Abstract
The MRN complex (Mre11/RAD50/NBS1) and ATM (ataxia telangiectasia, mutated) are critical for the cellular response to DNA damage. ATM disruption causes ataxia telangiectasia (A-T), while MRN dysfunction can lead to A-T-like disease (ATLD) or Nijmegen breakage syndrome (NBS). Neuropathology is a hallmark of these diseases, whereby neurodegeneration occurs in A-T and ATLD while microcephaly characterizes NBS. To understand the contrasting neuropathology resulting from Mre11 or Nbs1 hypomorphic mutations, we analyzed neural tissue from Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB mice after genotoxic stress. We found a pronounced resistance to DNA damage-induced apoptosis after ionizing radiation or DNA ligase IV (Lig4) loss in the Mre11ATLD1/ATLD1 nervous system that was associated with defective Atm activation and phosphorylation of its substrates Chk2 and p53. Conversely, DNA damage-induced Atm phosphorylation was defective in Nbs1ΔB/ΔB neural tissue, although apoptosis occurred normally. We also conditionally disrupted Lig4 throughout the nervous system using Nestin-cre (Lig4Nes-Cre), and while viable, these mice showed pronounced microcephaly and a prominent age-related accumulation of DNA damage throughout the brain. Either Atm−/− or Mre11ATLD1/ATLD1 genetic backgrounds, but not Nbs1ΔB/ΔB, rescued Lig4Nes-Cre microcephaly. Thus, DNA damage signaling in the nervous system is different between ATLD and NBS and likely explains their respective neuropathology.
Keywords: NBS, ATLD, A-T, DNA damage, neurodegeneration
The developing nervous system is highly susceptible to DNA damage, and many human syndromes caused by DNA repair deficiency present with pronounced neuropathlogy (Katyal and McKinnon 2007; McKinnon and Caldecott 2007; Rass et al. 2007). The DNA double-strand break (DSB) is a particularly deleterious DNA lesion in the developing nervous system and is repaired by either nonhomologous end joining (NHEJ) or homologous recombination (HR) (Lees-Miller and Meek 2003; Lieber et al. 2003; West 2003; Wyman and Kanaar 2006). Coincident with repair, DNA DSBs also activate cell cycle checkpoints to halt cellular proliferation, thereby ensuring fidelity of repair and the maintenance of genomic integrity (Zhou and Elledge 2000; Kastan and Bartek 2004; Shiloh 2006). An alternative outcome to DNA repair, and one that occurs frequently during neural development, is the activation of apoptosis to eliminate DNA-damaged cells (Lee and McKinnon 2007).
MRN (Mre11/Rad50/NBS1) and ATM (ataxia telangiectasia, mutated) are required to coordinate DNA DSB signaling responses (Kastan and Bartek 2004; Stracker et al. 2004; Shiloh 2006; Lavin 2008). The MRN complex rapidly localizes to DNA DSBs, where it recruits and activates the ATM kinase to initiate DNA damage signaling (Carson et al. 2003; Uziel et al. 2003; Kitagawa et al. 2004; Lee and Paull 2004; Difilippantonio et al. 2005). ATM initiates a signaling cascade via phosphorylation of multiple substrates that regulate cell cycle checkpoints, DNA repair, or apoptosis (Shiloh 2003; Lavin 2008). In the nervous system, p53 and Chk2 are particularly important in vivo for Atm-dependent apoptosis in immature neural cells (Lee et al. 2001; Takai et al. 2002). However, apoptosis regulated by ATM and Chk2 can also be independent and function in parallel, indicating tissue and cell-type specificity in this pathway (Hirao et al. 2000; Keramaris et al. 2003; Stracker et al. 2008). MRN regulation of ATM activation involves protein–protein interactions with the NBS1 C terminus (Falck et al. 2005), although the ATM–MRN interplay clearly requires additional levels of interactions in controlling the outcome of DNA DSB signaling (Difilippantonio et al. 2007; Stracker et al. 2007).
Neurological diseases result from mutations that render ATM, MRE11, or NBS1 dysfunctional, including ataxia telangiectasia (A-T), A-T-like disease (ATLD), or the Nijmegen breakage syndrome (NBS) respectively, underscoring the importance of the DSB response in the nervous system (Kobayashi et al. 2004; McKinnon 2004; Taylor et al. 2004). Unlike ATM, NBS1 and Mre11 are essential for mammalian development (Xiao and Weaver 1997; Zhu et al. 2001; Buis et al. 2008); accordingly, NBS and ATLD result from hypomorphic mutations (Kobayashi et al. 2004; Taylor et al. 2004; Frappart and McKinnon 2006). A-T is primarily a neurodegenerative syndrome, which presents with cerebellar ataxia associated with Purkinje cell degeneration, but also immunodeficiency, radiosensitivity, sterility, and cancer predisposition (McKinnon 2004). ATLD is also characterized by neurodegeneration and radiosensitivity (Taylor et al. 2004), while NBS features microcephaly, immunodeficiency, radiosensitivity, and cancer predisposition (Kobayashi et al. 2004). The similarities between these syndromes presumably reflect functional interactions between ATM and the MRN complex, while the different neuropathology indicates that the hypomorphic MRN mutations leading to these diseases must ultimately result from different downstream signaling in the nervous system. How this occurs is unknown.
Using mouse models for ATLD and NBS we undertook an in vivo analysis of the functional consequence of these hypomorphic mutations in the nervous system. We induced various genotoxic stresses to evaluate the relative roles of Atm, Mre11, and Nbs1 mutations during neural development to identify signaling defects that account for the distinct neuropathology characteristic of these diseases. Our data show that the disease-causing mutations in MRN lead to defective ATM-dependent DNA damage-induced signal transduction in the nervous system, whereby the relative activation of apoptosis accounts for the differential neuropathology of these syndromes.
Results
Ionizing radiation (IR)-induced apoptosis is defective in the Mre11ATLD1/ATLD1 brain
To determine the basis for the differential neuropathology associated with ATLD and NBS we used mice harboring hypomorphic alleles (Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB) reflecting those present in the human diseases (Carney et al. 1998; Varon et al. 1998; Stewart et al. 1999). The ATLD mutation (633R → STOP) results in a truncated Mre11 protein, while the Nbs1ΔB allele has a mutation that produces an 80-kd Nbs1 protein lacking the N-terminal FHA and BRCT domains (Williams et al. 2002; Theunissen et al. 2003). These mice recapitulate many aspects of the respective human syndromes, including radiosensitivity and cell cycle checkpoint defects, although they do not develop overt neuropathology (Williams et al. 2002; Theunissen et al. 2003).
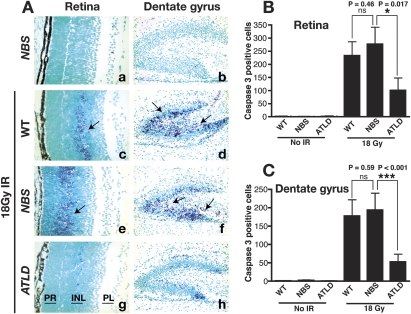
While many features of NBS and ATLD can be attributed to a defect in ATM activation (Carson et al. 2003; Uziel et al. 2003; Difilippantonio et al. 2005), the disparate neuropathology suggests differences in DNA damage signaling in the nervous system. To investigate DNA damage responses in the developing nervous system of ATLD and NBS mice, we used IR to induce genotoxic stress. Because apoptosis is a primary readout of IR-induced DNA damage in the developing nervous system (Lee and McKinnon 2007), we therefore analyzed representative regions including the postnatal retina and dentate gyrus using immunohistochemistry to assess caspase-3 activation. In the absence of radiation very little apoptosis was seen, as judged by caspase-3-positive cells (Fig. 1A–C). However after radiation, wild-type and Nbs1ΔB/ΔB mice showed an equivalent susceptibility to IR-induced apoptosis (Fig. 1A [panels c–f], B,C). In contrast, there was a marked resistance to apoptosis in the Mre11ATLD1/ATLD1 mice (Fig. 1A [panels g,h], B,C). IR-induced cell death was substantially reduced throughout immature post-mitotic regions of the Mre11ATLD1/ATLD1 developing nervous system, including the inner nuclear layer of the retina and the dentate gyrus of the hippocampus (Fig. 1A [panels g,h], B,C). The resistance of these particular cell populations is similar to that observed in the same regions in the early postnatal Atm−/− brain, as it is restricted to regions undergoing postnatal development (i.e., cerebellum, dentate gyrus, and retina) (Herzog et al. 1998; Lee et al. 2001). Studies in P5 were thus confined to these developing areas, whereas embryonic studies were primarily focused in the forebrain. IR treatment of Mre11ATLD1/ATLD1 (and Atm−/−) embryos at embryonic day 13.5 (E13.5) and E15.5 also showed resistance to apoptosis in immature, post-mitotic neurons akin to the P5 tissues, while in Nbs1ΔB/ΔB embryos IR-induced apoptosis was indistinguishable from wild-type embryos (data not shown). Additionally, IR-induced apoptosis in the proliferating ventricular zone of the embryonic nervous system occurred normally in Mre11ATLD1/ATLD1 (and Nbs1ΔB/ΔB) as it does in Atm−/− tissue (data not shown). Thus, ATLD but not NBS mutations measurably influence DNA damage-induced apoptosis in specific regions of the developing brain.
Figure 1.
Radiation-induced apoptosis in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB CNS. (A) Wild-type (WT), Mre11ATLD1/ATLD1, and Nbs1ΔB/ΔB mice at P5 were treated with 8Gy of γ-irradiation (panels c–h) and collected 6 h post-IR. Nonirradiated sections are shown in panels a and b. Immunostaining was performed against active-caspase 3. (Panels a,c,e,g) Images of the retina are at 400× magnification. (Panels b,d,f,h) Images of the dentate gyrus are at 200× magnification. Retinal photoreceptor layer (PR), inner nuclear layer (INL), and plexiform layer (PL) are indicated. Arrows point to immunopositive cells. The number of activated caspase-3-positive cells in the retina (B) or dentate gyrus (C) were quantified; error bars represent standard deviation.
Atm signaling in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB brain
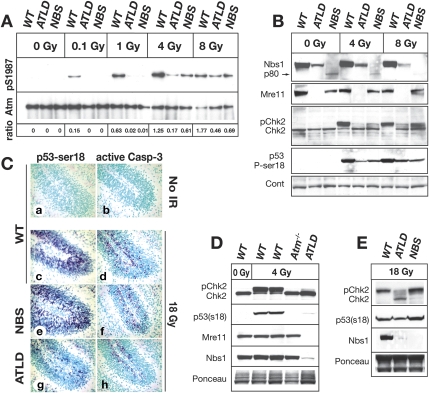
Because the MRN complex can modulate ATM kinase activity, we determined if the resistance to IR-induced apoptosis in the Mre11ATLD1/ATLD1 tissue was related to defective Atm activation. We assayed Atm ser1987 phosphorylation (equivalent to ser1981 of human ATM) as a marker for DNA damage-induced Atm activation in vivo in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB P5 cerebellum, a tissue significantly affected in both diseases. At doses of 1 Gy or below, wild-type cerebellum showed clear Atm ser1987 phosphorylation while very little Atm phosphorylation was observed in either Mre11ATLD1/ATLD1 or Nbs1ΔB/ΔB tissue (Fig. 2A). However, at higher radiation levels (>4 Gy) Atm phosphorylation was evident in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB tissue, albeit reduced compared with wild type (Fig. 2A). These data indicate that both Mre11 and Nbs1 mutations result in reduced Atm ser1987 phosphorylation, indicating that MRN modulates Atm signaling in the developing brain.
Figure 2.
DNA damage responses in the Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB nervous system at postnatal day 5. (A) Wild-type (WT), Mre11ATLD1/ATLD1, and Nbs1ΔB/ΔB mice were treated with indicated doses (Gy) of IR, and cerebella were harvested 30 min post-IR. Total Atm was immunoprecipitated from cerebellar lysates followed by immunoblotting for Atm pSer1987. Total Atm served as the loading control. Ratios represent the relative increase of phosphorylated Atm compared with total Atm protein. (B) Wild-type (WT), Mre11ATLD1/ATLD1, and Nbs1ΔB/ΔB mice were treated with IR and cerebella were harvested 3 h post-IR. Cerebellar lysates were probed using antibodies recognizing Nbs1, Mre11, and Chk2. Phosphorylated Chk2 (pChk2) was detected as an upward mobility shift. β-actin was used as a loading control. (C) Immunostaining 1 h post-IR (18 Gy) of wild-type, Mre11ATLD1/ATLD1, and Nbs1ΔB/ΔB mice to detect pSer18 of p53 (p53-ser18) and active-caspase-3 (200× magnification). (D) Wild type (WT), Atm−/−, and Mre11ATLD1/ATLD1 were treated with 4 Gy of IR to determine comparative Chk2 and p53 phosphorylation after 1 h in Atm−/− cerebellum. (E) Wild-type (WT), Mre11ATLD1/ATLD1, and Nbs1ΔB/ΔB mice were treated with 18 Gy of IR and collected 3 h post-IR. Cerebellar lysates in D and E were probed for Chk2, p53 (pSer18), and Nbs1 or Mre11. Ponceau staining is shown to indicate relative protein loading.
As Nbs1ΔB/ΔB neural tissue underwent DNA damage-induced apoptosis similar to wild type (Fig. 1), defective Atm phosphorylation was somewhat unexpected. Therefore, we confirmed the MRN complex was disrupted in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB tissue. Full-length Nbs1 was not present in the Nbs1ΔB/ΔB samples (Fig. 2B), although we did detect the 80-kd product translated from the Nbs1 mutant message (Maser et al. 2001). Similarly, the C terminus of Mre11 was not detected in Mre11ATLD1/ATLD1 tissue, and levels of Nbs1 were also substantially reduced (Fig. 2B). Consequently, we determined other readouts of Atm signaling relevant to apoptosis. Chk2 is a key DNA damage signaling effector that is activated by Atm-dependent phosphorylation (Ahn et al. 2000; Hirao et al. 2000; Matsuoka et al. 2000) and is essential for radiation-induced apoptosis in the developing nervous system (Takai et al. 2002). In both Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB tissues there was clearly less Chk2 modification (evident as reduced band mobility) compared with wild type after irradiation. However, despite this, Chk2 activation was substantially greater in the Nbs1ΔB/ΔB tissue compared with Mre11ATLD1/ATLD1, particularly after 8 Gy. We also evaluated ser18 phosphorylation of p53 (equivalent to ser15 of human p53) after radiation of P5 cerebellum using immunohistochemistry and using Western blot analysis. While both wild-type and Nbs1ΔB/ΔB tissue had robust induction of p53 ser18 phosphorylation and caspase-3 activation (Fig. 2C, panels c–f), both were markedly reduced in Mre11ATLD1/ATLD1 tissue after 18 Gy of IR (Fig. 2C, panels g,h). Furthermore, we also determined the relative level of Chk2 activation in Atm−/− P5 cerebellum, as a comparison with the MRN mutants, and found no detectable Chk2 activation or p53ser18 phosphorylation occurred after 4 Gy of IR, consistent with apoptosis being Atm-dependent in this tissue (Fig. 2D). Additionally, at higher radiation doses (18 Gy) the difference between Chk2 activation in Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB is more apparent, as is phosphorylation of p53ser18 (Fig. 2C,E). The abundant apoptosis and Chk2 modification observed in the Nbs1ΔB/ΔB mice indicate that the hypomorphic Nbs1 allele can activate Atm sufficiently to engage apoptosis in the CNS after DNA damage. We also observed defects in Atm signaling in the Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB thymus, with reduced apoptosis in ATLD tissue (Supplemental Fig. 1), consistent with recent data using Mre11ATLD1/ATLD1 thymocytes (Stracker et al. 2008). Thus, the MRN complex is required for ATM activation, and this is defective in both Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB tissue. However, the Mre11ATLD1/ATLD1 mutation results in a more substantial block in Atm activity and subsequent abrogation of apoptosis.
DNA damage signaling by Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB after Lig4 loss
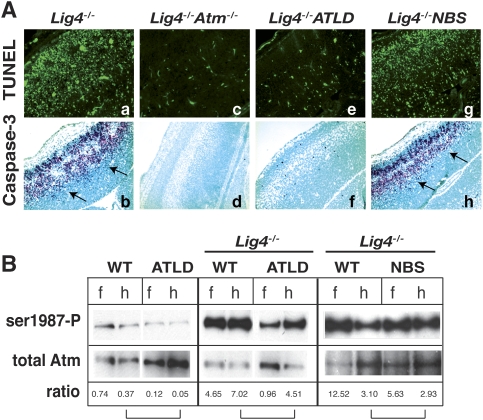
We further addressed the consequences of NBS and ATLD mutations in the context of chronic endogenous genotoxic stress, compared with acute damage from radiation, by using DNA Ligase IV (Lig4)-deficient mice (Barnes et al. 1998). In these DNA repair-deficient mice, Lig4 loss leads to mid-gestational embryonic lethality associated with neuraxis-wide Atm-dependent apoptosis (Lee et al. 2000; Sekiguchi et al. 2001). Therefore, we assessed the impact of Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB mutations by interbreeding with Lig4−/− mice to generate Lig4−/−;Mre11ATLD1/ATLD1 or Lig4−/−;Nbs1ΔB/ΔB compound mutants and appropriate controls. We found that in the context of Lig4 loss, the ATLD mutation but not the NBS1 mutation resulted in an almost complete block of apoptosis in neural tissue, similar to that found in Lig4−/−;Atm−/− tissue. As shown in the E15.5 neopallial cortex of the developing forebrain, Lig4−/− embryos showed widespread apoptosis throughout the developing nervous system (Fig. 3A, panels a,b). However, apoptosis was substantially abrogated in Lig4−/−;Atm−/− (Fig. 3A, panels c,d) or Lig4−/−;Mre11ATLD1/ATLD1 embryos (Fig. 3A, panels e,f) while levels of apoptosis similar to Lig4−/− were present in Lig4−/−;Nbs1ΔB/ΔB embryos (Fig. 3A, panels g,h). In other regions of the nervous system including the telencephalon, metencephalon, spinal cord, or the dorsal root ganglia, apoptosis was also largely absent in Lig4−/−;Mre11ATLD1/ATLD1 embryos, while the Nbs1ΔB/ΔB mutation did not affect apoptosis after Lig4 loss (Supplemental Fig. 2). Similar to Lig4−/−;Atm−/− mice, Lig4−/−;Mre11ATLD1/ATLD1 mice also survived until birth but died perinatally. In contrast, Lig4−/−;Nbs1ΔB/ΔB embryos died around mid-gestion similar to Lig4−/− embryos. Therefore, the DNA damage response resulting from Lig4 inactivation in the developing nervous system is differentially affected by Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB mutations.
Figure 3.
DNA damage signaling in ATLD and NBS after Lig4 loss. (A) TUNEL analysis and immunostaining against active caspase-3 was performed on E15.5 cryosections of Lig4−/−, Lig4−/−;Atm−/−, Lig4−/−;Mre11ATLD1/ATLD1, and Lig4−/−;Nbs1ΔB/ΔB. Arrows indicate immunopositive signal. Images of the neopallial cortex were captured at a 200× magnification. (B) Tissue lysates were prepared from E13.5 forebrain (f) and hindbrain (h) collected from the indicated genotypes. Total Atm was immunoprecipitated from lysates followed by immunoblotting for phosphorylated Atm (pser1987). Quantitative analysis depicted below the blot indicates the relative ratio between phospho ser1987 and total Atm; brackets indicate matched brain regions.
To gauge Atm activation in neural tissue from Lig4−/−, Lig4−/−;Nbs1ΔB/ΔB, and Lig4−/−;Mre11ATLD1/ATLD1 embryos, we assayed embryonic forebrain and hindbrain for phosphorylated Atm (ser1987). Atm phosphorylation was substantially enhanced in Lig4−/− tissue extract, but reduced in Lig4−/−;Mre11ATLD1/ATLD1 samples. In contrast, Lig4−/−;Nbs1ΔB/ΔB embryos exhibited levels of autophosphorylated Atm intermediate to that seen in Lig4−/− (Fig. 3B). These data further highlight the different Atm signaling that results from Mre11 and Nbs1 hypomorphic mutations after DNA damage.
Lig4Nes-Cre mice accumulate DNA damage in the brain
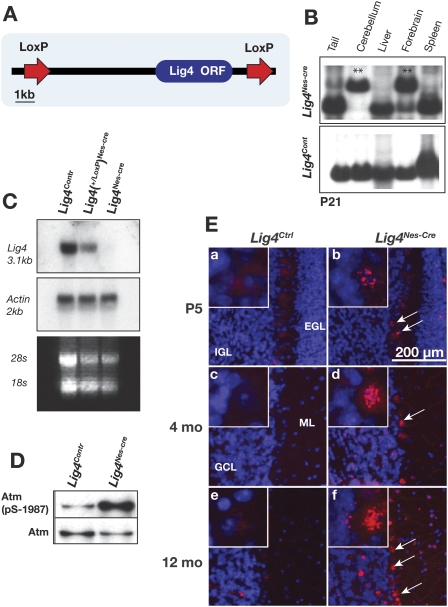
To expand analysis of DNA damage signaling resulting from Mre11ATLD1/ATLD1 or Nbs1ΔB/ΔB mutations, we generated a conditional Lig4 mutant mouse. We did this to bypass the embryonic lethality resulting from germ line deletion of Lig4, thereby providing a more refined model to investigate MRN function in the CNS. To generate a Lig4 conditional allele we floxed the Lig4 ORF to generate Lig4loxP/loxP mice and intercrossed these with mice expressing cre throughout the nervous system via a Nestin promoter to obtain Lig4loxP/loxP;Nestin-cre (Lig4Nes-Cre) mice (Fig. 4A). In all experiments, either Lig4+/+; Nestin-cre or Lig4+/loxP; Nestin-cre mice were used as controls and are collectively referred to as Lig4Ctrl. Southern blot analysis confirmed inactivation of genomic Lig4 and showed that extensive deletion had occurred in the cerebellum and forebrain by 4 wk (Fig. 4B), while Northern blot analysis confirmed the absence of the Lig4 transcript in Lig4Nes-Cre cerebellum (Fig. 4C). Consistent with Lig4 loss resulting in chronic genotoxic stress, we found increased Atm(ser1987) phosphorylation in the Lig4Nes-Cre brain (Fig. 4D).
Figure 4.
Generation and analysis of conditional Lig4-null mice. (A) The conditional Lig4 allele was generated by flanking the Lig4 ORF by LoxP sites. (B) Deletion of the Lig4 allele throughout the brain was done using Nestin-cre. Southern blot analysis of DNA from 1-mo-old Lig4Nes-Cre or Lig4Cont mice shows the deleted Lig4 allele (1.6 kb) in cerebellum and forebrain (**) but not in liver or spleen. (C) RNA was extracted from adult cerebella for Northern analysis. A full-length Lig4 cDNA probe was used to detect expression of Lig4 mRNA. (D) Cerebella were collected from P5 Lig4Nes-Cre and Lig4Ctrl animals. Atm was immunoprecipitated from tissue lysates, then immunoblotted for Atm pSer1987 and total Atm. (E) Lig4Nes-Cre and Lig4Ctrl tissues were collected at the indicated ages and immunostained for γH2AX. Representative images of cerebella from each age and genotype are shown (200× magnification). Arrows indicate that after Lig4 deletion Purkinje cells contain γH2AX; inset panels show Purkinje cells with γH2AX foci at 800× magnification.
While loss of Lig4 in the germ line leads to neuraxis-wide apoptosis and mid-gestational lethality (Barnes et al. 1998; Frank et al. 1998; Gao et al. 1998), Lig4Nes-Cre mice were viable up to a year of age. To assess the consequence of Lig4 loss in the mature brain, we surveyed neural tissue from mice between P5 and 12 mo of age. Using γH2AX staining to identify DNA DSBs, we found a widespread and progressive accumulation of γH2AX (Fig. 4E) and 53BP1 foci (data not shown) throughout the Lig4Nes-Cre brain. Additionally, Lig4Nes-Cre mice were unable to repair DNA breaks in the brain induced by IR up to 1 wk after radiation, whereas after 24 h few detectable strand breaks were present in control animals (Supplemental Fig. 3). Therefore, Lig4 is essential for the prevention of accumulated unrepaired DNA DSBs in post-mitotic cells in the nervous system.
Microcephaly after Lig4 loss requires Atm signaling
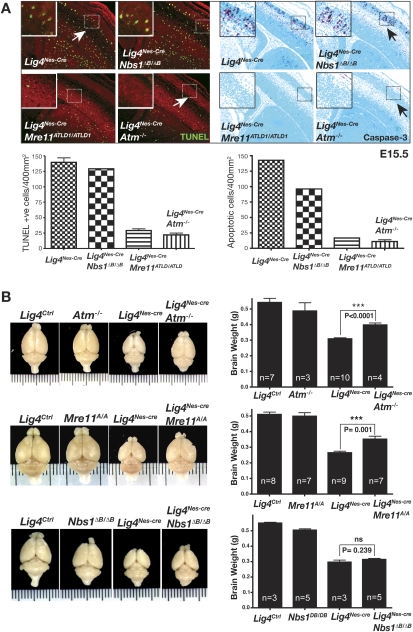
In comparison with age- and sex-matched littermates, Lig4Ctrl, Lig4Nes-Cre animals exhibit pronounced microcephaly, as evidenced by an ∼40% reduction in brain size (Fig. 5). Notably, microcephaly is a characteristic of LIG4 syndrome patients with hypomorphic mutations in LIG4 (O'Driscoll et al. 2004). To determine the cause of the microcephaly in the Lig4Nes-Cre mice we assessed apoptosis at various developmental stages. We found that from E13.5 onward there was markedly increased apoptosis in Lig4Nes-Cre neural tissue as judged by TUNEL and caspase 3 staining (Fig. 5), with a peak around E15.5 somewhat similar to germ line disruption of Lig4. Thus, microcephaly in Lig4Nes-Cre mice likely results from apoptotic cell loss during development. Therefore we generated Lig4Nes-Cre;Atm−/−, Lig4Nes-Cre;Mre11ATLD1/ATLD1, or Lig4Nes-Cre;Nbs1ΔB/ΔB mice to produce viable mice with which to monitor longer term consequences of MRN mutations. Initially, we confirmed that both MRN mutations produced effects in the Lig4 conditional mutant similar to those we observed after germ line disruption of Lig4. We found that apoptosis was abrogated in Lig4Nes-Cre;Mre11ATLD1/ATLD1 and Lig4Nes-Cre;Atm−/− neural tissue but occurred to the same level as Lig4Nes-Cre in Lig4Nes-Cre;Nbs1ΔB/ΔB mice (Fig. 5A).
Figure 5.
Impact of ATLD and NBS mutations in Lig4Nes-Cre. (A) TUNEL analysis and immunostaining for active caspase-3 were performed on E15.5 embryos from indicated genotypes. The total number of positive cells per 400 μm2 was quantified; error bars represent standard deviation. Micrographs encompass ganglionic eminence and neopallial cortex (200× magnification). (B) The effect of A-T, ATLD, and NBS mutations on Lig4Nes-Cre brain size. Brain weights were collected from sex-matched cohorts at 4 mo of age. No significant difference was observed between Lig4Ctrl groups and Atm−/− or Mre11ATLD1/ATLD1 controls; however a statistically significant decrease in Nbs1ΔB/ΔB brain weight was observed compared with controls (P = 0.013). A statistically significant recovery in Lig4Nes-Cre brain weight was observed in Lig4Nes-Cre;Atm−/− and Lig4Nes-Cre;Mre11ATLD1/ATLD1 but not in Lig4Nes-Cre;Nbs1ΔB/ΔB mice. Error bars represent standard deviation.
To assess the long-term consequences of MRN dysfunction after chronic DNA damage in the CNS, we monitored adult Lig4Nes-Cre;Atm−/−, Lig4Nes-Cre;Mre11ATLD1/ATLD1, or Lig4Nes-Cre;Nbs1ΔB/ΔB mice. Inactivation of either Atm or Mre11 significantly rescued microcephaly, whereas the Nbs1ΔB/ΔB mutation had no obvious affect toward microcephaly in Lig4Nes-Cre mice (Fig. 5B). Excluding microcephaly, no other significant morphologic aberrations were found in the Lig4Nes-Cre brain. However, with age, adult Lig4Nes-Cre animals developed a severe hindlimb ataxia of unknown etiology from 7 to 9 mo onward. This phenotype was observed with 100% penetrance in Lig4Nes-cre animals allowed to reach that age span (n = 23) (data not shown) and was not seen in an equivalent number of age-matched Lig4+/loxP;Nestin-cre mice. Neither Atm−/−, Mre11ATLD1/ATLD1, nor Nbs1ΔB/ΔB background affected the latency or severity of this phenotype, suggesting that this latter pathway is independent of MRN and ATM, highlighting the importance of this pathway during development. Thus, the microcephalic phenotype in Lig4Nes-Cre mice is attributable to increased apoptosis via ATM-dependent DNA damage signaling.
Discussion
Defective responses to DNA damage resulting from dysfunction of ATM or the MRN complex can cause the human syndromes, A-T, ATLD, and NBS. Although these diseases share similar phenotypes including radiosensitivity and chromosomal instability, A-T and ATLD exhibit neurodegeneration while NBS is characterized by microcephaly. This suggests that the respective disease-causing mutations impact DNA damage signaling differently within the brain. To test this, we analyzed the DNA damage response in the nervous systems of ATLD (Mre11ATLD1/ATLD1) and NBS (Nbs1ΔB/ΔB) mice and found that different effects upon ATM activation and downstream apoptotic signaling distinguish these MRN mutations. Therefore, we hypothesize that the different neuropathology in ATLD and NBS is a result of the relative levels of ATM activation by MRN after DNA damage, most probably in response to DNA DSBs (Fig. 6).
Figure 6.
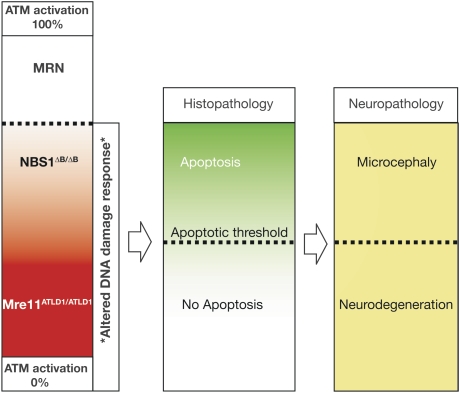
Neuropathology in ATLD and NBS. Full ATM activation and activity require the MRN complex. Both Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB mice show defective Atm phosphorylation. However, Atm activity is substantially higher in the Nbs1ΔB/ΔB nervous system compared with the Mre11ATLD1/ATLD1 nervous system and is sufficient to activate apoptosis. The different apoptotic response observed between the Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB mice suggests a threshold level of Atm activity is required to induce apoptosis during neurogenesis. In the human brain, NBS mutations elevate DNA damage, leading to increased apoptosis and microcephaly. In contrast, ATLD mutations cannot sufficiently activate ATM, thereby failing to engage apoptosis and eliminate DNA damaged cells. Similar to A-T, damaged cells may become incorporated into the nervous system and at later times will malfunction and die, resulting in progressive neurodegeneration.
While both Mre11ATLD1/ATLD1 and Nbs1ΔB/ΔB are defective in Atm phosphorylation after DNA damage, only Mre11ATLD1/ATLD1 mutations block the subsequent activation of apoptosis by Atm. In this manner, the Mre11ATLD1/ATLD1 mutation creates a scenario similar to the Atm−/− nervous system (Lee et al. 2001), whereby defective ATM signaling inhibits apoptosis of DNA-damaged neural cells. Therefore our data provide a mechanistic explanation for the similar neuropathology between A-T and ATLD. We suggested previously that the loss of ATM prevents apoptosis, leading to incorporation of cells harboring genomic damage into the mature nervous system, and over time these cells presumably succumb to this damage and die (Herzog et al. 1998; Lee et al. 2001). Thus, attenuation of DNA damage-dependent apoptosis in ATLD would mimic A-T, resulting in neurodegeneration characteristic of each disease.
Conversely, in NBS, DNA damage accumulates due to dysfunctional MRN, but NBS1 mutations do not block Atm-dependent apoptosis, and therefore we predict that increased apoptosis occurs during development, contributing to microcephaly. This is consistent with defective NBS1 leading to increased genotoxic stress, cancer, and radiosensitivity (Kobayashi et al. 2004). As opposed to the Nbs1ΔB/ΔB mouse, complete disruption of Nbs1 in the murine nervous system leads to microcephaly from DNA damage-induced apoptosis and decreased proliferation (Frappart et al. 2005). In contrast to the microcephaly observed in LIG4 syndrome and modeled in the Lig4Nes-cre mice, neuropathology associated with NBS could be due to damage accumulation in either post-mitotic or proliferating cells, as NBS1 is also important during HR (Tauchi et al. 2002; Limbo et al. 2007; Sartori et al. 2007). The Nbs1ΔB allele results from deletion of exons 4 and 5, producing a protein that lacks the N-terminal FHA and BRCT domains encoded within exons 1 through 5. In contrast to the Nbs1ΔB/ΔB mouse, other models that lack a C terminus (Nbs1ΔC/ΔC and Nbs1tr735) showed reduced apoptosis in some tissues consistent with a defect in ATM activation (Difilippantonio et al. 2007; Stracker et al. 2007). Additionally, deletion of the Nbs1 C terminus in the context of the Nbs1ΔB protein (the Nbs1ΔBC allele) results in apoptotic defects in thymocytes but does not rescue gestational lethality associated with Lig4 loss (Supplemental Fig. 4), which is linked to neuraxis-wide apoptosis, suggesting that modulation of apoptosis by Nbs1 is tissue-specific.
As NBS individuals harbor mutations in the FHA and BRCT domains, not the C terminus, the pathology associated with NBS involves an intact C terminus. Preservation of the C terminus presumably allows interaction of the resulting NBS1 protein with ATM and Mre11 and the activation of apoptosis. Although the level of Nbs1 is reduced in Mre11ATLD1/ATLD1 cells, there is still a relatively higher level than that of the NBS1 p80 protein found in Nbs1ΔB/ΔB tissue. This suggests that the apoptotic defect we observe in the Mre11ATLD1/ATLD1 nervous system directly involves the Mre11 mutant protein rather than an indirect effect upon Nbs1. Given the multiple contacts ATM makes with the MRN complex (Lee and Paull 2004), it is likely that ATLD mutations directly and selectively impact ATM–MRN interactions to preclude effective ATM activation.
Why, then, do A-T and NBS have many phenotypic similarities outside of the nervous system? These similarities encompass immunodeficiency, cancer, and radiosensitivity, and reflect the interrelated roles of MRN and ATM during DNA rearrangements in the immune system and maintenance of genomic integrity in proliferating cells. For example, ATM and NBS1 are required to ensure the fidelity of programmed rearrangements and elimination of cells with inappropriate genomic rearrangements in the immune system (Kracker et al. 2005; Reina-San-Martin et al. 2005; Bredemeyer et al. 2006; Callen et al. 2007; Jankovic et al. 2007). Additionally, both ATM and NBS1 can function in stem and progenitor populations in the hematopoeitic and germ cell compartments (Kang et al. 2002; Ito et al. 2004; Takubo et al. 2008). These requirements for ATM and NBS1 contrast the situation in the nervous system where ATM fulfills an indispensable role in DNA damage-induced apoptosis in post-mitotic neurons, but not in proliferating neural progenitors where ATR or DNA-PK may be the more important kinases (Lee et al. 2001; Orii et al. 2006).
In summary, our data demonstrate that the respective MRN mutations that lead to NBS and ATLD impact differently upon ATM signaling. These data illuminate the mechanistic underpinnings of the different neuropathology present in ATLD and NBS, suggesting they result from alterations in the modulation of ATM-dependent apoptosis after DNA damage.
Materials and methods
Animals
Mice carrying germ line mutations for Mre11, Nbs1, DNA ligase IV, and Atm have been described (Barnes et al. 1998; Lee et al. 2000; Williams et al. 2002; Theunissen et al. 2003). To generate a conditional DNA Ligase IV allele we used a murine BAC genomic clone (strain 129Ola) to isolate a 13-kb BamHI fragment encompassing the Lig4 genomic locus. We placed a LoxP sequence into a unique MluI site 4.8 kb upstream of the Lig4 ORF and a floxed NeoTK cassette downstream as a HindIII fragment to generate pLig4LoxP. Embryonic stem (ES) cells (W9.5; 129S1/SvImJ) were electroporated with linearized pLig4LoxP, selected with G418 antibiotic, and targeted cells were identified by Southern Blot analysis. ES clones were then electroporated with pMC-cre, followed by selection in FIAU (5-iodo-2′-fluoro-2′deoxy-1-arabinofuranosyluracil) to kill cells containing TK. ES cells with a correct LoxP configuration were determined by PCR and were then injected into blastocysts and the resultant chimeric mice assessed for germ line transmission of the floxed Lig4 allele. Nestin-cre mice [B6.Cg-Tg(Nes-cre)1Kln/J; JAX #003771] were used to delete Lig4 throughout the nervous system.
Animals were housed in an Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facility and were maintained in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The St. Jude Children's Research Hospital IACUC approved all procedures for animal use.
Histology and Immunohistochemistry
Mice were transcardially perfused with 4% paraformaldehyde (PFA), and collected tissues were cryoprotected in 25% PBS-buffered sucrose solution and then embedded in O.C.T. (Tissue-Tek). Antigen retrieval was used for all immunohistochemistry using the following antibodies: anti-p53 (CM5; 1:500; Vector Laboratories); anti-p53 phospho-Ser-15 (1:500; Cell Signaling); anti-active caspase 3 (1:500; BD Biosciences); anti-phospho-H2AX antibodies (1:500, rabbit; Abcam). Immunohistochemistry for activated caspase-3 or p53-ser15 was performed overnight at room temperature after quenching endogenous peroxidase with 0.6% hydrogen peroxide in methanol. Immunoreactivity was visualized with a VIP substrate kit (Vector Laboratories) according to the manufacturer's directions after tissues were treated with biotinylated secondary antibody and avidin DH-biotinylated horseradish peroxidase-H complex (Vectastain Elite kit; Vector Laboratories). Sections were counterstained with 0.1% methyl green (Vector Laboratories), dehydrated, and mounted in DPX reagent. Quantitation of TUNEL- or caspase-3-positive cells was determined by counting positive signal from images equivalent in size and magnification between genotypes.
For immunofluorescence with γH2AX, sections were subsequently immunolabeled with Cy3-conjugated goat anti-rabbit (1:500; Jackson Immunoresearch) and mounted with Vectashield anti-fade reagent containing DAPI (Vector Laboratories). Apoptotic cells were detected with ApopTag Fluorescein in situ apoptosis detection kit (Chemicon International) according to the manufacturer's instructions. Images were captured using an Axioskop 2.0 microscope (Carl Zeiss) and a SPOT camera (Diagnostic Instruments, Inc.).
Protein extraction and Western blot analysis
Tissues were resuspended in 500 μL of lysis buffer (100 mM Tris-HCl, 150 mM NaCl at pH 7.5, 0.5% SDS, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM EGTA, 0.5 mM ZnCl2, 0.02% NaN3, 10% glycerol, 0.1% β-mercaptoethanol, 0.2 mg/mL PMSF, Complete protease inhibitor cocktail [Roche]) and dissociated by passage through a 23-G needle. Protein concentrations of whole-cell extracts were quantified using Bradford reagent (Bio-Rad). Proteins (50 μg per lane) were separated through an 8% SDS–polyacrylamide gel and transferred onto nitrocellulose membrane (Bio-Rad). Blots were immunostained with antibodies recognizing NBS1 (1:500, Cell Signaling Technology), Mre11 (1:500, BD Transduction Laboratories), and CHK2 (1:800, Upstate Biotechnologies/Millipore) followed by horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:2000; GE Healthcare) and detected using ECL Plus chemiluminescence reagent (GE Healthcare). Immunostaining with goat polyclonal anti-actin (1:500; Santa Cruz Biotechnologies) antibody served as a protein loading control.
For assessment of Atm phosphorylation, Atm was immunoprecipitated from postnatal brain or embryonic neural tissues with Atm antibody D1611 (obtained from Dr. Michael Kastan) using protein A/G agarose beads. Tissue lysates for immunoprecipitation were prepared in 50 mM Tris (pH 7.5) containing 150 mM NaCl, 50 mM NaF, 1% Tween 20, 0.2% NP-40, 1 mM AEBSF (Roche), 1 mM DTT, and 1× protease inhibitor mixture (Roche). Beads were rinsed with RIPA buffer (PBS containing 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 2 mM EDTA), split 1:2, and run on two separate 3%–8% NOVEX gels and electroblotted to PDVF membranes. Phosphorylated Atm was visualized using anti-Ser-1981 Atm antibody (Rockland Immunochemicals), and MAT3 antibody (obtained from Dr. Yosef Shiloh) was used to detect total Atm. The ratio of phosphorylated Atm to total Atm was determined from densitometric scans of autoradiographs using NIH Image J version 1.14n software.
Acknowledgments
We thank the Hartwell Center and the Transgenic Core at SJCRH for their support of this work. These studies were supported by the National Institutes of Health (NIH) (NS-37956 and CA-21765) and the CCSG (P30 CA21765), and the American Lebanese and Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital. J.H.J.P was funded by grants from the NIH and the Joel and Jean Smilow Initiative. T.H.S. was funded by an NIH National Research Service Award and is a Leukemia and Lymphoma Society Special Fellow.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1746609.
Supplemental material is available at http://www.genesdev.org.
References
- Ahn J.Y., Schwarz J.K., Piwnica-Worms H., Canman C.E. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- Barnes D.E., Stamp G., Rosewell I., Denzel A., Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J.M., Chang S., Ferguson D.O. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E., Jankovic M., Difilippantonio S., Daniel J.A., Chen H.T., Celeste A., Pellegrini M., McBride K., Wangsa D., Bredemeyer A.L., et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Carney J.P., Maser R.S., Olivares H., Davis E.M., Le Beau M., Yates J.R., 3rd, Hays L., Morgan W.F., Petrini J.H. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Carson C.T., Schwartz R.A., Stracker T.H., Lilley C.E., Lee D.V., Weitzman M.D. The Mre11 complex is required for ATM activation and the G(2)/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Fernandez-Capetillo O., Chen H.T., Reina San Martin B., Van Laethem F., Yang Y.P., Petukhova G.V., Eckhaus M., Feigenbaum L., et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat. Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S., Celeste A., Kruhlak M.J., Lee Y., Difilippantonio M.J., Feigenbaum L., Jackson S.P., McKinnon P.J., Nussenzweig A. Distinct domains in Nbs1 regulate irradiation-induced checkpoints and apoptosis. J. Exp. Med. 2007;204:1003–1011. doi: 10.1084/jem.20070319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Frank K.M., Sekiguchi J.M., Seidl K.J., Swat W., Rathbun G.A., Cheng H.L., Davidson L., Kangaloo L., Alt F.W. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Frappart P.O., McKinnon P.J. Ataxia-telangiectasia and related diseases. Neuromolecular Med. 2006;8:495–511. doi: 10.1385/NMM:8:4:495. [DOI] [PubMed] [Google Scholar]
- Frappart P.O., Tong W.M., Demuth I., Radovanovic I., Herceg Z., Aguzzi A., Digweed M., Wang Z.Q. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat. Med. 2005;11:538–544. doi: 10.1038/nm1228. [DOI] [PubMed] [Google Scholar]
- Gao Y., Sun Y., Frank K.M., Dikkes P., Fujiwara Y., Seidl K.J., Sekiguchi J.M., Rathbun G.A., Swat W., Wang J., et al. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell. 1998;95:891–902. doi: 10.1016/s0092-8674(00)81714-6. [DOI] [PubMed] [Google Scholar]
- Herzog K.H., Chong M.J., Kapsetaki M., Morgan J.I., McKinnon P.J. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science. 1998;280:1089–1091. doi: 10.1126/science.280.5366.1089. [DOI] [PubMed] [Google Scholar]
- Hirao A., Kong Y.Y., Matsuoka S., Wakeham A., Ruland J., Yoshida H., Liu D., Elledge S.J., Mak T.W. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N., et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Jankovic M., Nussenzweig A., Nussenzweig M.C. Antigen receptor diversification and chromosome translocations. Nat. Immunol. 2007;8:801–808. doi: 10.1038/ni1498. [DOI] [PubMed] [Google Scholar]
- Kang J., Bronson R.T., Xu Y. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J. 2002;21:1447–1455. doi: 10.1093/emboj/21.6.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Katyal S., McKinnon P.J. DNA repair deficiency and neurodegeneration. Cell Cycle. 2007;6:2360–2365. doi: 10.4161/cc.6.19.4757. [DOI] [PubMed] [Google Scholar]
- Keramaris E., Hirao A., Slack R.S., Mak T.W., Park D.S. Ataxia telangiectasia-mutated protein can regulate p53 and neuronal death independent of Chk2 in response to DNA damage. J. Biol. Chem. 2003;278:37782–37789. doi: 10.1074/jbc.M304049200. [DOI] [PubMed] [Google Scholar]
- Kitagawa R., Bakkenist C.J., McKinnon P.J., Kastan M.B. Phosphorylation of SMC1 is a critical downstream event in the ATM–NBS1–BRCA1 pathway. Genes & Dev. 2004;18:1423–1438. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J., Antoccia A., Tauchi H., Matsuura S., Komatsu K. NBS1 and its functional role in the DNA damage response. DNA Repair (Amst.) 2004;3:855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- Kracker S., Bergmann Y., Demuth I., Frappart P.O., Hildebrand G., Christine R., Wang Z.Q., Sperling K., Digweed M., Radbruch A. Nibrin functions in Ig class-switch recombination. Proc. Natl. Acad. Sci. 2005;102:1584–1589. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M.F. Ataxia-telangiectasia: From a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Lee Y., McKinnon P.J. Responding to DNA double strand breaks in the nervous system. Neuroscience. 2007;145:1365–1374. doi: 10.1016/j.neuroscience.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee Y., Barnes D.E., Lindahl T., McKinnon P.J. Defective neurogenesis resulting from DNA ligase IV deficiency requires Atm. Genes & Dev. 2000;14:2576–2580. doi: 10.1101/gad.837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Chong M.J., McKinnon P.J. Ataxia telangiectasia mutated-dependent apoptosis after genotoxic stress in the developing nervous system is determined by cellular differentiation status. J. Neurosci. 2001;21:6687–6693. doi: 10.1523/JNEUROSCI.21-17-06687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller S.P., Meek K. Repair of DNA double strand breaks by non-homologous end joining. Biochimie. 2003;85:1161–1173. doi: 10.1016/j.biochi.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- Limbo O., Chahwan C., Yamada Y., de Bruin R.A., Wittenberg C., Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser R.S., Zinkel R., Petrini J.H. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat. Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S.J. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon P.J. ATM and ataxia telangiectasia. EMBO Rep. 2004;5:772–776. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon P.J., Caldecott K.W. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- O'Driscoll M., Gennery A.R., Seidel J., Concannon P., Jeggo P.A. An overview of three new disorders associated with genetic instability: LIG4 syndrome, RS-SCID and ATR-Seckel syndrome. DNA Repair (Amst.) 2004;3:1227–1235. doi: 10.1016/j.dnarep.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Orii K.E., Lee Y., Kondo N., McKinnon P.J. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. 2006;103:10017–10022. doi: 10.1073/pnas.0602436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U., Ahel I., West S.C. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- Reina-San-Martin B., Nussenzweig M.C., Nussenzweig A., Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc. Natl. Acad. Sci. 2005;102:1590–1595. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J., Ferguson D.O., Chen H.T., Yang E.M., Earle J., Frank K., Whitlow S., Gu Y., Xu Y., Nussenzweig A., et al. Genetic interactions between ATM and the nonhomologous end-joining factors in genomic stability and development. Proc. Natl. Acad. Sci. 2001;98:3243–3248. doi: 10.1073/pnas.051632098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: Taking shape. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Stewart G.S., Maser R.S., Stankovic T., Bressan D.A., Kaplan M.I., Jaspers N.G., Raams A., Byrd P.J., Petrini J.H., Taylor A.M. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stracker T.H., Theunissen J.W., Morales M., Petrini J.H. The Mre11 complex and the metabolism of chromosome breaks: The importance of communicating and holding things together. DNA Repair (Amst.) 2004;3:845–854. doi: 10.1016/j.dnarep.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Stracker T.H., Morales M., Couto S.S., Hussein H., Petrini J.H. The carboxy terminus of NBS1 is required for induction of apoptosis by the MRE11 complex. Nature. 2007;447:218–221. doi: 10.1038/nature05740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker T.H., Couto S.S., Cordon-Cardo C., Matos T., Petrini J.H. Chk2 suppresses the oncogenic potential of DNA replication-associated DNA damage. Mol. Cell. 2008;31:21–32. doi: 10.1016/j.molcel.2008.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Naka K., Okada Y., Watanabe M., Harada N., Saito S., Anderson C.W., Appella E., Nakanishi M., Suzuki H., et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K., Ohmura M., Azuma M., Nagamatsu G., Yamada W., Arai F., Hirao A., Suda T. Stem cell defects in ATM-deficient undifferentiated spermatogonia through DNA damage-induced cell-cycle arrest. Cell Stem Cell. 2008;2:170–182. doi: 10.1016/j.stem.2007.10.023. [DOI] [PubMed] [Google Scholar]
- Tauchi H., Kobayashi J., Morishima K., van Gent D.C., Shiraishi T., Verkaik N.S., vanHeems D., Ito E., Nakamura A., Sonoda E., et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–98. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- Taylor A.M., Groom A., Byrd P.J. Ataxia-telangiectasia-like disorder (ATLD)—Its clinical presentation and molecular basis. DNA Repair (Amst.) 2004;3:1219–1225. doi: 10.1016/j.dnarep.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Theunissen J.W., Kaplan M.I., Hunt P.A., Williams B.R., Ferguson D.O., Alt F.W., Petrini J.H. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell. 2003;12:1511–1523. doi: 10.1016/s1097-2765(03)00455-6. [DOI] [PubMed] [Google Scholar]
- Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon R., Vissinga C., Platzer M., Cerosaletti K.M., Chrzanowska K.H., Saar K., Beckmann G., Seemanova E., Cooper P.R., Nowak N.J., et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Williams B.R., Mirzoeva O.K., Morgan W.F., Lin J., Dunnick W., Petrini J.H. A murine model of nijmegen breakage syndrome. Curr. Biol. 2002;12:648–653. doi: 10.1016/s0960-9822(02)00763-7. [DOI] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. DNA double-strand break repair: All's well that ends well. Annu. Rev. Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Weaver D.T. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J. The DNA damage response: Putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- Zhu J., Petersen S., Tessarollo L., Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr. Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]