biol. 1994;14:2159–2169. doi: 10.1128/mcb.14.3.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel S., Pizov G., Bergman Y., Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Glahn F., Schmidt-Heck W., Zellmer S., Guthke R., Wiese J., Golka K., Hergenroder R., Degen G.H., Lehmann T., Hermes M., et al. Cadmium, cobalt and lead cause stress response, cell cycle deregulation and increased steroid as well as xenobiotic metabolism in primary normal human bronchial epithelial cells which is coordinated by at least nine transcription factors. Arch. Toxicol. 2008;82:513–524. doi: 10.1007/s00204-008-0331-9. [DOI] [PubMed] [Google Scholar]
- Heckman C.A., Duan H., Garcia P.B., Boxer L.M. Oct transcription factors mediate t(14;18) lymphoma cell survival by directly regulating bcl-2 expression. Oncogene. 2006;25:888–898. doi: 10.1038/sj.onc.1209127. [DOI] [PubMed] [Google Scholar]
- Herr W., Sturm R.A., Clerc R.G., Corcoran L.M., Baltimore D., Sharp P.A., Ingraham H.A., Rosenfeld M.G., Finney M., Ruvkin G., et al. The POU domain: A large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes & Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- Hitomi T., Matsuzaki Y., Yasuda S., Kawanaka M., Yogosawa S., Koyama M., Tantin D., Sakai T. Oct-1 is involved in the transcriptional repression of the p15(INK4b) gene. FEBS Lett. 2007;581:1087–1092. doi: 10.1016/j.febslet.2007.01.092. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K., Yamada Y., Beard C., Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Jin T., Branch D.R., Zhang X., Qi S., Youngson B., Goss P.E. Examination of POU homeobox gene expression in human breast cancer cells. Int. J. Cancer. 1999;81:104–112. doi: 10.1002/(sici)1097-0215(19990331)81:1<104::aid-ijc18>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Kim Y.M., Ko C.B., Park Y.P., Kim Y.J., Paik S.G. Octamer motif is required for the NF-κB-mediated induction of the inducible nitric oxide synthase gene expression in RAW 264.7 macrophages. Mol. Cell. 1999;9:99–109. [PubMed] [Google Scholar]
- Klemm J.D., Rould M.A., Aurora R., Herr W., Pabo C.O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- LeBowitz J.H., Kobayashi T., Staudt L., Baltimore D., Sharp P.A. Octamer-binding proteins from B or HeLa cells stimulate transcription of the immunoglobulin heavy-chain promoter in vitro. Genes & Dev. 1988;2:1227–1237. doi: 10.1101/gad.2.10.1227. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Luo X., Huang Y., Sheikh M.S. Cloning and characterization of a novel gene PDRG that is differentially regulated by p53 and ultraviolet radiation. Oncogene. 2003;22:7247–7257. doi: 10.1038/sj.onc.1207010. [DOI] [PubMed] [Google Scholar]
- Magne S., Caron S., Charon M., Rouyez M.C., Dusanter-Fourt I. STAT5 and Oct-1 form a stable complex that modulates cyclin D1 expression. Mol. Cell. Biol. 2003;23:8934–8945. doi: 10.1128/MCB.23.24.8934-8945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meighan-Mantha R.L., Riegel A.T., Suy S., Harris V., Wang F.H., Lozano C., Whiteside T.L., Kasid U. Ionizing radiation stimulates octamer factor DNA binding activity in human carcinoma cells. Mol. Cell. Biochem. 1999;199:209–215. doi: 10.1023/a:1006958217143. [DOI] [PubMed] [Google Scholar]
- Murayama A., Sakura K., Nakama M., Yasuzawa-Tanaka K., Fujita E., Tateishi Y., Wang Y., Ushijima T., Baba T., Shibuya K., et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nie J., Sakamoto S., Song D., Qu Z., Ota K., Taniguchi T. Interaction of Oct-1 and automodification domain of poly(ADP-ribose) synthetase. FEBS Lett. 1998;424:27–32. doi: 10.1016/s0014-5793(98)00131-8. [DOI] [PubMed] [Google Scholar]
- Nieto L., Joseph G., Stella A., Henri P., Burlet-Schiltz O., Monsarrat B., Clottes E., Erard M. Differential effects of phosphorylation on DNA binding properties of N Oct-3 are dictated by protein/DNA complex structures. J. Mol. Biol. 2007;370:687–700. doi: 10.1016/j.jmb.2007.04.072. [DOI] [PubMed] [Google Scholar]
- Nishio K., Masaike Y., Ikeda M., Narimatsu H., Gokon N., Tsubouchi S., Hatakeyama M., Sakamoto S., Hanyu N., Sandhu A., et al. Development of novel magnetic nano-carriers for high-performance affinity purification. Colloids Surf. B Biointerfaces. 2008;64:162–169. doi: 10.1016/j.colsurfb.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Pfeuffer I., Klein-Hessling S., Heinfling A., Chuvpilo S., Escher C., Brabletz T., Hentsch B., Schwarzenbach H., Matthias P., Serfling E. Octamer factors exert a dual effect on the IL-2 and IL-4 promoters. J. Immunol. 1994;153:5572–5585. [PubMed] [Google Scholar]
- Qin X.F., Luo Y., Suh H., Wayne J., Misulovin Z., Roeder R.G., Nussenzweig M.C. Transformation by homeobox genes can be mediated by selective transcriptional repression. EMBO J. 1994;13:5967–5976. doi: 10.1002/j.1460-2075.1994.tb06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A., Tomilin A., Pohl E., Lins K., Philippsen A., Reinbold R., Scholer H.R., Wilmanns M. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell. 2001;8:569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- Reymann S., Borlak J. Transcription profiling of lung adenocarcinomas of c-myc-transgenic mice: Identification of the c-myc regulatory gene network. BMC Syst. Biol. 2008;2:46. doi: 10.1186/1752-0509-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan A.K., Rosenfeld M.G. POU domain family values: Flexibility, partnerships, and developmental codes. Genes & Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- Schild-Poulter C., Pope L., Giffin W., Kochan J.C., Ngsee J.K., Traykova-Andonova M., Hache R.J. The binding of Ku antigen to homeodomain proteins promotes their phosphorylation by DNA-dependent protein kinase. J. Biol. Chem. 2001;276:16848–16856. doi: 10.1074/jbc.M100768200. [DOI] [PubMed] [Google Scholar]
- Schild-Poulter C., Shih A., Tantin D., Yarymowich N.C., Soubeyrand S., Sharp P.A., Hache R.J. DNA-PK phosphorylation sites on Oct-1 promote cell survival following DNA damage. Oncogene. 2007;26:2980–2988. doi: 10.1038/sj.onc.1210165. [DOI] [PubMed] [Google Scholar]
- Segil N., Roberts S.B., Heintz N. Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science. 1991;254:1814–1816. doi: 10.1126/science.1684878. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu X., DiRenzo J., Lazar M.A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Sharov A.A., Masui S., Sharova L.V., Piao Y., Aiba K., Matoba R., Xin L., Niwa H., Ko M.S. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genomics. 2008;9:269. doi: 10.1186/2471-2164-9-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N., Sugimoto K., Tang J., Nishi T., Sato I., Hiramoto M., Aizawa S., Hatakeyama M., Ohba R., Hatori H., et al. High-performance affinity beads for identifying drug receptors. Nat. Biotechnol. 2000;18:877–881. doi: 10.1038/78496. [DOI] [PubMed] [Google Scholar]
- Sive H.L., Heintz N., Roeder R.G. Multiple sequence elements are required for maximal in vitro transcription of a human histone H2B gene. Mol. Cell. Biol. 1986;6:3329–3340. doi: 10.1128/mcb.6.10.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Saito S., Ohtani N., Sakai T. Involvement of the Oct-1 regulatory element of the gadd45 promoter in the p53-independent response to ultraviolet irradiation. Cancer Res. 2001;61:1187–1195. [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tantin D., Tussie-Luna M.I., Roy A.L., Sharp P.A. Regulation of immunoglobulin promoter activity by TFII-I class transcription factors. J. Biol. Chem. 2004;279:5460–5469. doi: 10.1074/jbc.M311177200. [DOI] [PubMed] [Google Scholar]
- Tantin D., Schild-Poulter C., Wang V., Hache R.J., Sharp P.A. The octamer binding transcription factor Oct-1 is a stress sensor. Cancer Res. 2005;65:10750–10758. doi: 10.1158/0008-5472.CAN-05-2399. [DOI] [PubMed] [Google Scholar]
- Tantin D., Gemberling M., Callister C., Fairbrother W. High-throughput biochemical analysis of in-vivo location data reveals novel distinct classes of POU5F1(Oct4)/DNA complexes. Genome Res. 2008;18:631–639. doi: 10.1101/gr.072942.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomilin A., Remenyi A., Lins K., Bak H., Leidel S., Vriend G., Wilmanns M., Scholer H.R. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell. 2000;103:853–864. doi: 10.1016/s0092-8674(00)00189-6. [DOI] [PubMed] [Google Scholar]
- Ushmorov A., Ritz O., Hummel M., Leithauser F., Moller P., Stein H., Wirth T. Epigenetic silencing of the immunoglobulin heavy-chain gene in classical Hodgkin lymphoma-derived cell lines contributes to the loss of immunoglobulin expression. Blood. 2004;104:3326–3334. doi: 10.1182/blood-2003-04-1197. [DOI] [PubMed] [Google Scholar]
- Voleti B., Agrawal A. Regulation of basal and induced expression of C-reactive protein through an overlapping element for OCT-1 and NF-κB on the proximal promoter. J. Immunol. 2005;175:3386–3390. doi: 10.4049/jimmunol.175.5.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yamamoto S., Hijiya N., Benveniste E.N., Gladson C.L. Transcriptional regulation of the human osteopontin promoter: Functional analysis and DNA-protein interactions. Oncogene. 2000;19:5801–5809. doi: 10.1038/sj.onc.1203917. [DOI] [PubMed] [Google Scholar]
- Wang B., Ma L., Tao X., Lipsky P.E. Triptolide, an active component of the Chinese herbal remedy Tripterygium wilfordii Hook F, inhibits production of nitric oxide by decreasing inducible nitric oxide synthase gene transcription. Arthritis Rheum. 2004;50:2995–3003. doi: 10.1002/art.20459. [DOI] [PubMed] [Google Scholar]
- Wang R.H., Yu H., Deng C.X. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc. Natl. Acad. Sci. 2004;101:17108–17113. doi: 10.1073/pnas.0407585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V.E., Tantin D., Chen J., Sharp P.A. B cell development and immunoglobulin transcription in Oct-1-deficient mice. Proc. Natl. Acad. Sci. 2004;101:2005–2010. doi: 10.1073/pnas.0307304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang V.E.H., Schmidt T., Chen J., Sharp P.A., Tantin D. Embryonic lethality, decreased erythopoiesis, and defective octamer-dependent promoter activation in Oct-1-deficient mice. Mol. Cell. Biol. 2004;24:1022–1032. doi: 10.1128/MCB.24.3.1022-1032.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G.D., Lai E.J., Huang N., Wen X. Oct-1 and CCAAT/enhancer-binding protein (C/EBP) bind to overlapping elements within the interleukin-8 promoter. The role of Oct-1 as a transcriptional repressor. J. Biol. Chem. 1997;272:2396–2403. [PubMed] [Google Scholar]
- Zhao H., Jin S., Fan F., Fan W., Tong T., Zhan Q. Activation of the transcription factor Oct-1 in response to DNA damage. Cancer Res. 2000;60:6276–6280. [PubMed] [Google Scholar]
- Zheng L., Roeder R.G., Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 2003;114:255–266. doi: 10.1016/s0092-8674(03)00552-x. [DOI] [PubMed] [Google Scholar]