Abstract
Local translation of asymmetrically enriched mRNAs is a powerful mechanism for functional polarization of the cell. In Drosophila, exclusive accumulation of Oskar protein at the posterior pole of the oocyte is essential for development of the future embryo. This is achieved by the formation of a dynamic oskar ribonucleoprotein (RNP) complex regulating the transport of oskar mRNA, its translational repression while unlocalized, and its translational activation upon arrival at the posterior pole. We identified the nucleo–cytoplasmic shuttling protein PTB (polypyrimidine tract-binding protein)/hnRNP I as a new factor associating with the oskar RNP in vivo. While PTB function is largely dispensable for oskar mRNA transport, it is necessary for translational repression of the localizing mRNA. Unexpectedly, a cytoplasmic form of PTB can associate with oskar mRNA and repress its translation, suggesting that nuclear recruitment of PTB to oskar complexes is not required for its regulatory function. Furthermore, PTB binds directly to multiple sites along the oskar 3′ untranslated region and mediates assembly of high-order complexes containing multiple oskar RNA molecules in vivo. Thus, PTB is a key structural component of oskar RNP complexes that dually controls formation of high-order RNP particles and translational silencing.
Keywords: Drosophila, PTB, oskar, translation, RNP assembly, hnRNP
In eukaryotic cells, transcription represents the first step of gene expression. However, a variety of nuclear and cytoplasmic post-transcriptional events determine the final fate of RNAs and thus regulate gene product diversity as well as the spatio–temporal pattern of gene expression.
In recent years, subcellular targeting of mRNAs, coupled to localized translation, has emerged as a powerful mechanism to spatially and temporally restrict protein synthesis. Furthermore, a genome-wide in situ hybridization analysis in Drosophila embryos suggests that RNA localization could represent a general mechanism for the establishment of cell polarity (Lecuyer et al. 2007). Consistent with this, functional studies have shown that local translation of asymmetrically enriched mRNAs is used by differentiated cells to generate functionally distinct compartments, or by developing organisms to partition cell fate determinants (St Johnston 2005; Du et al. 2007). In several species, the asymmetric distribution in unfertilized eggs of maternal RNAs encoding cytoplasmic determinants controls embryonic body axis specification. In Drosophila, oskar mRNA encodes the posterior determinant and is transported to the posterior pole of the oocyte, where it is specifically translated. This precise spatio–temporal control is critical for embryonic patterning, as mutant oocytes in which oskar is not expressed at the posterior pole develop into embryos lacking abdominal structures and germ cells (Ephrussi et al. 1991; Kim-Ha et al. 1991). Conversely, ectopic translation of unlocalized oskar causes patterning defects characterized by a loss of anterior structures, and in extreme cases, duplication of posterior structures (Ephrussi and Lehmann 1992; Kim-Ha et al. 1995).
RNA localization relies on the formation of functional ribonucleoprotein (RNP) complexes that precisely control and coordinate multiple steps including motor-based transport of the mRNA along the cytoskeleton and translational repression of the localizing mRNA, as well as its translation activation and anchoring upon arrival at the final destination (St Johnston 2005). These complexes contain different RNA-associated proteins, including heterogeneous nuclear RNPs (hnRNPs) that regulate various RNA processing events (Dreyfuss et al. 2002; Glisovic et al. 2008). Furthermore, RNPs seem to assemble in vivo into large particles containing several RNA molecules and associated proteins, as evidenced by light microscopy visualization or by biochemical sedimentation techniques (Kiebler and Bassell 2006; Lange et al. 2008). Although the in vivo functional relevance of such high-order structures remains unknown, their formation has been proposed to represent a crucial step in the establishment of transport-competent RNP complexes.
The composition, architecture, and dynamics of RNP complexes dictate the specific behavior of target RNAs within the cell. Importantly, RNP complexes undergo an extensive remodeling upon export into the cytoplasm, yet their cytoplasmic fate is highly connected to their nuclear history (Kress et al. 2004; St Johnston 2005; Giorgi and Moore 2007; Lewis and Mowry 2007). For example, in the case of oskar mRNA, nuclear recruitment of the exon–junction complex upon splicing is necessary for oskar mRNA transport to the posterior pole of the oocyte (Hachet and Ephrussi 2001, 2004). Furthermore, oskar translational repression is controlled by nucleo–cytoplasmic shuttling RNA-binding proteins such as Bruno and Hrp48 (Kim-Ha et al. 1995; Yano et al. 2004; Snee et al. 2008). While these proteins likely associate with the RNA in the nucleus, how important the nuclear recruitment of these proteins to their target RNA is remains to be functionally tested.
In a visual protein-trap screen, we identified Drosophila polypyrimidine tract-binding protein (PTB) as a protein colocalizing with oskar mRNA. PTB belongs to the hnRNP family of RNA-binding proteins and regulates various nuclear and cytoplasmic RNA processing events in vertebrates (Auweter and Allain 2008). Specifically, PTB plays a predominant role in the regulation of alternative splicing (Valcarcel and Gebauer 1997; Spellman et al. 2005), and has also been shown to regulate internal ribosome entry site (IRES)-dependent translation initiation (Stoneley and Willis 2004; Jang 2006; Semler and Waterman 2008) and mRNA localization (Cote et al. 1999; Ma et al. 2007). In our in vivo study, we show that Drosophila PTB is a new component of the oskar RNP complex and that PTB regulates translational repression of oskar mRNA. Surprisingly, while PTB strongly accumulates in germ cell nuclei and thus, potentially, associates with oskar in this compartment, its recruitment to oskar RNP in the germ cell cytoplasm is sufficient for efficient repression. Consistent with a direct role in oskar translation regulation, PTB binds to multiple binding sites in the oskar 3′ untranslated region (UTR). Finally, we show that PTB is a trans-acting factor required in vivo for oskar mRNA oligomerization. Our study thus reveals that Drosophila PTB is a key structural component of oskar RNP complexes and suggests a functional link between the assembly of high-order RNP particles and the mechanism of oskar translational repression.
Results
PTB colocalizes with oskar mRNA during Drosophila oogenesis
To identify new proteins involved in oskar mRNA regulation, we performed a protein-trap screen that relies on GFP-tagging of proteins by random mobilization of a GFP-containing transposable element in the genome (Morin et al. 2001; Bonin and Mann 2004). Among the recovered GFP fusion proteins colocalizing with oskar mRNA, one showed an enrichment in the oocyte cytoplasm during early oogenesis, followed by an exclusive accumulation at the posterior pole from mid-oogenesis onward (Fig. 1A,B; see Spradling 1993 for a detailed description of ovarian cell types and developmental stages). Additionally, the GFP-tagged protein localizes to the nuclei of both somatic and germ cells, as well as in the cytoplasm of the follicular epithelial cells surrounding the oocyte, at late stages of oogenesis.
Figure 1.
Distribution of PTB during Drosophila oogenesis. (A,B) GFP expression pattern in an ovariole (A) and a stage 10 egg chamber (B) from homozygous GFP-PTB protein-trap females. An ovariole contains egg chambers of different developmental stages, from stage 1 in the germarium, until stage 14 (not shown). The oocyte and its 15 sibling nurse cells develop as a syncytium. The 16-cell germline cyst is surrounded by a layer of somatic epithelial cells. Egg chambers are oriented anterior to the left, posterior to the right. The oocyte, whose size dramatically increases as oogenesis proceeds, is the most posterior cell of the germline cyst. Bars, 60 μm. (C) Schematic representation of Drosophila PTB domain organization indicating the percentage of amino acid identity of the RRM domains compared with PTB orthologs, as well as the insertion site of the GFP sequence (red triangle) in the protein-trap line. (D) Western blot of ovarian extracts from w females (first lane), females heterozygous (protein-trap/+; second lane) or homozygous (protein-trap; third lane) for the protein-trap insertion, probed with rat anti-PTB antibodies. (E–G) w stage 5 (E), stage 8, (F) and stage 9 (G) egg chambers triple-stained with anti-PTB antibodies (top panel, red in the overlay), anti-Staufen antibodies (middle panel, blue in the overlay), and phalloidin (green in the overlay). Note that PTB is expressed both in the germline and in the somatic epithelium. In contrast, Staufen exclusively accumulates in the oocyte. Bar, 60 μm.
Using inverse PCR, we mapped the insertion site of the protein-trap cassette to the hephaestus locus (CG31000), encoding the unique Drosophila homolog of mammalian PTB, also known as hnRNP-I (Dansereau et al. 2002; Davis et al. 2002). Similar to its mammalian ortholog, Drosophila PTB contains a putative N-terminal bipartite nuclear localization signal (NLS) and four conserved RNA recognition motifs (RRM1–4) (Fig. 1C). Western-blot analysis using anti-PTB antibody raised against the Drosophila protein revealed a 65-kDa doublet in wild-type ovarian extracts (Fig. 1D), which is shifted to 95 kDa in the protein-trap line as a result of the GFP insertion upstream of RRM1 (Fig. 1C).
Staining of wild-type ovaries with anti-PTB antibodies revealed a localization pattern of endogenous PTB identical to that of GFP-PTB fusions (Fig. 1E–G). Similarly to the oskar-associated protein Staufen, PTB accumulates in the cytoplasm of young oocytes as early as in the germarium (Fig. 1E–G″; data not shown), transiently in the center of the oocyte at stage 7/8 (Fig. 1F–F″), and at the posterior pole of the oocyte from stage 9 onward (Fig. 1G–G″; see Supplemental Fig. S1C for a specificity control). The perfect colocalization of PTB with Staufen in the oocyte cytoplasm throughout oogenesis suggested that PTB is transported together with oskar mRNA to the posterior pole of the oocyte.
PTB is a component of the oskar RNP complex
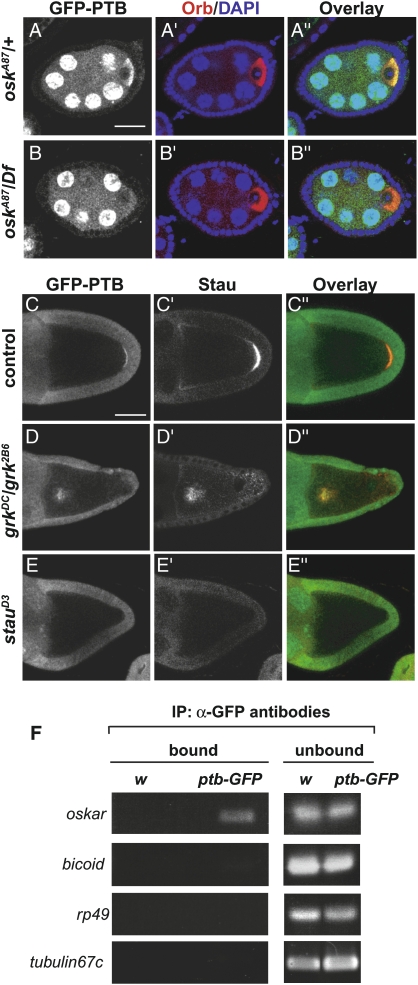
To confirm the in vivo association of PTB with oskar mRNA, we tested if the distribution of PTB in the oocyte depends on oskar mRNA. Strikingly, the cytoplasmic enrichment of PTB in young oocytes is no longer detected in egg chambers lacking oskar mRNA (osk A87/Df(3R)pXT03) (Fig. 2B), indicating that the initial transport of PTB into the oocyte cytoplasm requires the presence of oskar mRNA. As oskar mRNA-null egg chambers do not develop past stage 7 (Jenny and Hachet et al. 2006), we analyzed the oskar dependence of PTB localization during mid-oogenesis, following the distribution of PTB in gurken (grk) and staufen mutant oocytes. In grk2E12/grk2B6 females, microtubule (MT) polarity is altered such that MT plus ends are enriched in the center of the oocyte instead of at the posterior pole (González-Reyes et al. 1995). In these mutants, oskar mRNA and Staufen are detected in the center of the oocyte, together with PTB (Fig. 2D–D″). Furthermore, PTB fails to localize at the posterior pole in stauD3 oocytes (Fig. 2E), which appear to have normal MT polarity but fail to enrich oskar mRNA at the posterior pole (St Johnston et al. 1991; Palacios and St Johnston 2002; S. López de Quinto, unpubl.). Thus, PTB accumulation at the posterior of the oocyte relies on that of oskar mRNA, suggesting that PTB is part of oskar RNP in vivo.
Figure 2.
PTB colocalizes and associates with oskar mRNA. (A,B) Stage 5 egg chambers from oskA87/+ (A) and oskA87/Df(3R)pXT03 (B) females expressing GFP-PTB constructs under the control of the mat-α4-tub promoter, and triple-stained for GFP (green in the overlay), DNA (blue, DAPI), and the oocyte cytoplasm marker Orb (red). Bar, 15 μm. (C–E) Distribution of the GFP-PTB protein-trap fusion in wild-type (C), grk2E12/grk2B6 (D), and stauD3 (E) oocytes. (Left) GFP signal. (Middle) Staufen protein. (Right) Overlay. Bar, 60 μm. (F, left panel) RT–PCR amplification of mRNAs recovered in fractions immunoprecipitated from GFP-PTB protein-trap or w control ovarian extracts using anti-GFP antibodies. (Right panel) Amplifications from unbound fractions are used as controls.
Consistent with this, semiquantitative RT–PCR showed that oskar mRNA is enriched in the fraction precipitated from GFP-PTB protein-trap ovaries using anti-GFP antibodies, but not in immunoprecipitates from w control females (Fig. 2F). No specific enrichment of mRNAs highly expressed in ovaries (rp49, tubulin67C) or of other mRNAs asymmetrically localized in the oocyte (bicoid) was observed in the GFP-PTB precipitated fractions. Altogether, these results indicate that PTB is a bona fide component of oskar RNP in vivo.
oskar mRNA localization to the posterior pole is delayed in ptb mutant oocytes
To investigate the role of PTB in oskar mRNA regulation, we analyzed the distribution of oskar mRNA in ptb mutant oocytes. Four different lethal alleles belonging to the same complementation group were used (Supplemental Fig. S1A). hephe1 and hephe2 are EMS-induced mutations, the first harboring an amino acid substitution (G → Q) in RRM1, and the second corresponding to a deletion covering RRM1, RRM2, and part of RRM3 (Dansereau et al. 2002). heph03429 and heph1545 are insertions of a P-element and a piggyBac transposon into the heph locus, respectively. hephe2 behaves as a null allele (Dansereau et al. 2002), and hephe1, heph03429, and heph1545 as hypomorphic mutants strongly affecting PTB expression levels (Supplemental Fig. S1A,B). Germ cells homozygous for hephe2 stop developing after formation of 16-cell cysts and never produce differentiated egg chambers, suggesting that PTB function is required for early germ cell maintenance (data not shown).
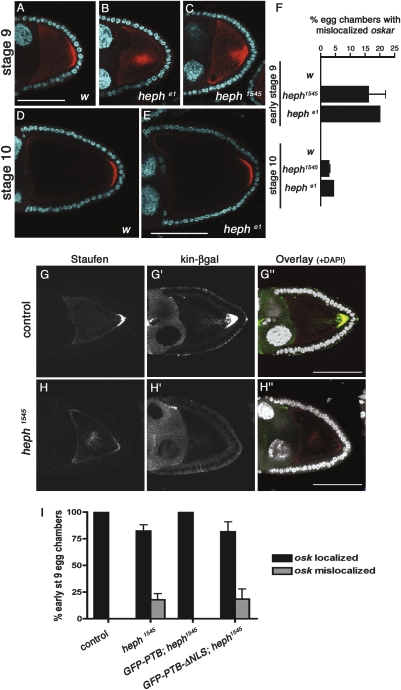
Three other alleles—heph03429, heph1545, and, to a lesser extent, hephe1—support development to later stages. While oskar mRNA accumulates normally in these mutant oocytes during early stages of oogenesis (data not shown), its transport to the posterior pole at mid-oogenesis is affected (Fig. 3B,C). In wild-type oocytes, oskar mRNA and Staufen transiently localize in the center of the oocyte at stages 7/8 and accumulate as a tight posterior crescent from early stage 9 onward (Figs.1E–G, 3A; data not shown). In contrast, oskar mRNA persists in the center of the oocyte or as a cloud near the posterior cortex, in ∼20% of early stage 9 ptb oocytes (Fig. 3B,C,F). This phenotype seems to reflect a transient delay in oskar mRNA transport to the posterior pole, as it is rarely observed in late stage 9 or stage 10 oocytes (Fig. 3E,F; data not shown). Since oskar mRNA localization relies on the polarity of the MT cytoskeleton, we assessed the distribution of the MT plus-end marker kin-β-gal (Clark et al. 1994) in ptb mutant oocytes at stage 9. In these oocytes, oskar mRNA mislocalization correlates with a strong reduction in the posterior accumulation of kin-β-gal (Fig. 3H–H′), suggesting that oskar mRNA localization defects may result from a delay in focusing the MT plus-ends at the posterior pole.
Figure 3.
PTB modulates oskar mRNA localization and oocyte MT polarity. (A–E) Double staining of early stage 9 (A–C) or stage 10 (D,E) oocytes with anti-Staufen antibodies (red) and DAPI (cyan). (A,D) w. (B,E) OvoD-selected hephe1 germline clone. (C) OvoD-selected heph1545 germline clone. Bars, 60 μm. (F) Percentage of early stage 9 or stage 10 oocytes in which Staufen was mislocalized (at least 56 oocytes were scored per genotype). (G,H) Stage 9 oocytes heterozygous (G) and homozygous (H) for heph1545, and triple-stained with anti-Staufen (G,H, red in the overlay), anti-βgal (G′,H′, green in the overlay), and DAPI (white in the overlay). Bar, 60 μm. (I) Graph showing the percentages of early stage 9 oocytes with mislocalized oskar mRNA in different mutant and rescue contexts. GFP-PTB and GFP-PTBΔNLS constructs were expressed under the control of the germline-specific promoter mat-α4-tub. Expression levels of the insertions chosen for the rescue experiments were similar, and comparable with endogenous levels (data not shown). The number of egg chambers counted per genotype is control (n = 62); heph1545 (n = 86); GFP-PTB; heph1545 (n = 56); GFP-PTBΔNLS; heph1545 (n = 56).
As shown in Figure 3I, the oskar localization phenotype was rescued upon expression of a wild-type GFP-PTB fusion in ptb mutant germ cells. Interestingly, a mutant version of PTB unable to enter the nucleus, but still able to associate with oskar mRNA in the cytoplasm (GFP-PTB-ΔNLS) (Supplemental Fig. S2), failed to rescue this phenotype (Fig. 3I). Our results thus indicate that PTB function is required in germ cell nuclei for efficient establishment of MT polarity at stages 8–9 of oogenesis, ultimately controlling localization of oskar mRNA. Given that mammalian PTB has been extensively implicated in the regulation of alternative splicing (Valcarcel and Gebauer 1997; Spellman et al. 2005), it is possible that nuclear Drosophila PTB regulates the splicing of genes involved in establishment of MT polarity in the oocyte.
oskar is prematurely translated in ptb mutant oocytes
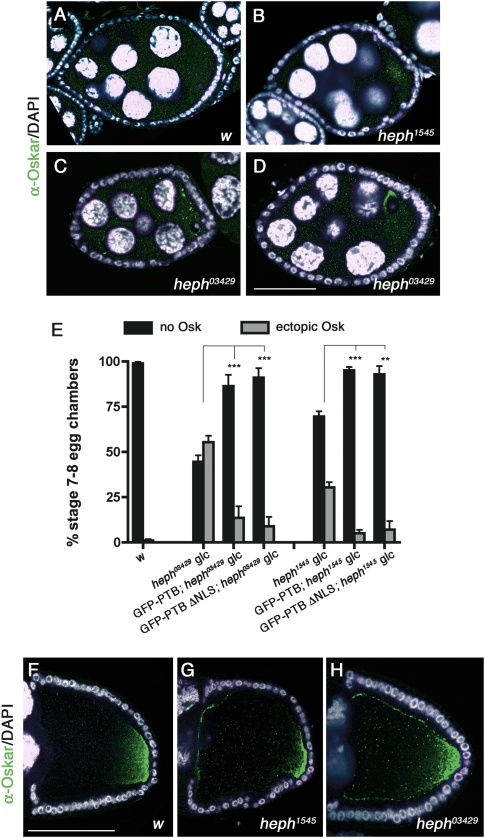
In wild-type females, oskar translation is repressed in early egg chambers and is activated only once the mRNA reaches the posterior pole of the oocyte (Markussen et al. 1995; Rongo et al. 1995). Thus, Oskar protein is not detected before late stage 8 (Fig. 4A) and accumulates exclusively at the oocyte posterior pole in later stages (Fig. 4F). Interestingly, oskar is prematurely translated in ptb mutant females, and ectopic Oskar is detected as early as in stages 5–6 ptb oocytes (data not shown). At stage 7, ∼50% of heph03429 and 30% of heph1545 mutant oocytes display ectopic Oskar expression (Fig. 4B–E), the protein accumulating as a poorly defined cloud in the center of the oocyte (Fig. 4B), more rarely as cytoplasmic aggregates (Fig. 4C), or close to the oocyte nucleus at the anterior-dorsal corner (Fig. 4D). Notably, premature oskar translation is suppressed by germline expression of wild-type GFP-PTB, as well as by expression of the GFP-PTB-ΔNLS mutant protein (Fig. 4E). These results indicate that PTB represses oskar mRNA translation during early oogenesis and that assembly of translationally silenced oskar RNP complexes does not require the recruitment of PTB in the nuclei of germ cells. Furthermore, the differential phenotypic rescue obtained when expressing GFP-PTB-ΔNLS rules out that the premature translation of oskar observed in ptb mutant oocytes is a consequence of MT polarity defects.
Figure 4.
ptb mutant oocytes display ectopic Oskar protein accumulation. (A–D) w (A), heph1545 (B), and heph03429 (C,D) stage 7 egg chambers double-stained with anti-Oskar antibodies (green) and DAPI (white). Bar, 30 μm. The different patterns of ectopic Oskar accumulation reflect the dynamic distribution of oskar mRNA at this stage, which is not altered in ptb mutant oocytes compared with wild-type oocytes (data not shown). (E) Graph showing the percentage of stage 7 oocytes with premature Oskar translation in different mutant and rescue backgrounds (see the legend for Fig. 3I for a description of the rescue constructs). (**) P < 0.005; (***) P < 0.001 in a χ2 test. (F–H) w (F), heph1545 (G), and heph03429 (H) stage 10 oocytes double-stained with anti-Oskar antibodies (green) and DAPI (white). Bar, 60 μm.
Ectopic Oskar accumulation is also detected in later stage 10 ptb oocytes, either at the lateral or at the anterior cortex (Fig. 4G,H). Since such a phenotype has been associated with embryonic anterior patterning defects, ranging from deletion of the head to duplication of posterior structures (Kim-Ha et al. 1995; Yoshida et al. 2004), we examined the cuticle pattern of embryos derived from heph03429 germline clones. The vast majority of these embryos stopped developing before cuticle formation (57.4%; 251/437) or exhibited severe, seemingly oskar-independent morphological defects that prevented the analysis of their head (27.7%; 121/437) (Supplemental Fig. S3). However, 12% (8/63) of the analyzable embryos lacked head structures, consistent with ectopic accumulation of Oskar protein.
To test if PTB might act by regulating the recruitment of other translational repressors, we analyzed the binding of the two best-characterized oskar repressors, namely, Bruno and Hrp48 (Kim-Ha et al. 1995; Gunkel et al. 1998; Yano et al. 2004), using both UV-cross-linking and affinity pull-down assays. As illustrated in Supplemental Figure S4, binding of these proteins to oskar 3′UTR is preserved in ptb mutant extracts. Taken together, these results suggest that PTB is a new oskar translational repressor and that it does not affect the interaction of the two best described oskar repressors with the RNA.
PTB binds directly and with high affinity to the oskar 3′UTR
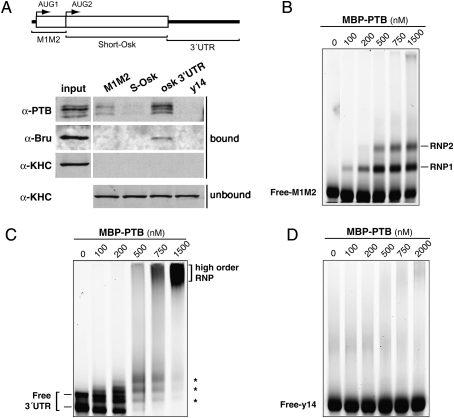
PTB is an RNA-binding protein that regulates several aspects of RNA metabolism through direct interaction with its target RNAs. Depending on the target, PTB binds either to coding sequences or to untranslated regions (Auweter and Allain 2008). We thus analyzed the capacity of endogenous PTB to interact with different regions of oskar mRNA. As shown in Figure 5A, ovarian PTB preferentially associates with the oskar 3′UTR, and to a lesser extent with the oskar 5′ regulatory region separating the two functional AUGs (M1M2) (Gunkel et al. 1998). In this assay, PTB poorly associated with oskar coding sequence, and no significant interaction with a nonrelated RNA (y14 coding sequence) was observed.
Figure 5.
Characterization of PTB binding to oskar RNA. (A) RNA affinity pull-down assay using biotinylated RNAs covering the entire oskar mRNA, and w ovarian extracts. The coding sequence of y14 was used as an unrelated RNA control. (Top) Diagram showing the three different probes used in the assay. Proteins in the bound or unbound fractions were visualized after Western blot analysis using the indicated antibodies. Bruno exclusively binds to the oskar 3′UTR, whereas Khc, a component of the oskar transport machinery without RNA-binding capacity, is not detected in the precipitated fraction. (B–D) EMSA analysis using 50 nM fluorescently labeled oskar M1M2 region (B), oskar 3′UTR (C), or y14 coding sequence (D) in the presence of increasing amounts of MBP-PTB (from 0 to 1500 nM) and of 5 μM of tRNA. The position of the unbound RNA (Free-RNA) and of the discrete RNP complexes is marked on the left and right, respectively.
To test if PTB can interact with oskar mRNA directly, we analyzed the binding of recombinant PTB to the oskar M1M2 and 3′UTR regions, in electrophoretic mobility shift assays (EMSA). Two complexes (RNP1 and RNP2) form upon incubation of M1M2 RNA with increasing amounts of maltose-binding protein (MBP)-PTB (Fig. 5B). This interaction is specific, as it is not observed when using another MBP-tagged RNA-binding protein (MS2-binding protein) (data not shown), or an unrelated RNA (y14 coding sequence) (Fig. 5D). However, the interaction of PTB with oskar M1M2 RNA differs from that observed with the 3′UTR. Under native conditions, the oskar 3′UTR runs as two bands, likely corresponding to RNA monomers and dimers (Fig. 5C). With increasing amounts of PTB, the discrete 3′UTR complexes detected at low protein concentrations (asterisks) shift into more heterogeneous complexes, and finally into higher-order RNP complexes (Fig. 5C), which are efficiently out-competed by addition of increasing amounts of unlabeled oskar 3′UTR competitor to the binding reaction, but not upon addition of unlabeled y14 RNA (Supplemental Fig. S5A). A similar binding profile was obtained using His-tagged recombinant protein (data not shown).
Filter-binding assays in the presence of increasing amounts of recombinant protein revealed that PTB/oskar 3′UTR complexes are formed very rapidly, within 1 min of incubation (data not shown), and indicate that PTB binds with an “apparent” dissociation constant (Kd) of ∼280 nM (Supplemental Fig. S5B). Furthermore, the sigmoidal shape of the binding curve, together with the Hill coefficient (2.4) obtained after Hill transformation, shows that the binding of PTB to the oskar 3′UTR is cooperative. In conclusion, this analysis indicates that PTB can interact directly with oskar mRNA and binds with high affinity, cooperatively, and specifically to the 3′UTR.
The oskar 3′UTR contains multiple PTB-binding sites
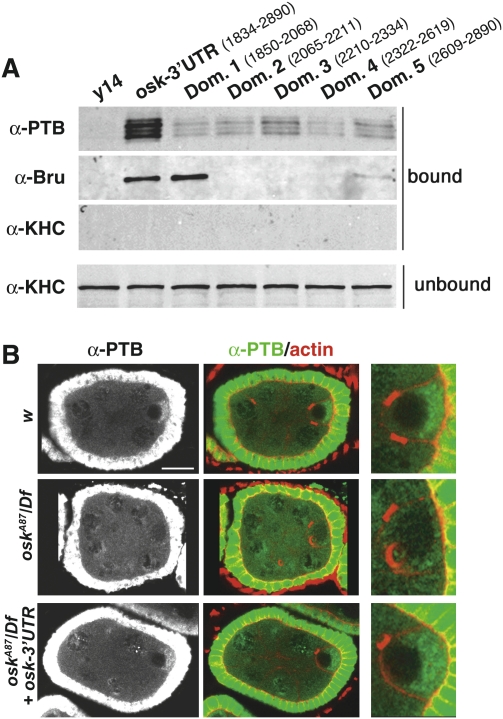
The results of the EMSA suggested the presence of multiple PTB-binding sites in the oskar 3′UTR. Although no strict consensus sequence for PTB binding has so far been defined, previous reports showed that mammalian PTB binds with high affinity to pyrimidine-rich sequences (Singh et al. 1995; Perez et al. 1997). Consistent with this, short RNA sequences containing two pyrimidine-rich motifs efficiently compete the formation of PTB/oskar 3′UTR complexes in EMSA assays (Supplemental Fig. S5A). Strikingly, we found a high number of phylogenetically conserved pyrimidine-rich sequences distributed throughout the oskar 3′UTR (Supplemental Fig. S6A). To see if PTB could interact in vivo with several polypyrimidine tract-containing subregions of the oskar 3′UTR, we divided the 3′UTR into five domains and used the corresponding RNAs in affinity pull-down assays. As shown in Figure 6A, PTB associates with each individual domain, although less strongly than with the entire 3′UTR. This binding pattern is distinct from that of other oskar regulatory proteins that bind to specific regions of the 3′UTR (see Bruno-binding pattern in Fig. 6A; Kim-Ha et al. 1995; Gunkel et al. 1998; Chang et al. 1999).
Figure 6.
PTB binds to multiple sites along the entire oskar 3′UTR. (A) Affinity pull-down assays comparing the binding of PTB to the whole oskar 3′UTR and to individual 3′UTR subdomains (see Supplemental Fig. S5). The conditions used were similar to those described in Figure 5A. (B) PTB (green) and actin (red) staining showing the accumulation of PTB in the cytoplasm of stage 6 w oocytes, or oocytes either lacking endogenous oskar mRNA (oskA87/Df) or expressing full-length oskar 3′UTR (see the Materials and Methods for complete genotypes). Bar, 15 μm.
Consistent with these data, PTB associates with oskar mRNA in vivo, most obviously in the context of the entire 3′UTR. Indeed, the accumulation of endogenous PTB in the cytoplasm of early oocytes is restored to wild-type levels upon sole expression of the oskar 3′UTR in oskar RNA-null ovaries (Fig. 6B). However, expression of subfragments of the 3′UTR at similar levels resulted in only mild accumulation of PTB in the oocyte (data not shown). Together, these results indicate that, both in vitro and in vivo, PTB binds to multiple sites distributed throughout the oskar 3′UTR.
PTB mediates in vivo oligomerization of oskar mRNA molecules
Previous studies showed that in wild-type oocytes, hybrid LacZ-oskar 3′UTR RNAs localize to the posterior pole by hitchhiking on endogenous oskar molecules (Hachet and Ephrussi 2004). This property seems to be mediated by the oskar 3′UTR, as a LacZ transcript fused to the oskar coding sequence, but lacking most of the oskar 3′UTR region, does not localize at the posterior pole (Kim-Ha et al. 1993). We therefore hypothesized that, in vivo, oskar mRNA might assemble via its 3′UTR into RNP particles containing multiple mRNA molecules.
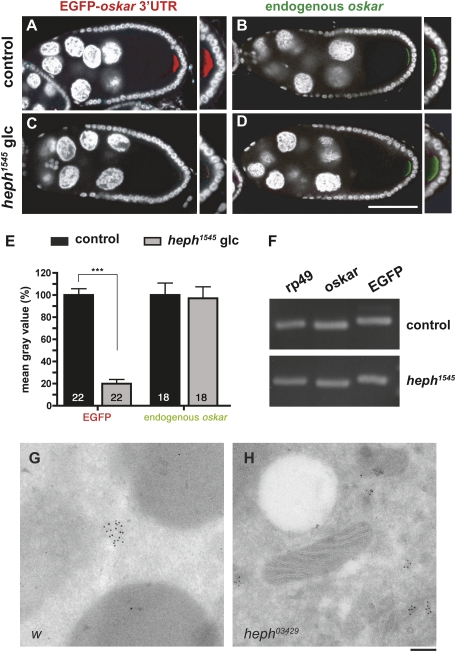
Mammalian PTB has been proposed to act as a chaperone promoting intra- and intermolecular RNA interactions (Mitchell et al. 2005; Oberstrass et al. 2005; Song et al. 2005; Auweter and Allain 2008). The binding of PTB to multiple regions of the oskar 3′UTR therefore prompted us to test if PTB might mediate the multimerization of oskar molecules in vivo. To do so, we analyzed the efficiency with which intronless oskar 3′UTR-containing RNAs hitchhike on endogenous oskar in oocytes expressing low levels of PTB. As shown in Figure 7, the amount of endogenous oskar RNA accumulating at the posterior pole of the oocyte (Fig. 7B,D), as well as the total amount of chimeric reporter EGFP-oskar 3′UTR RNA expressed in the ovary (Fig. 7F), are comparable in heph1545 and the control. However, while the EGFP-oskar 3′UTR reporter RNA accumulates at the posterior pole of control oocytes at late stage 9 (Fig. 7A), only low levels of EGFP-oskar 3′UTR RNA are detected at the posterior pole of heph1545 oocytes at this stage (Fig. 7A–E). A similar reduction in the amount of M1M2-LacZ-oskar 3′UTR mRNA (Gunkel et al. 1998) hitchhiking to the posterior pole was observed in heph1545 oocytes (Supplemental Fig. S7). These results indicate that the oskar 3′UTR is not sufficient for efficient multimerization of oskar mRNA in vivo and that the trans-acting factor PTB plays an essential role in this process.
Figure 7.
PTB promotes copackaging of oskar mRNA molecules in vivo. In situ hybridization of heterozygous (A,B) and homozygous (C,D) heph1545 late stage 9 egg chambers expressing a UAS-EGFP-osk 3′UTR construct under the control of the mat-α4-tub-Gal4 driver. (A,C) Egg chambers double-stained for EGFP transcripts (red) and DAPI (white). Images A and C were taken using identical settings. (B,D) Egg chambers double-stained for endogenous oskar transcripts (green) and DAPI (white). Endogenous oskar was detected using a probe exclusively covering the coding region. Pictures B and D were taken using identical settings. Bar, 60 μm. (E) Quantification of the in situ hybridization signal detected at the posterior pole of late stage 9 oocytes. Intensity values obtained using ImageJ are expressed as a percentage of control oocytes. (***) P < 0.001 in a Student's t-test. (F) Expression levels of rp49, endogenous oskar, and EGFP-oskar 3′UTR transcripts in both heterozygous and homozygous heph1545 backgrounds, as measured by semiquantitative RT–PCR. (G,H) Electron micrographs of w (G) and heph03429 (H) early stage 9 oocytes hybridized with an oskar antisense probe. oskar mRNA was detected using 10-nm gold particles, and the central region of the oocyte is shown. Note that in heph03429 oocytes, the slightly electron-dense oskar RNP complexes are smaller and more numerous. Bar, 150 nm.
To assess the role of PTB in assembly of endogenous oskar RNP particles, we analyzed their size in early stage 9 ptb oocytes using in situ hybridization coupled to immuno-electron microscopy (IEM). As illustrated in Figure 7, G and H, this ultrastructural analysis revealed that oskar particles are significantly smaller in heph03429 oocytes compared with wild type (52 ± 1 nm and 149 ± 6 nm, respectively), indicative of a defective copackaging of oskar mRNA molecules. Thus, both the presence of oskar 3′UTR cis-regulatory sequences and the activity of the trans-acting factor PTB are required in vivo for the formation of high-order RNP complexes.
Discussion
PTB promotes formation of high-order oskar RNP complexes
The finding that exogenous RNAs fused to the oskar 3′UTR hitchhike on endogenous oskar molecules for their localization at the posterior pole of the oocyte revealed the capacity of oskar to oligomerize in vivo and assemble into high-order RNP particles containing multiple mRNA molecules (Hachet and Ephrussi 2004). Importantly, the oskar 3′UTR is not only sufficient, but also required for in vivo oligomerization, as exogenous RNAs harboring deletions in this region fail to hitchhike on endogenous oskar (Kim-Ha et al. 1993).
We identified PTB as a trans-acting factor required for formation of high-molecular-weight complexes in vitro (Fig. 5C), and for efficient copackaging of both 3′UTR-containing reporters and endogenous oskar mRNAs in vivo (Fig. 7). This property correlates with the strong binding of PTB to multiple sites dispersed throughout the 3′UTR. Interestingly, a chaperone activity has been proposed for the vertebrate PTB, based on its capacity to bridge two separate regions of the FMDV IRES (Song et al. 2005), on the conformational changes in RNA induced upon its binding (Mitchell et al. 2003; Pickering et al. 2004), as well as on its role in remodeling of the Vg1 RNP complex (Lewis et al. 2008). This function is further supported by a structural analysis revealing that RRM3 and RRM4 of human PTB adopt a fixed and atypical orientation in which the RNA-binding surfaces of these domains are positioned away from each other (Oberstrass et al. 2005). As a consequence, RRM3 and RRM4 have the capacity to bring distantly located tracts into close proximity and thus, induce looping of the bound RNA. Given that the amino acid composition of these two RNA-binding domains and their linker region is highly conserved (F. Besse and S. López de Quinto, unpubl.), Drosophila PTB likely folds and functions similarly to its mammalian counterpart. In the context of oskar mRNA, PTB binding may therefore induce specific RNA folding required to establish the RNA–RNA or RNA–protein interactions essential for multimerization of oskar mRNA. Alternatively, PTB may itself bridge different oskar RNA molecules and nucleate the assembly of multimolecular complexes.
PTB represses oskar translation
Repression of oskar mRNA translation is a complex process involving both cap-dependent and cap-independent mechanisms (Nakamura et al. 2004; Chekulaeva et al. 2006), as well as the presence of 5′ and 3′ regulatory regions (Gunkel et al. 1998). The phenotype of oocytes with reduced PTB levels indicates that Drosophila PTB, while dispensable for the transport of oskar mRNA, is required for translational repression of the localizing mRNA. Mammalian PTB is already known to regulate translation, mainly by promoting cap-independent translation initiation. PTB binds the IRES located on the 5′UTR of cellular and viral RNAs, thus enhancing the recruitment of trans-acting factors and ribosomes (Stoneley and Willis 2004; Jang 2006; Semler and Waterman 2008). Recent reports indicate that PTB can also promote translation of specific mRNAs when bound to their 3′UTRs (Reyes and Izquierdo 2007; Galban et al. 2008). However, PTB does not exclusively act as a translational activator, as its interaction with the IRES present in unr and bip mRNAs has been shown to down-regulate their activity (Kim et al. 2000; Cornelis et al. 2005). Our results suggest that Drosophila PTB also acts as a translational repressor when bound to the 3′UTR of a target mRNA.
The mechanism by which PTB enhances or represses translation of target mRNAs remains elusive. The most accepted hypothesis, which may also explain the multiple roles of PTB in RNA regulation, is its capacity to act as a chaperone molecule, promoting the folding of RNAs into specific conformations, thereby modulating the binding of other regulatory proteins (Mitchell et al. 2003; Pickering et al. 2004; Song et al. 2005; Lewis et al. 2008). Thus, depending on the specific structure adopted by the RNA, the cellular context, and the binding of other regulatory trans-acting factors, PTB may either promote or inhibit recruitment of the translation machinery. In the case of oskar mRNA, however, our results show that PTB is not required for the binding of the two best-characterized oskar translational repressors, namely, Bruno and Hrp48 (Kim-Ha et al. 1995; Gunkel et al. 1998; Yano et al. 2004). Although we cannot exclude that PTB influences the activity of these proteins, PTB function seems beyond the simple recruitment of oskar main translation repressors.
Significantly, our results establish an in vivo link between oskar translation repression and multimerization of oskar 3′UTR-containing RNAs. An attractive possibility is that PTB promotes the formation of densely packed oskar RNP particles, thereby rendering the mRNA inaccessible to the translation machinery. A similar cap-independent function has been proposed for the translational repressor Bruno, based on its ability to promote in vitro oligomerization of a 3′UTR fragment containing duplicated Bruno response elements (BREs), together with its capacity to nucleate the assembly of heavy silencing particles (Chekulaeva et al. 2006). However, we observed that BREs are neither strictly required nor sufficient for the hitchhiking of oskar 3′UTR-containing RNAs on endogenous oskar in vivo (M. Chekulaeva and F. Besse, unpubl.). Therefore, other trans-acting factors and cis-acting sequences likely contribute to the formation of high-order oskar RNPs in the oocyte. In this context, the chaperone activity of PTB may be essential to promote multimerization of oskar molecules, which would ultimately contribute to their complete translational repression. In contrast, the oskar phenotype of ptb mutant oocytes suggests that the assembly of high-order RNP complexes does not play a significant role in oskar mRNA posterior transport.
Association of PTB with oskar in the cytoplasm is sufficient to repress translation
The assembly of RNP complexes competent for mRNA localization and precise translational control has been suggestedto occur in a stepwise manner, some factors associating with the mRNA in the nucleus, and being essential for the subsequent recruitment of other components in the cell cytoplasm (Hachet and Ephrussi 2001, 2004; Huynh et al. 2004; Kress et al. 2004; St Johnston 2005; Giorgi and Moore 2007; Lewis and Mowry 2007; Snee et al. 2008). For example, nucleolar association of the RNA-binding protein She2p with its mRNA target ash1 was recently proposed to be an essential step in the assembly of translationally silenced localizing ash1 RNP complexes in yeast (Du et al. 2008). Some oskar translation repressors have been shown to localize both in the nucleus and in the cytoplasm of germ cells (Huynh et al. 2004; Yano et al. 2004; Snee et al. 2008). However, whether these regulators are recruited to the oskar complex in the nucleus and whether nuclear association of these factors is required for subsequent translation silencing have not been tested so far.
PTB belongs to the hnRNP family of nucleo–cytoplasmic shuttling RNA-binding proteins, which regulate different aspects of RNA metabolism both in the nucleus and in the cytoplasm of eukaryotic cells (Dreyfuss et al. 2002; Glisovic et al. 2008). Consistent with this, we observed that Drosophila PTB not only colocalizes with oskar in the oocyte cytoplasm, but also strongly accumulates in the nuclei of germ cells. Given that the nuclear association of PTB with Vg1 mRNA has been proposed to control the subsequent localization of these transcripts in the cytoplasm of Xenopus oocytes (Kress et al. 2004), we tested if the association of Drosophila PTB with oskar mRNA in the nuclei of germ cells is required for its translation repression activity. Notably, we found that a cytoplasmic version of PTB localizes to the posterior pole of wild-type and ptb mutant oocytes and that this localization is oskar-dependent (Supplemental Fig. S2; data not shown), strongly suggesting that it is still able to associate with oskar mRNA. More importantly, the mutant GFP-PTB-ΔNLS is competent in oskar translation repression (Fig. 4E). Although it is possible that endogenous PTB is loaded onto oskar RNP complexes in the nucleus of germ cells, our data suggest that nuclear recruitment of PTB is not a prerequisite for the formation of translationally silenced complexes. Our analysis supports a model in which the complex behavior of RNP particles is controlled by the independent association of specific protein modules in different cell compartments. It also provides further evidence for the reorganization of RNP complexes upon translocation into the cytoplasm.
Materials and methods
Identification of GFP-PTB protein-trap line
The GFP protein-trap screen and inverse PCR mapping were described previously (Besse et al. 2007). In the PTB protein-trap line, the piggyBac transposon is inserted at position 27,699,986 in the genomic scaffold AE014297. Females homozygous for the protein-trap insertion are homozygous viable and do not show oskar-related phenotypes.
Fly stocks
w1118 flies (w) served as wild-type controls. The P(PZ}heph[03429] (Bloomington Stock Center #11589) and PBac(WH}heph[f01545] (Exelixis Collection, Harvard Medical School) were recombined onto the FRT82B chromosome. The FRT82B-recombined EMS mutant alleles hephe1 and hephe2 were characterized by Dansereau et al. (2002). Germline clone analysis was carried out using the dominant female sterile technique (Chou et al. 1993). Other mutant stocks used were osk A87 and Df(3R)pXT03 (Jenny and Hachet et al. 2006), grk2B6 (Schüpbach 1987), grk2E12 (Neuman-Silberberg and Schüpbach 1993), and stau D3 (Schüpbach and Wieschaus 1986). The MT polarity marker kin-β-gal was obtained from the stock y w KZ32; Ly/TM3Sb (Clark et al. 1994). Rescue of the oskar RNA-null phenotype using an UAS-oskar 3′UTR transgene was performed as described (Jenny and Hachet et al. 2006). For the hitchhiking assay described in Figure 7, a mat-α4-tub-Gal4VP16 insertion (Bloomington stock center #7062) was recombined with a UASp-EGFP-oskar 3′UTR transgene (H. Jambor and A. Ephrussi, unpubl.).
Transgenes
For rescue analyses, a full-length clone (RE56755) was N-terminal-tagged with EGFP and expressed using the germline-specific tubulin67c promoter. Briefly, the PTB coding sequence was PCR-amplified using the PTB-sense and PTB-antisense primers (Supplemental Table 1) and ligated into pCR II-TOPO (Invitrogen). To generate the PTBΔNLS protein, lysines at positions 56, 57, and 59 were mutated into glutamic acid using a nested-PCR site-directed mutagenesis approach with the mutagenic primer Mut-NLS-II together with PTB-sense and Internal-as as external primers (Supplemental Table 1). The SspI–NruI fragment covering the mutated positions was exchanged in the wild-type pCR II-TOPO-PTB construct, and both wild-type and ΔNLS-mutant pCR II-TOPO-PTB clones were fully sequenced. To generate EGFP fusions of PTB and PTBΔNLS, the 1.45-kb BamHI–XhoI fragment was introduced into a GFP-containing BamHI–XhoI backbone of GFP:par-1(N1S) plasmid (Shulman et al. 2000).
Generation of Drosophila anti-PTB antisera and Western blot analysis
The BDGP clone LD03185 was PCR-amplified using PTB-GST-sense and PTB-GST-as primers (Supplemental Table 1). The PCR fragment was subsequently ligated into pCR II-TOPO (Invitrogen) and fully sequenced, and the ∼1.4 BamHI–XhoI fragment was inserted into the pGEX5-5X-2 plasmid (GenBank accession no. U13857) resulting in the N-terminal fusion of GST to PTB coding sequence. The recombinant protein was expressed in Escherichia coli, purified using standard conditions, and injected into rabbits and rats.
Immunofluorescence
Ovaries from females mated for 1–2 d at 25°C on fresh food were dissected in PBS and processed as described (Vanzo and Ephrussi 2002). Samples were incubated overnight at 4°C with mouse anti-βGal (Promega; 1:2000), rabbit anti-Staufen (St Johnston et al. 1991), rabbit anti-Oskar (Vanzo and Ephrussi 2002), rat anti-PTB (1:1000), and rabbit anti-Bruno (1:5000) (R. Matthiesen and A. Ephrussi, unpubl.). Alexa-conjugated secondary antibodies (1:750; Invitrogen) were used. For detection of ectopic Oskar, we used the protocol for in situ hybridization coupled to immunofluorescence described already (Vanzo and Ephrussi 2002) in combination with preadsorbed anti-Oskar and Alexa488-conjugated antibodies. Ovaries were stained with phalloidin to label F-actin (Invitrogen) and/or with 3 μg/mL DAPI to label nuclei. Images were captured by a confocal microscope (Leica TCS SP2 AOBS) using a 40× PL APO oil-immersion lens (N.A. 1.25) and processed with Adobe Photoshop.
RNA in situ hybridization
RNA in situ hybridization was performed as described previously (Vanzo and Ephrussi 2002). Probes corresponding to either oskar or EGFP coding sequences were detected using sheep HRP-conjugated anti-DIG antibody (1:200; Roche) followed by Cy3-tyramide signal amplification reaction (Perkin Elmer).
Immunoprecipitation and RT–PCR analysis
The protocol was adapted from Munro et al. (2006) with the following modifications: protein extracts were precleared with protein A-agarose beads (Roche) and incubated without prior cross-linking with 1.5 μg of mouse anti-GFP antibodies (Molecular Probes) coupled to protein A-agarose beads (Roche) for 3 h at 4°C. RNAs were released in 100 μL of extraction buffer by incubation for 20 min at 65°C. Total RNA was extracted using Trizol (Invitrogen), DNase-treated, and used as template for RT. All RNA recovered from the bound fraction and 2.5% of that in the unbound fraction was used for cDNA synthesis in combination with Superscript III (Invitrogen) and Oligo(dT). One percent of the RT product was then PCR-amplified. Twenty-two cycles were carried out at an annealing temperature of 55°C using specific primers (Supplemental Table 1).
Affinity pull-down assays
Ovaries of w1118 females were dissected in cold PBS and extracted using a pestle in 20 μL of hypotonic buffer [10 mM HEPES at pH 7.4, 10 mM KOAc, 1.5 mM Mg(OAc)2, 2.5 mM DTT]. Typically, 20 pairs of ovaries were used per condition. The protein extract was cleared by centrifugation at 12,000 rpm for 5 min, and the supernatant was incubated with magnetic streptavidin beads (Roche) previously coupled to equimolecular amounts of in vitro transcribed UTP-biotinylated-RNA probes (ranging from 0.5 to 2 μg) in binding buffer (10 mM HEPES-KOH at pH 7.9, 3 mM MgCl2, 40 mM KCl, 5 mM EDTA, 5% glycerol, 2 mM DTT, 0.5% IGEPAL, 3 μg/μL Heparin, and 0.5 μg/μL tRNA) during 1 h at 4°C. After two washes in binding buffer followed by two more washes in binding buffer containing 150 mM KCl, the bound proteins were eluted in SDS-PAGE loading buffer and subjected to electrophoresis and Western blotting using rabbit anti-PTB (1:2000), rat anti-Bruno (1:4000) (R. Matthiesen and A. Ephrussi) and rabbit anti-Khc (1:10.000; Cytoskeleton).
Subcloning of different oskar mRNA regions
All oskar regions were cloned into pBS-II-KS+ (Stratagene) downstream from the T7 promoter. Nucleotides flanking each fragment described below have been numbered based on their position in the oskar cDNA sequence (osk-RA; Flybase), and all primer sequences are specified in Supplemental Table 1. Using a genomic oskar clone as template (Ephrussi et al. 1991), the M1M2 (nucleotides 1–434) and 3′UTR (nucleotides 1834–2890) regions were PCR-amplified with primers M1M2-sense, and M1M2-as, or 3UTR-s and 3UTR-as, respectively, and subsequently inserted into the SacI–HindIII pBS fragment. A similar strategy was used for subcloning the individual 3′UTR domains with the following pair of primers: Domain1 (nucleotides 1850–2068), Dom1-sense and Dom1-as; Domain2 (nucleotides 2065–2211), Dom2-Long-s and Dom2-as; Domain3 (nucleotides 2210–2334), Dom3-Long-s and Dom3-as; Domain4 (nucleotides 2322–2619), Dom4-sense and Dom4-as and Domain5 (nucleotides 2609–2890), Dom5-sense and 3UTR-as. The region corresponding to Short Oskar coding sequence (nucleotides 427–1838) was PCR-amplified using ShortOsk-Not and ShortOsk-as as primers and an oskar cDNA clone as template (Ephrussi et al. 1991). The PCR fragment was ligated into pCR II-TOPO (Invitrogen), and the EcoRI fragment was inserted into EcoRI-linearized pBS-II-SK+ (Stratagene).
Recombinant PTB protein
Full-length PTB was N-terminal-tagged by MBP via PCR amplification of the DGC clone RE56755 with GW-PTB-sense and GW-PTB-as primers (Supplemental Table 1). The PCR product was ligated into pENTRY/D-TOPO vector (Invitrogen), fully sequenced, and subsequently recombined into the Gateway destination vector pETG-40A (A. Geerlof). The protein was expressed in E. coli (BL21 DE3) and purified under standard conditions. The MS2-binding protein—MBP-expressing plasmid (Zhou et al. 2002) was a gift from Reinhard Lührmann.
EMSA
RNA transcripts were fluorescently labeled with ATTO-680 Aminoallyl-UTP (Jena Bioscience) via in vitro transcription (T7 megascript kit; Ambion). Full-length RNA molecules were gel-purified and incubated with different amounts of recombinant MBP-PTB in the presence of 20 mM Tris-HCl (pH 8.0), 60 mM NaCl, 3 mM MgCl2, 1 mM DTT, 8% glycerol, 5 μM tRNA during 15 min at 4°C. Complexes were resolved in native 0.8% agarose gels containing 0.5× TBE buffer and 5% (v/v) glycerol, run at 100 V constant at 4°C. Gels were visualized and quantitated using the Odyssey Infrared Imaging System (LI-COR).
IEM
Ovaries were fixed in 4% paraformaldehyde and processed as described (Delanoue and Herpers et al. 2007), with minor modifications. Digoxygenin-labeled oskar antisense probes were detected using sheep anti-Dig antibody (1:650; Roche), followed by rabbit anti-sheep secondary antibody (1:500; Dako) and protein A-coupled gold particles (10 nm). For quantification, the largest diameter of electron-dense oskar-containing particles present in the center of the oocyte was measured using Image J. At least 200 particles from three different oocytes were analyzed for each genotype.
Acknowledgments
We thank Helena Jambor, William Brook, Daniel St Johnston, Reinhard Lührmann, and Paul McDonald for flies and reagents. We also thank the EMBL Laboratory Animal Resources for antibody production and the EMBL Electron Microscopy Core Facility for technical support. We thank Eva Löser and Anna Cyrklaff for excellent technical assistance. We are grateful to members of the Ephrussi group for discussions and to Vladimir Rybin for advice. F.B. was supported by fellowships from the Federation of European Biochemical Societies (FEBS) and the Human Frontier Science Program Organization. S.L.Q. was supported by a Long Term EMBO Fellowship, a Marie Curie Intra-European Fellowship, and a RCUK Fellowship in Translational Medicine. V.M. was supported by a fellowship from the Fondation pour la Recherche Médicale and A.T. was supported by a FEBS fellowship.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.505709.
Supplemental material is available at http://www.genesdev.org.
References
- Auweter S.D., Allain F.H. Structure–function relationships of the polypyrimidine tract binding protein. Cell. Mol. Life Sci. 2008;65:516–527. doi: 10.1007/s00018-007-7378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F., Mertel S., Kittel R.J., Wichmann C., Rasse T.M., Sigrist S.J., Ephrussi A. The Ig cell adhesion molecule Basigin controls compartmentalization and vesicle release at Drosophila melanogaster synapses. J. Cell Biol. 2007;177:843–855. doi: 10.1083/jcb.200701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonin C.P., Mann R.S. A piggyBac transposon gene trap for the analysis of gene expression and function in Drosophila . Genetics. 2004;167:1801–1811. doi: 10.1534/genetics.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.S., Tan L., Schedl P. The Drosophila CPEB homolog, Orb, is required for Oskar protein expression in oocytes. Dev. Biol. 1999;215:91–106. doi: 10.1006/dbio.1999.9444. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M., Hentze M.W., Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chou T.-B., Noll E., Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Clark I., Giniger E., Ruohola-Baker H., Jan L.Y., Jan Y.N. Transient posterior localization of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol. 1994;4:289–300. doi: 10.1016/s0960-9822(00)00068-3. [DOI] [PubMed] [Google Scholar]
- Cornelis S., Tinton S.A., Schepens B., Bruynooghe Y., Beyaert R. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 2005;33:3095–3108. doi: 10.1093/nar/gki611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote C.A., Gautreau D., Denegre J.M., Kress T.L., Terry N.A., Mowry K.L. A Xenopus protein related to hnRNPI has a role in cytoplasmic RNA localization. Mol. Cell. 1999;4:431–437. doi: 10.1016/s1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- Dansereau D.A., Lunke M.D., Finkielsztein A., Russell M.A., Brook W.J. Hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster . Development. 2002;129:5553–5566. doi: 10.1242/dev.00153. [DOI] [PubMed] [Google Scholar]
- Davis M.B., Sun W., Standiford D.M. Lineage-specific expression of polypyrimidine tract binding protein (PTB) in Drosophila embryos. Mech. Dev. 2002;111:143–147. doi: 10.1016/s0925-4773(01)00595-0. [DOI] [PubMed] [Google Scholar]
- Delanoue R., Herpers B., Soetaert J., Davis I., Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev. Cell. 2007;13:523–538. doi: 10.1016/j.devcel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V.N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Du T.G., Schmid M., Jansen R.P. Why cells move messages: The biological functions of mRNA localization. Semin. Cell Dev. Biol. 2007;18:171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Du T.G., Jellbauer S., Muller M., Schmid M., Niessing D., Jansen R.P. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson L.K., Lehmann R. oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Galban S., Kuwano Y., Pullmann R., Jr, Martindale J.L., Kim H.H., Lal A., Abdelmohsen K., Yang X., Dang Y., Liu J.O., et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1α. Mol. Cell. Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C., Moore M.J. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Reyes A., Elliott H., St Johnston D. Polarization of both major body axes in Drosophila by gurken–torpedo signalling. Nature. 1995;375:654–658. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- Gunkel N., Yano T., Markussen F.-H., Olsen L.C., Ephrussi A. Localization-dependent translation requires a functional interaction between the 5′ and 3′ ends of oskar mRNA. Genes & Dev. 1998;12:1652–1664. doi: 10.1101/gad.12.11.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- Hachet O., Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Huynh J.R., Munro T.P., Smith-Litiere K., Lepesant J.A., St Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev. Cell. 2004;6:625–635. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- Jang S.K. Internal initiation: IRES elements of picornaviruses and hepatitis c virus. Virus Res. 2006;119:2–15. doi: 10.1016/j.virusres.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Jenny A., Hachet O., Zavorszky P., Cyrklaff A., Weston M.D., Johnston D.S., Erdelyi M., Ephrussi A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- Kiebler M.A., Bassell G.J. Neuronal RNA granules: Movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Hahm B., Jang S.K. Polypyrimidine tract-binding protein inhibits translation of bip mRNA. J. Mol. Biol. 2000;304:119–133. doi: 10.1006/jmbi.2000.4179. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Smith J.L., Macdonald P.M. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell. 1991;66:23–35. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Webster P.J., Smith J.L., Macdonald P.M. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- Kim-Ha J., Kerr K., Macdonald P.M. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Kress T.L., Yoon Y.J., Mowry K.L. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J. Cell Biol. 2004;165:203–211. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Katayama Y., Schmid M., Burkacky O., Brauchle C., Lamb D.C., Jansen R.P. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic. 2008;9:1256–1267. doi: 10.1111/j.1600-0854.2008.00763.x. [DOI] [PubMed] [Google Scholar]
- Lecuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., Krause H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lewis R.A., Mowry K.L. Ribonucleoprotein remodeling during RNA localization. Differentiation. 2007;75:507–518. doi: 10.1111/j.1432-0436.2007.00159.x. [DOI] [PubMed] [Google Scholar]
- Lewis R.A., Gagnon J.A., Mowry K.L. PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol. Cell. Biol. 2008;28:678–686. doi: 10.1128/MCB.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Liu G., Sun Y., Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim. Biophys. Acta. 2007;1773:912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Markussen F.-H., Michon A.-M., Breitwieser W., Ephrussi A. Translational control of oskar generates Short OSK, the isoform that induces pole plasm assembly. Development. 1995;121:3723–3732. doi: 10.1242/dev.121.11.3723. [DOI] [PubMed] [Google Scholar]
- Mitchell S.A., Spriggs K.A., Coldwell M.J., Jackson R.J., Willis A.E. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 2003;11:757–771. doi: 10.1016/s1097-2765(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Mitchell S.A., Spriggs K.A., Bushell M., Evans J.R., Stoneley M., Le Quesne J.P., Spriggs R.V., Willis A.E. Identification of a motif that mediates polypyrimidine tract-binding protein-dependent internal ribosome entry. Genes & Dev. 2005;19:1556–1571. doi: 10.1101/gad.339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila . Proc. Natl. Acad. Sci. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro T.P., Kwon S., Schnapp B.J., St Johnston D. A repeated IMP-binding motif controls oskar mRNA translation and anchoring independently of Drosophila melanogaster IMP. J. Cell Biol. 2006;172:577–588. doi: 10.1083/jcb.200510044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A., Sato K., Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F.S., Schüpbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFα-like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- Oberstrass F.C., Auweter S.D., Erat M., Hargous Y., Henning A., Wenter P., Reymond L., Amir-Ahmady B., Pitsch S., Black D.L., et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Palacios I.M., St Johnston D. Kinesin light chain-independent function of the Kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 2002;129:5473–5485. doi: 10.1242/dev.00119. [DOI] [PubMed] [Google Scholar]
- Perez I., Lin C.H., McAfee J.G., Patton J.G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Pickering B.M., Mitchell S.A., Spriggs K.A., Stoneley M., Willis A.E. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol. Cell. Biol. 2004;24:5595–5605. doi: 10.1128/MCB.24.12.5595-5605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes R., Izquierdo J.M. The RNA-binding protein PTB exerts translational control on 3′-untranslated region of the mRNA for the ATP synthase β-subunit. Biochem. Biophys. Res. Commun. 2007;357:1107–1112. doi: 10.1016/j.bbrc.2007.04.088. [DOI] [PubMed] [Google Scholar]
- Rongo C., Gavis E.R., Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development. 1995;121:2737–2746. doi: 10.1242/dev.121.9.2737. [DOI] [PubMed] [Google Scholar]
- Schüpbach T. Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster . Cell. 1987;49:699–707. doi: 10.1016/0092-8674(87)90546-0. [DOI] [PubMed] [Google Scholar]
- Schüpbach T., Wieschaus E. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila . Dev. Biol. 1986;113:443–448. doi: 10.1016/0012-1606(86)90179-x. [DOI] [PubMed] [Google Scholar]
- Semler B.L., Waterman M.L. IRES-mediated pathways to polysomes: Nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Shulman J., Benton R., St Johnston D. The Drosophila homolog of C.elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Singh R., Valcarcel J., Green M.R. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Snee M., Benz D., Jen J., Macdonald P.M. Two distinct domains of Bruno bind specifically to the oskar mRNA. RNA Biol. 2008;5:1–9. [PubMed] [Google Scholar]
- Song Y., Tzima E., Ochs K., Bassili G., Trusheim H., Linder M., Preissner K.T., Niepmann M. Evidence for an RNA chaperone function of polypyrimidine tract-binding protein in picornavirus translation. RNA. 2005;11:1809–1824. doi: 10.1261/rna.7430405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R., Rideau A., Matlin A., Gooding C., Robinson F., McGlincy N., Grellscheid S.N., Southby J., Wollerton M., Smith C.W. Regulation of alternative splicing by PTB and associated factors. Biochem. Soc. Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. Developmental genetics of oogenesis. In: Bate M., Martinez-Arias A., editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. pp. 1–70. [Google Scholar]
- St Johnston D. Moving messages: The intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 1993;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Beuchle D., Nüsslein-Vorhard C. staufen, a gene required to localize maternal RNAs in Drosophila eggs. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- Stoneley M., Willis A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Valcarcel J., Gebauer F. Post-transcriptional regulation: The dawn of PTB. Curr. Biol. 1997;7:R705–R708. doi: 10.1016/S0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- Vanzo N.F., Ephrussi A. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development. 2002;129:3705–3714. doi: 10.1242/dev.129.15.3705. [DOI] [PubMed] [Google Scholar]
- Yano T., López de Quinto S., Matsui Y., Shevchenko A., Ephrussi A. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev. Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]
- Yoshida S., Muller H.A., Wodarz A., Ephrussi A. PKA-R1 spatially restricts Oskar expression for Drosophila embryonic patterning. Development. 2004;131:1401–1410. doi: 10.1242/dev.01034. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Sim J., Griffith J., Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc. Natl. Acad. Sci. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]