Abstract
MicroRNA (miRNA) silencing fine-tunes protein output and regulates diverse biological processes. Argonaute (Ago) proteins are the core effectors of the miRNA pathway. In lower organisms, multiple Agos have evolved specialized functions for distinct RNA silencing pathways. However, the roles of mammalian Agos have not been well characterized. Here we show that mouse embryonic stem (ES) cells deficient for Ago1–4 are completely defective in miRNA silencing and undergo apoptosis. In miRNA silencing–defective ES cells, the proapoptotic protein Bim, a miRNA target, is increased, and up-regulation of Bim is sufficient to induce ES cell apoptosis. Expression of activated Akt inhibits Bim expression and partially rescues the growth defect in Ago-deficient ES cells. Furthermore, reintroduction of any single Ago into Ago-deficient cells is able to rescue the endogenous miRNA silencing defect and apoptosis. Consistent with this, each Ago is functionally equivalent with bulged miRNA duplexes for translational repression, whereas Ago1 and Ago2 appear to be more effective at utilizing perfectly matched siRNAs. Thus, our results demonstrate that mammalian Agos all contribute to miRNA silencing, and individual Agos have largely overlapping functions in this process.
Keywords: Argonaute, microRNA, siRNA, stem cells, apoptosis
RNA silencing is a conserved gene regulatory mechanism present in almost all eukaryotic organisms (Hannon 2002; Mello and Conte 2004; Zaratiegui et al. 2007). siRNAs and microRNAs (miRNAs) can inhibit gene expression in a sequence-specific manner via mRNA degradation or translation repression. In the siRNA pathway, long dsRNAs are processed by multifunctional Dicer ribonucleases into 21- to 24-nucleotide (nt) siRNAs. siRNAs share perfect or near-perfect matches to their targets and primarily mediate the sequence-specific cleavage of target mRNAs. All known plant miRNAs and some recently discovered endogenous animal siRNAs direct cleavage of target RNAs (Du and Zamore 2005; Okamura and Lai 2008). In contrast, most animal miRNAs are partially complementary to their targets (Bartel 2004; Lai 2005). miRNAs primarily mediate their silencing effect via translation repression and sometimes coupled to mRNA deadenylation (Filipowicz et al. 2008; Wu and Belasco 2008). Animal miRNAs are processed from stem–loop-containing primary transcripts (pri-miRNAs) in two steps (Kim 2005). In the first step, pri-miRNAs are processed within the nucleus by “Microprocessor” complexes that consist of the core RNase III enzyme Drosha, into pre-miRNAs. Once exported from the nuclear pores, pre-miRNAs are cleaved by Dicer to produce miRNA/miRNA* (miRNA* refers to miRNA's partner strand) duplexes. It is estimated that miRNAs may regulate as many as one-third of human genes and miRNAs have been shown to regulate diverse biological processes (for review, see Bartel 2004; Bushati and Cohen 2007; Stefani and Slack 2008).
Both classes of small RNAs associate with Argonaute (Ago) proteins to form the core effector complexes, known as RNA-induced silencing complexes (RISCs) (Hutvagner and Simard 2008). Small RNAs guide RISCs to complementary target sequences to mediate different silencing effects. Ago proteins consist of four distinct domains: the N-terminal, PAZ, Mid, and PIWI domains. Structural analyses have revealed that a binding pocket within the Mid domain of Argonaute proteins interfaces with the 5′-phosphate of a small RNA, leaving internal nucleotides for base-pairing interactions with target mRNAs (Ma et al. 2005; Y. Wang et al. 2008). The PIWI domain has a fold that resembles RNase H-like enzymes (Song et al. 2004). Some Ago family members retain a functional catalytic center and have cleavage activity, whereas many others do not (Tolia and Joshua-Tor 2007). Multicellular organisms usually have multiple Ago family proteins (Hutvagner and Simard 2008). Studies from genetic model systems such as Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana have uncovered that different members of Ago family proteins are functionally specialized in distinct RNA silencing pathways. For example, in C. elegans, RDE-1 is required for the siRNA pathway (Tabara et al. 1999), and ALG-1 and ALG-2 are required for miRNA-mediated silencing (Grishok et al. 2001). In D. melanogaster, Ago2 is essential for siRNA-mediated target cleavage, whereas Ago1 is required for miRNA silencing (Hammond et al. 2001; Okamura et al. 2004). Therefore, the silencing effect of RISC is not only determined by the degree of complementarity between a small RNA to its target, but also affected by the specialized function that an Ago protein may possess. It has been recently shown that siRNAs and miRNAs with distinct structural features are actively sorted into specific Ago-RISCs. In D. melanogaster and C. elegans, perfectly matched siRNA duplexes are actively loaded into the Ago2-RISCs specialized for the siRNA pathway, whereas bulged miRNA precursor duplexes (miRNA/miRNA*) are sorted into Ago1-RISCs that function in the miRNA pathway (Forstemann et al. 2007; Steiner et al. 2007; Tomari et al. 2007). In A. thaliana, a different small RNA sorting mechanism has been found. The identity of the 5′-terminal nucleotide in a small RNA determines its loading into a particular Ago effector complex (Mi et al. 2008; Montgomery et al. 2008). Thus, it appears that abundant small RNA species and diverse Ago family members have evolved different sorting mechanisms to ensure effective RNA silencing in animal and plant kingdoms.
Human and mouse genomes encode four closely related Ago subfamily proteins (Ago1–4) and three PIWI-related subfamily members (Hutvagner and Simard 2008). The four Ago proteins are ubiquitously expressed, and they share extensive sequence homology. For instance, both human Ago3 and Ago2 share the same conserved motif in the catalytic center for cleavage. However, experimental evidence clearly shows that only Ago2 is capable of catalyzing the cleavage of the target mRNA by an siRNA/miRNA with perfect complementarity to the target (Liu et al. 2004; Meister et al. 2004; Rand et al. 2004). Ago2 is therefore unique and has an essential role in the siRNA pathway in mammals. In Ago2-deficient cells, miRNA-mediated translational silencing remains unaffected (Liu et al. 2004). It is therefore possible that other nonnucleolytic Ago proteins may predominate in miRNA silencing in mammals, as in C. elegans or D. melanogaster. Human Ago3 appears to be the most potent translational repressor when tethered to a reporter mRNA (Wu et al. 2008). Alternatively, all mammalian Agos may have overlapping functions in the miRNA pathway. Consistent with this idea, all four Ago proteins when overexpressed have been shown to bind endogenous miRNAs indistinguishably (Liu et al. 2004; Meister et al. 2004). More recently, high-throughput pyrosequencing showed miRNA profiles associated with the endogenous hAgo2 and hAgo3 largely overlaps, while a small set of miRNAs is preferentially loaded onto either hAgo2 or hAgo3 (Azuma-Mukai et al. 2008). In human cells, miRNA-containing RISCs can mediate both translation repression and cleave complementary target mRNAs (Hutvagner and Zamore 2002); however, it is unknown whether different Ago-RISCs are utilized for two distinct silencing effects. When artificially tethered to the 3′ untranslated region (UTR) of a reporter mRNA by λN22 peptide tag, human Ago1, Ago2, Ago3, and Ago4 are able to mimic the inhibitory effect of miRNAs on the translation of reporter mRNAs (Pillai et al. 2004; Wu et al. 2008). Nevertheless, definitive genetic evidence that differentiates these two alternatives is not available.
In this study, we used a genetic approach to address the questions of whether mouse Ago subfamily members (Ago1–4) are required for miRNA silencing and whether individual Agos have specialized or overlapping functions in this process. We demonstrate that mammalian Ago subfamily proteins are essential for translational silencing and individual Agos have overlapping and distinct functions in this process.
Results
Generation of an inducible ES cell line deficient for mouse Argonautes 1–4
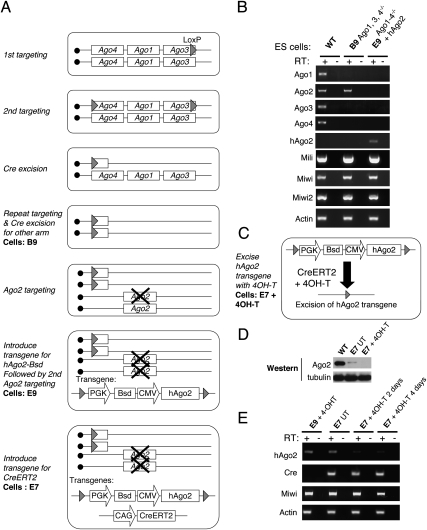
We began to investigate mammalian Argonaute functions by generating an inducible embryonic stem (ES) cell line that is deficient for Ago1–4. Mouse Ago1, Ago3, and Ago4 are clustered in tandem on chromosome 4, and Ago2 is on chromosome 15. We used a combination of gene-targeting and Cre-mediated chromosomal deletion strategies to generate Ago1, Ago3, Ago4 −/− ES cells (B9) (Fig. 1A). We then inactivated both alleles of endogenous Ago2 by gene targeting to generate an ES cell line (E9). In anticipation that the inactivation of the miRNA pathway might have a negative effect on ES cell growth, we introduced a floxed hAgo2 transgene before targeting the second mAgo2 allele. Finally, we introduced a CreERT2 transgene and generated an inducible knockout cell line for all Agos (E7) (Fig. 1A). Upon 4-hydroxy-tamoxifen (4OH-T) treatment, Cre-mediated deletion of the floxed hAgo2 generates a null for all Agos (Fig. 1C). RT–PCR analyses show that endogenous Ago1–4 mRNAs are absent in E9 ES cells (Fig. 1B). Upon 4OH-T induction, Ago2 protein and mRNA are deleted in E7 cells (Fig. 1D,E). Therefore, we successfully established an inducible Ago1–4 knockout ES cell line for functional analyses.
Figure 1.
Generation of inducible knockout ES cells for all four Ago proteins. (A) A genetic strategy to generate inducible ES cells deficient for Ago1–4. Multiple steps of gene targeting and Cre-mediated excision were employed to first generate B9 ES cells (Ago1, Ago3, Ago4−/−). In the presence of floxed hAgo2 transgene, both mAgo2 alleles were deleted by targeting in E9 ES cells. A CreERT2 transgene was finally introduced to generate the inducible E7 line. (B) RT–PCR analyses confirm that the endogenous Ago transcripts are absent in mutant ES cell lines. Note that related PIWI transcripts remain unchanged. (C) Schematic of the 4OH-T-induced excision of the hAgo2 transgene from E7 cells. (D) Western blot analysis showing that hAgo2 is expressed at a lower level in E7 cells compared with the endogenous Ago2 in control ES cells and hAgo2 is absent after 4OH-T induction. (E) RT–PCR analysis on E7 cells showing a decrease in hAgo2 after 2–4 d of 4OH-T treatment. The residual hAgo2 expression reflects an incomplete Cre excision. Based on calculating the number of drug (Bsd)-resistant colonies, we estimated that >80% of E7 cells excised hAgo2 by CreERT2 after 48-h 4OHT treatment and the excision efficiency increases with longer treatments. (UT) Untreated.
Ago subfamily proteins are essential for miRNA-mediated translational repression
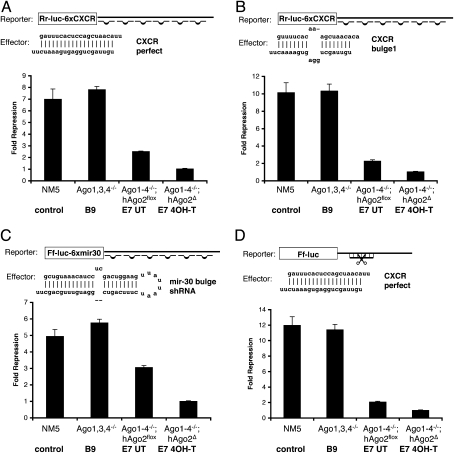
To demonstrate the role of Agos in miRNA silencing, we performed miRNA translational repression assays using the different Ago mutant cell lines described above (Fig. 2). We chose luciferase reporters that contain six copies of an imperfect binding site for mir-CXCR (an artificial miRNA) or mir-30 to assay miRNA silencing effect (Zeng et al. 2002; Doench et al. 2003). Endogenous mir-CXCR does not exist, and only a low level of endogenous mir-30 is expressed in ES cells (Calabrese et al. 2007). The silencing effect is therefore predominantly the result of transfected miRNAs. As shown in Figure 2, Ago1, Ago3, Ago4 −/− B9 ES cells have normal function in miRNA-mediated repression, suggesting that Ago2 alone can compensate for other Agos in miRNA silencing. Consistent with this, hypomorphic E7 cells that express a lower level of Ago2 (Fig. 1D) have compromised functions in siRNA- as well as miRNA-mediated silencing (Fig. 2A–D, comparing E7 cells with control and B9 cells). Most importantly, Ago-null ES cells are unable to perform miRNA-mediated repression after 4OH-T treatment using two different reporter systems (Fig. 2A–C). siRNA-mediated cleavage of target mRNA is also lost in treated E7 cells (Fig. 2D), which serves as functional proof that hAgo2 is deleted in the majority of cells treated with 4OH-T. It has been previously shown that miRNA translation repression is normal in Ago2 −/− mouse embryonic fibroblasts (Liu et al. 2004). Taken together, these results demonstrate that Ago1–4 are collectively required for miRNA-mediated gene silencing in mouse ES cells.
Figure 2.
Ago1–4 are required for miRNA-mediated translational repression. (A–C) miRNA-mediated translation repression is defective in Ago1-4−/−; hAgo2Δ ES cells. Shown are luciferase assays using two different reporters and three different effectors indicated in A–C. (D) siRNA-mediated mRNA cleavage is defective in Ago1-4−/−; hAgo2Δ E7 ES cells. Shown are assays using a firefly luciferase reporter with a single perfect complementary antisense CXCR-binding site in the 3′UTR of Ff-luc using CXCR dsRNA as an effector. All results are shown as means ± SEM from six independent transfections.
The mouse has an additional Argonaute like protein Ago5 on the X chromosome. ES cells deficient for Ago1–4 are completely defective in miRNA silencing, thus suggesting that Ago5 is not required for this function. In fact, Ago5 is missing in the human and other vertebrate genomes. A more detailed sequence alignment identifies internal deletions of conserved residues in PAZ, Mid, and PIWI domains (Supplemental Fig. S1). Most noticeably, the majority of conserved amino acids (in the Mid domain) that have been shown recently to be critical for nucleic acid binding are missing in mouse Ago5 (Y. Wang et al. 2008), arguing that Ago5 might be quite different from other Ago proteins (Supplemental Fig. S1).
Argonautes are essential for the survival of ES cells
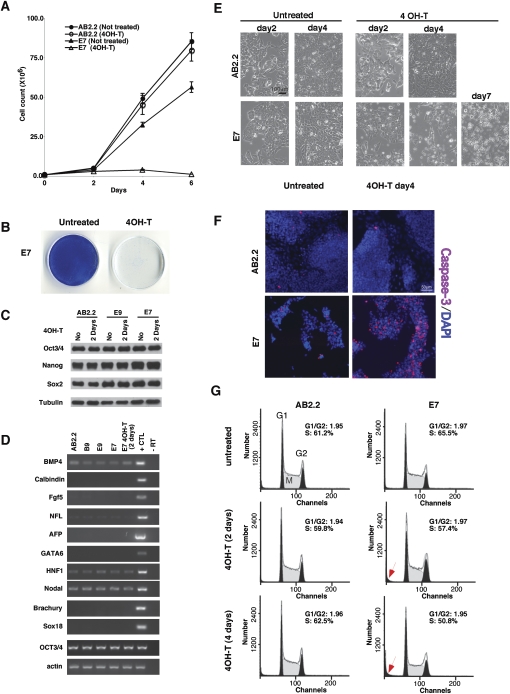
To examine the long-term effect of Ago depletion on ES cell maintenance, we first measured the growth rate of Ago-deficient ES cells. Although E7 cells (untreated) have compromised miRNA function in the reporter assays (Fig. 2A–C), these cells grew only slightly slower compared with a wild-type ES cell line, AB2.2 (Fig. 3A). This might reflect the fact that the endogenous miRNA pathways are at least partially functional in untreated E7 cells (Fig. 2; Supplemental Fig. S2). Upon 4OH-T treatment, loss of Ago activity and miRNA function (treated E7) block stem cell self-renewal (Fig. 3A,B). After five consecutive passages in the presence of 4OH-T, Ago-depleted ES cells were essentially lost (Fig. 3B). Thus, Ago function is required for ES cell self-renewal.
Figure 3.
Ago1–4 are essential for the survival of mouse ES cells. (A) Growth curves showing a severe growth defect in Ago1-4−/−; hAgo2Δ E7 ES cells. (B) E7 ES cells are not viable after 4OH-T-induced Ago depletion. Shown are methylene blue stains of both untreated and 4OH-T-treated E7 ES cells after five consecutive passages at a ratio of 1:4. (C) The pluripotent transcription factors Oct3/4, Sox2, and Nanog are not down-regulated in Ago mutant ES cell lines E9 and E7 as compared with wild-type (AB2.2). β-tubulin serves as a loading control. (D) RT–PCR analyses on various cell lineage and differentiation markers demonstrate that no abnormal differentiation occurs in Ago1–4 mutant cells. The control is a mixture of RNAs from differentiated embryoid bodies and E12.5 mouse embryos. (E) Phase-contrast images of wild-type and E7 ES cells treated with 4OH-T at various time points. E7 cells begin to die and detach (seen as rounded up bright cells) after 4OH-T treatment. (F) Active caspase-3 staining on wild-type and E7 cells untreated and treated with 4OH-T shows a significant increase of apoptosis in E7-treated cells. (G) FACScan analysis of the cell cycle profiles of wild-type and E7-untreated and -treated ES cells. Note an increase of dying cells (arrow) and a decrease of S-phase cells in 4-d 4OH-T-treated E7 cells.
Several factors might contribute to loss of ES cell proliferation and pluripotency in Ago-deficient cells. We first examined whether pluripotent transcription factors were down-regulated in Ago-depleted cells. Western blot analyses show that Oct3/4, Nanog, and Sox2 were not decreased in Ago-hypomorphic (E9 and untreated E7) or Ago-depleted ES cells (treated E7) (Fig. 3C). Next, we tested whether the loss of Agos leads to a precocious differentiation and exit from cell proliferation. RT–PCR assays showed that a variety of cell lineage and differentiation markers were not induced upon depletion of Agos (Fig. 3D). We then tested whether Agos play a role in maintaining ES cell survival. We observed that some Ago-depleted cells began to die and detach from the culture surface after a 4-d 4OH-T treatment, and the majority of cells were dying after a 7-d 4OH-T treatment (Fig. 3E). Apoptotic cells were visualized with an antibody against active caspase-3, confirming that ES cells underwent apoptosis in the absence of Ago proteins (Fig. 3F). Analysis of cell cycle profiles of Ago-depleted ES cells demonstrated that Ago depletion did not significantly alter the cell cycle profiles, although a slight decrease of S-phase cells and a corresponding increase of dying cells (Fig. 3G, arrow) were observed after a 4-d 4OH-T treatment (Fig. 3G). The decrease of S-phase cells correlates with the increase in cell death, suggesting that the proliferating cells in S phase are more vulnerable to the loss of Ago proteins and the miRNA regulatory pathway. Collectively, our data demonstrate that Argonaute proteins play an essential role in maintaining the survival of ES cells.
Proapoptotic Bim is an endogenous miRNA target in ES cells
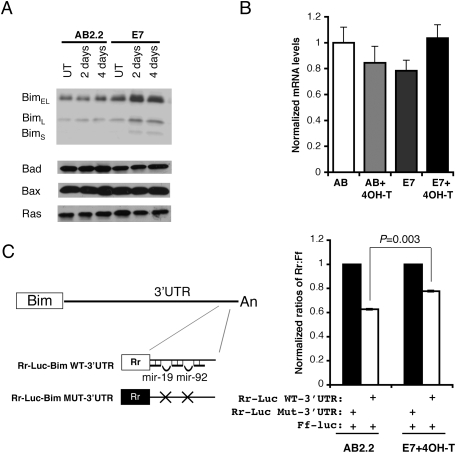
ES cells lacking Dicer and DGCR8, critical components in miRNA biogenesis, are viable but display severe defects in growth and differentiation (Kanellopoulou et al. 2005; Murchison et al. 2005; Wang et al. 2007). In contrast, Dicer-deficient stem cells are depleted in the developing mouse embryo, and conditional ablation of Dicer in ES cells causes a dramatic cell loss (Bernstein et al. 2003; Murchison et al. 2005). In this study, ES cells deficient for Ago1–4 undergo apoptosis. Therefore, it is not clear whether the different phenotypes are due to compensatory genetic changes in Dicer-null cells, miRNA-independent functions, or both. Therefore, we investigated whether the abnormal regulation of miRNA function could in part contribute to the apoptotic phenotype in Ago-deficient cells. Several major miRNA species including mir-21, mir-17–92, mir-15b, mir-16, and mir-290–295 are abundantly expressed in mouse ES cells (Calabrese et al. 2007). RT–quantitative PCR analyses on levels of mature miRNAs that include mir-19, mir-130, mir-21, and mir-290 confirmed that these miRNAs are expressed in ES cells. In hypomorphic and Ago-deficient E7 cells (untreated and 4OH-T-treated), levels of four analyzed miRNAs are decreased significantly but returned to higher levels in Ago2-rescued E7 cells (Supplemental Fig. S3), showing that Ago proteins are required for maintaining the stability of miRNA species. To test whether miRNA target proteins are abnormally up-regulated in Ago-depleted cells, we examined several candidate proteins, including PTEN (Lewis et al. 2003; Xiao et al. 2008), E2F1 (O'Donnell et al. 2005), Bim/Bcl2L11 (Ventura et al. 2008; Xiao et al. 2008), Ras (Johnson et al. 2005), and PDCD4 (Lu et al. 2008), which are known miRNA targets. Three isoforms of Bim (BimEL, BimL, and BimS) were reproducibly up-regulated in Ago mutant ES cells, and no significant changes for Ras and PDCD4 were detected (Fig. 4A; Supplemental Fig. S4). We did not detect endogenous E2F1 in ES cells and found PTEN protein levels increased only in a subset of samples (Supplemental Figs. S4, S6). miRNAs can affect target gene expression through either translational repression or mRNA destabilization (Filipowicz et al. 2008). To distinguish these alternatives, quantitative RT–PCR was used to analyze the levels of Bim mRNA in Ago-depleted cells. The data showed that Bim mRNA is not significantly altered after Ago depletion (Fig. 4B), suggesting that miRNAs repress Bim expression mainly through repressing translation. Bioinformatics analyses suggested that there might be >150 miRNA-binding sites within the entire 3-kb UTR of Bim (http://www.microrna.org). It has been reported that the 3′UTR of Bim mRNA consists of mir-19- and mir-92-binding sites that are required for clustered mir-17–92-mediated regulation (Ventura et al. 2008). Therefore, we compared mir-17–92-mediated repression of a luciferase reporter that contains a 138-base-pair (bp) fragment of Bim 3′UTR in both control and Ago-depleted ES cells (Fig. 4C). The result confirmed that endogenous miRNAs repress Bim expression in an Ago-dependent manner. The loss of Ago activity has a relatively mild effect on the Bim 3′UTR reporter activity compared with a significant increase of the endogenous Bim proteins (Fig. 4A). This could be due to the fact that only a small portion of Bim 3′UTR was tested in this assay. Additional uncharacterized miRNA target sites might account for this difference. Taken together, our data confirmed that Bim is an endogenous miRNA target in mouse ES cells.
Figure 4.
Bim is an endogenous miRNA target in mouse ES cells. (A) Bim protein is up-regulated in Ago-depleted ES cells. Western blots show that three Bim isoforms are increased in miRNA-defective E7 cells. Note other proapoptotic proteins such as Bad, Bax, and miRNA let-7 target Ras remain unchanged in Ago mutant ES cells. (B) RT–quantitative PCR for Bim show no significant change in mRNA levels in Ago-depleted ES cells. (C) The 3′UTR of Bim contains functional binding sites for mir-19 and mir-92, two miRNAs expressed in ES cells. miRNA-mediated repression of a Rr-luc reporter containing a segment of the 3′UTR of Bim was compared with that of a Rr-luc reporter containing a mutant 3′UTR from Bim in both control and Ago-depleted E7 cells. E7 cells were unable to repress the Rr-luc reporter containing the wild-type Bim 3′UTR to the same level as wild-type control (P < 0.003). The data shown are from 12 independent transfections.
miRNA target protein Bim induces ES cell apoptosis
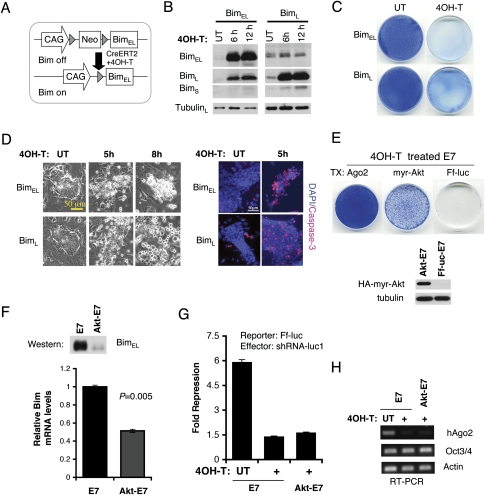
Bim has proapoptotic activity in some cells (O'Connor et al. 1998), and thus Bim up-regulation might contribute to the apoptosis of miRNA-defective ES cells. Consistent with this, constitutive expression of different Bim isoforms negatively regulates ES cell growth in colony formation assays (Supplemental Fig. S5). To demonstrate the apoptotic role of Bim in ES cells, we generated inducible BimEL or BimL expression in ES cell lines (Fig. 5A). Upon 4OH-T induction, Bim proteins were rapidly induced (Fig. 5B), and the ES cells underwent apoptosis (Fig. 5C,D). To examine whether abnormal Bim up-regulation in Ago-depleted cells contributes to the apoptosis of ES cells, we performed genetic rescue by expressing Bcl2 (Supplemental Fig. S6). Coimmunoprecipitation confirmed that Bcl2 interacts with Bim (O'Connor et al. 1998). Overexpression of Bcl2 partially rescued the apoptosis of E7 under 4OH-T treatment (Supplemental Fig. S6B). However, the Bcl2-E7 (4OH-T-treated) cells appeared to be differentiated and looked like fibroblasts. Interestingly, miRNA target proteins, Bim and PTEN, were significantly up-regulated in Bcl2-expressing E7 cells even before 4OH-T treatment (Supplemental Fig. S6C). One explanation is that Bcl2 overexpression prevented a subpopulation of E7 cells from undergoing apoptosis due to increased levels of Bim and PTEN. To test the effect of PTEN overexpression on ES cell growth, we established PTEN-inducible ES cells (Supplemental Fig. S6D). Upon 4OH-T induction, PTEN-expressing ES cells grew significantly slower (Supplemental Fig. S6E). Therefore, the miRNA targets, both Bim and PTEN, could negatively regulate ES cell growth and contribute to the phenotype of Ago-deficient ES cells. To antagonize both Bim and PTEN function, we expressed a constitutively active myristylated Akt in E7 cells. In addition to its well-established function in the PTEN-PI3 kinase pathway, an activated Akt phosphorylates and inactivates FOXO transcription factors to down-regulate Bim transcription in hematopoietic cells (Dijkers et al. 2002). Akt can also directly phosphorylate BimEL and antagonize its apoptotic activity (X.J. Qi et al. 2006). We found that the overexpression of myr-Akt can partially rescue the apoptosis and growth defect in Ago-depleted ES cells by down-regulating Bim gene expression (Fig. 5E,F). We also found a small number of Akt-rescued cells always express hAgo2 by a RT–PCR assay, although the majority of rescued cells do not express hAgo2 and are unable to silence the reporter with an shRNA (Fig. 5G,H). We attempted to isolate a pure population of Akt-rescued Ago-deficient ES cells by expanding single ES cells at clonal density. After analyzing nearly 400 colonies, we failed to isolate any pure Ago-deficient clones. Thus, Ago-deficient cells might have growth defects at clonal density and/or paracrine signals from Ago-positive cells might be necessary for the survival of Akt-rescued Ago-deficient cells. Taken together, our data demonstrate that the repression of Bim and possibly PTEN protein synthesis by Argonautes and miRNAs is a critical regulatory mechanism in the maintenance of normal ES cells.
Figure 5.
Regulation of ES cell survival by Bim. (A) Schematic of an inducible Bim ES cell line. Upon 4OH-T treatment, CreERT2 excises Neo selection marker and activates Bim transcription. (B) Western blots show that BimEL and BimL proteins are rapidly induced after 4OH-T treatment in two independent inducible ES cell lines. (C) Methylene blue stains show that most Bim-expressing cells are lost 2 d after 4OH-T induction. (D) Bim overexpression induces apoptosis in ES cells. Phase-contrast images and active caspase-3 stains are shown. (E) An activated Akt (myr-Akt) partially rescues cell growth defects of Ago1-4−/−; hAgo2Δ E7 cells after five passages with 4OH-T. Western blotting shows that HA-tagged myr-Akt is expressed in rescued E7 cells. Ago2-expressing E7 cells serve as the positive control for a complete rescue. (F) myr-Akt down-regulates Bim transcription in 4OH-T-treated E7 ES cells. Bim mRNA and protein levels are decreased in myr-Akt-rescued E7 cells. Shown are quantitative PCR and Western blot results. (G) The luciferase assay confirms loss of Ago2 and inability to use shRNA to silence a Ff-Luc reporter in myr-Akt-rescued cells. RT–PCR analysis shows that the majority of rescued E7 cells no longer express the floxed hAgo2 as a pool. The residual PCR signal from the pool might reflect a small population of rescued cells that still express hAgo2.
Genetic rescue of Ago-deficient ES cells with individual Agos
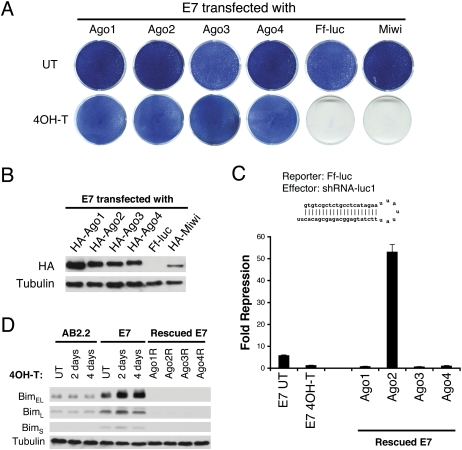
Thus far, our analysis has focused on Ago function as a whole but not that of individual Agos. To evaluate the role of individual Agos, we performed genetic rescue experiments using inducible Ago knockout E7 cells. We used Piggy-Bac transposons to generate ES cells that stably express a single HA-tagged Ago protein (Fig. 6A,B). Western blotting shows that individual Ago proteins are expressed at similar levels in stably transfected cells (Fig. 6B). The siRNA-mediated cleavage assay shows that the floxed hAgo2 is deleted in rescued E7 cells and confirms that only a single Ago is expressed in the rescued cells (Fig. 6C). E7 ES cells that express individual Ago proteins are viable and proliferate indistinguishably from wild-type ES cells. This demonstrates that the expression of any single Ago is capable of rescuing the cell death phenotype of 4OH-T-treated E7 cells, whereas firefly luciferase or the Ago-related MIWI protein is unable to rescue (Fig. 6A). Thus, the four Ago proteins share redundant functions in supporting ES cell survival and that a PIWI subfamily protein does not compensate for the specific Ago function in ES cells. Consistent with overlapping functions of Agos in ES cell survival, individual Ago proteins are also indistinguishable from their ability to rescue the endogenous miRNA silencing defect. Using the endogenous miRNA target-Bim as an example, we demonstrated that re-expression of any single Ago in E7 cells down-regulates Bim protein in rescued ES cells (Fig. 6D). Thus, individual Ago proteins share significant overlap in the endogenous miRNA silencing pathway.
Figure 6.
Genetic rescue reveals overlapping functions of individual Ago proteins. (A) Individual Agos rescue 4OH-T-treated E7 cells from apoptosis, whereas controls Ff-luc and Miwi are unable to rescue. Shown are methylene blue stains of indicated ES cells after five passages with or without 4OH-T. (B) Individual HA-tagged Ago1–4 are expressed at a similar level in rescued cell lines with β-tubulin included as a loading control. (C) A cleavage assay shows that the floxed hAgo2 is deleted and confirms that only a single Ago is expressed in each rescued cell line. The shRNA-luc1-mediated mRNA cleavage is measured by dual luciferase assay using Rr-Luc as an internal control. Compared with untreated E7 cells, Ago1-, Ago3-, and Ago4-rescued E7 cells have no cleavage activity, demonstrating that the floxed hAgo2 allele is completely deleted in these cells. Shown are means ± SEM from six independent transfections. (D) Individual Ago proteins are capable of rescuing the endogenous miRNA silencing defect in E7 cells. For the endogenous miRNA target protein Bim, Western blots show that three Bim isoforms are increased in miRNA-defective cells and return to a basal level in individual Ago-rescued E7 cells. Tubulin serves as a loading control.
Individual Agos have different preferences for miRNAs and siRNAs
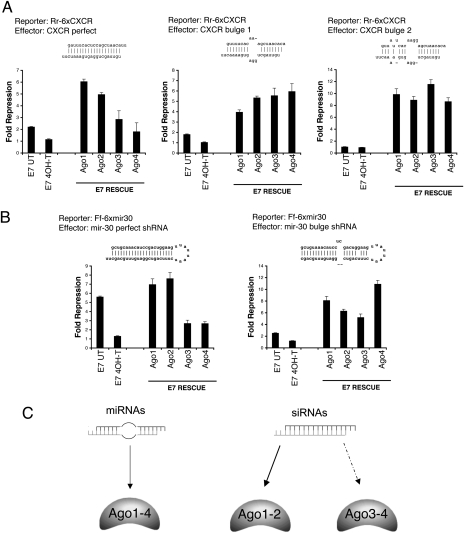
Although individual Agos function redundantly in ES cells, an important remaining question is whether individual Ago proteins have identical activities in miRNA silencing. In flies and worms, miRNA precursor RNA duplexes are preferentially loaded into Argonaute-containing complexes to mediate translational repression, whereas siRNA duplexes are specifically recruited to the siRNA pathway to cleave target mRNAs (Forstemann et al. 2007; Steiner et al. 2007; Tomari et al. 2007). miRNA precursors from fly to human share the same structural feature—a central bulge in RNA duplexes (Khvorova et al. 2003; Han et al. 2006). Therefore, we examined whether individual mammalian Ago proteins have a preference for siRNAs or miRNAs. Using ES cells that express a single Ago protein (Fig. 6B), we performed miRNA-mediated translational repression assays with different combinations of luciferase reporters and miRNA/siRNA effectors (Fig. 7A,B). First, we generated one perfectly matched and two bulged CXCR RNA duplexes to compare their ability to repress a Rr-Luc-6xCXCR reporter (Fig. 7A). Because these RNA duplexes have different thermodynamic properties and the efficiency of duplex annealing might be different for each duplex, we evaluated whether any ssRNA molecules (guide or passenger strand) could efficiently repress a Rr-luc reporter. Our results show that any contribution from ssRNA species is negligible and RNA duplexes are predominant effectors in our assays (Supplemental Fig. S7). Second, we used the mouse U6 promoter to express shRNAs that encode perfectly matched mir-30 and bulged mir-30 precursors (Fig. 7B) because in vitro annealed RNA duplexes bypass miRNA biogenesis. For RNA duplexes mimicking endogenous miRNA/miRNA* precursors (mir-30-bulge, CXCR-bulge1, and CXCR-bulge2), individual Ago-rescued ES cells have a similar repression of luciferase reporters. For perfectly matched RNA duplexes that resemble siRNAs, including mir-30-perfect and CXCR-perfect, Ago1- and Ago2-mediated luciferase repression is significantly more robust in comparison with Ago3 and Ago4. Taken together, these results demonstrate that all Agos can equally utilize miRNA precursor duplexes to mediate the silencing effect, whereas only Ago2 and Ago1 efficiently incorporate perfectly matched siRNA duplexes. Thus, redundancy among the four Agos in the endogenous miRNA silencing pathway is not surprising given the fact that each efficiently utilizes miRNA/miRNA* duplexes containing a central bulge, the most common form of endogenous miRNA precursors in mammals. In addition to their shared preference for miRNAs, we demonstrate that individual Agos differ significantly in their preference for siRNAs, in the order Ago1 and Ago2 > Ago3 and Ago4. Therefore, the relative abundance of different Ago isoforms could affect the efficacy and specificity of RNAi.
Figure 7.
Individual Ago proteins have different preferences for miRNAs and siRNAs. (A,B) Different combinations of effectors and luciferase reporters were used in the translational repression assay to compare the loading preference of individual Ago proteins for small RNA duplexes with distinct secondary structures. Shown above each graph are the reporter used and the secondary structure of dsRNA effector. Ago2 and Ago1 are more effective than Ago3 and Ago4 in utilizing siRNA duplexes, while all Agos can repress with miRNA effectors. The experiments in A and B used mir-CXCR and mir-30 reporters, respectively. Shown are means ± SEM from six independent transfections. (C) A summary illustrates the partition of miRNA and siRNA duplexes among different mammalian Ago proteins.
Discussion
In this study, we provide in vivo evidence that mammalian Ago subfamily proteins as a whole are essential for miRNA-mediated silencing. We found that individual Agos have redundant functions in the miRNA pathway, and any single Ago is sufficient to confer wild-type miRNA function in mouse ES cells. Furthermore, individual Agos appear functionally specialized in recruiting structurally distinct small RNA duplexes for the silencing effect. siRNA duplexes are preferentially utilized by Ago1 and Ago2, while bulged miRNA*/miRNA duplexes equally partition among all Agos (Fig. 7C). Therefore, our results, together with previous studies in flies and worms (Forstemann et al. 2007; Steiner et al. 2007; Tomari et al. 2007), demonstrate that mechanisms of sorting distinct small RNAs into different Ago-effector complexes are partially conserved throughout the animal kingdom.
Argonautes are the core effectors of RNA silencing pathways. Previous studies in C. elegans, D. melanogaster, and A. thaliana have demonstrated that different Ago proteins have become specialized for the siRNA or miRNA pathway (Tabara et al. 1999; Grishok et al. 2001; Hammond et al. 2001; Okamura et al. 2004; Vaucheret 2008). In addition to the functional specification of Agos, other upstream or downstream components specific for one particular pathway have also been found in these systems. In C. elegans, RDE-1 is specific for the siRNA pathway and recruits amplification machinery to generate secondary siRNAs (Sijen et al. 2001). In D. melanogaster, Dicer-1 and Dicer-2 serve distinct functions in processing and loading of miRNAs and siRNAs into specific Ago complexes, respectively (Lee et al. 2004). In contrast, the siRNA and miRNA pathways intersect at several levels in mammals. For instance, a single Dicer enzyme processes dsRNA or stem–loop precursors to produce siRNAs and miRNAs (Kanellopoulou et al. 2005; Murchison et al. 2005). While previous work demonstrated that Let-7 miRNA can be loaded into Ago2-RISCs for target cleavage (Hutvagner and Zamore 2002), we showed that siRNAs can be efficiently incorporated into Ago1/2-RISC for translational silencing. Recent miRNA profiling also demonstrated that endogenous human Ago2 and Ago3 bind to largely overlapping subsets of miRNAs in Jurkat cells (Azuma-Mukai et al. 2008). The significant overlaps between the siRNA and miRNA pathways and the functional redundancy among different Argonaute family members imply a vital role for endogenous miRNAs in mammalian species. Consistent with this view, miRNAs are implicated in every aspect of mammalian biology, and loss of general miRNA pathway components often gives rise to devastating phenotypes in animals.
In other species, the siRNA and miRNA pathways diverge in part due to sorting mechanisms that direct distinct small RNA duplexes into specific Ago-RISCs (Forstemann et al. 2007; Steiner et al. 2007; Tomari et al. 2007; Mi et al. 2008; Montgomery et al. 2008). However, such sorting mechanisms appear to be partially lost in mammals. Ago1–4 appear to be indistinguishable for miRNAs, while only Ago1 and Ago2 prefer siRNAs. As a consequence, siRNAs are fully functional as miRNAs and cause widespread off-target effects (Scacheri et al. 2004). Such off-target effects may be intrinsic to the machinery for RNA silencing in mammals. A bioinformatics approach to avoid sequence homology within the seed region might help to alleviate this problem. Another type of off-target effect is due to the overloading of the endogenous miRNA pathway (Grimm et al. 2006). Our observation that Ago2 preferentially utilizes siRNAs suggests that one could tilt the balance of the siRNA and miRNA pathways by altering the relative level of Ago2 to other Ago proteins in a cell. Therefore, our results provide a rationale for using Ago2 overexpression in RNAi applications since Ago2 can perform both miRNA and siRNA silencing and minimize off-target effects due to the saturation of endogenous Agos. In fact, such practice has recently been demonstrated to enhance the efficacy of RNAi (Diederichs et al. 2008).
Our study also demonstrates an essential role for Argonaute proteins in the maintenance of ES cells. Several different mutant ES cell lines in the miRNA pathway have been generated, including Dicer and DGCR8 (Kanellopoulou et al. 2005; Murchison et al. 2005; Wang et al. 2007). Despite the fact that all these mutant ES cells have miRNA silencing defects, different mutant ES cells exhibit distinct phenotypes. DGCR8 is a dsRNA-binding protein that facilitates the cleavage of pri-miRNA by Drosha during the miRNA biogenesis (Kim 2005). The production of pre-miRNAs and mature miRNAs is blocked in DGCR8 −/− ES cells (Wang et al. 2007). DGCR8 −/− ES cells have a subtle proliferation defect and are partially defective in differentiation. Like DGCR8 −/− cells, Dicer-deficient ES cells are unable to produce mature miRNAs but exhibit a complete block of ES cell differentiation (Kanellopoulou et al. 2005). In addition, conditional ablation of Dicer in ES cells also revealed profound survival and proliferation defects in Dicer-depleted ES cells (Murchison et al. 2005). The majority of Dicer-deficient cells were depleted in the cell population after transient Cre expression. In this study, we showed that upon depletion of all Agos, ES cells failed to mediate miRNA silencing and underwent dramatic apoptosis. Consistent with the idea that miRNA defects could contribute to ES cell apoptosis, we demonstrated that Argonaute-mediated miRNA silencing down-regulates Bim translation. Furthermore, up-regulation of Bim is sufficient to induce ES cell apoptosis, and down-regulation of Bim by an activated Akt could partially rescue apoptosis in Ago-deficient ES cells. Taken together, these results support a critical role for miRNA silencing in the regulation of ES cell survival, in addition to its role in the proliferation and differentiation of ES cells. A more recent study of Dicer's role in B-cell development also strongly supports the notion that miRNA mediated regulation of Bim plays a critical role in cell survival (Koralov et al. 2008). Dicer depletion during B-cell development led to the excessive apoptosis of pre-B cells and significant up-regulation of Bim. In a similar fashion, Dicer ablation-induced B-cell apoptosis could be partially rescued by ablation of Bim (Koralov et al. 2008).
In both Ago-depleted ES cells and Dicer-deficient B cells, the apoptotic phenotypes can be rescued in part by down-regulating critical miRNA targets such as Bim, strongly suggesting that the miRNA-dependent function of Dicer and Agos plays a critical role in this process. It is likely that other miRNA targets such as PTEN could also contribute to the phenotype observed in Ago-depleted ES cells. Nevertheless, the different phenotypes among different miRNA silencing-defective ES cells suggest that additional miRNA-independent functions of Dicer and Agos might play a role in the maintenance of ES cells. It is well known that both Dicer and the Agos are both involved in small RNA-mediated transcriptional silencing. Defects in centromeric and heterochromatin silencing have been described in one Dicer-deficient ES cell line as well as Dicer-deficient chicken–human hybrid DT40 cells (Fukagawa et al. 2004; Kanellopoulou et al. 2005). Studies in A. thaliana demonstrated that Ago4 is important for RNA-directed heterochromatic silencing (Zilberman et al. 2003; Y. Qi et al. 2006). Human Ago1 has been implicated in siRNA-induced heterochromatin formation, and C. elegans ALG-1 is required for transcriptional silencing of a transgene (Grishok et al. 2005; Kim et al. 2006). Piwi proteins, another subfamily of Ago-related proteins, have also been shown to play critical roles in heterochromatin silencing (Saito et al. 2006; Yin and Lin 2007). Therefore, it is likely that Ago-deficient cells may have transcriptional silencing defects in addition to the miRNA silencing defect. If so, the transcriptional silencing defect could also contribute to the apoptosis in the ES cells. In comparison with the well-characterized post-transcriptional silencing pathways, molecular mechanisms of how small RNAs mediate transcriptional silencing are relatively uncharacterized in mammalian cells. Our inducible Ago knockout ES cells provide a valuable tool to study the effects of small RNA-mediated transcriptional silencing in future studies.
Materials and methods
BAC recombineering and gene targeting
To generate Ago1–4-deficient ES cells, we constructed Ago gene targeting vectors using the BAC recombineering technique (Liu et al. 2003). The original S129 BAC clones containing mouse Ago genes were obtained from the Welcome Trust Sanger Institute. To achieve high targeting efficiency, we used a Blm-deficient parental ES cell line, NM5, for all gene targeting experiments (Guo et al. 2004). The detailed targeting vectors and selection scheme are depicted in Supplemental Figures S8–S13. All targeting events were identified by long-range PCR screens with the primers labeled in Supplemental Figures S8–S13 and confirmed by Southern blot. The primer sequences are listed in Supplemental Table 1.
Piggy-Bac transgenics in ES cells
All cDNA expression vectors used in this study were constructed as derivatives of the Piggy-Bac transposon (pCyl50) with a CAG promoter to express genes of interest and a PGK or Pol2 promoter to express drug resistance genes (Hygro, Puro, and Bsd) (W. Wang et al. 2008). The CAG-CreERT2 and CAG-myr-HA-Akt1 transposons were modified from constructs obtained from Addgene. The N22HA-tagged hAgo1, hAgo2, mAgo3, mAgo4, and MIWI were initially constructed using DNA templates from either IMAGE clones or PCR-amplified cDNAs. To construct the full-length hAgo2 cDNA, a GC-rich 5′cDNA fragment was synthesized and ligated with the rest of the cDNA. All constructs were verified by complete or partial sequencing. To establish stable ES cell lines expressing the proteins of interest, we electroporated ES cells with both the PB transposons and CAG-PBase (pCYL96) and selected for stable ES cells by the drug resistance marker of the transposons.
siRNA, miRNA dsRNA duplexes, and shRNA expression vectors
The following RNA oligos were obtained from IDT and Dharmacon according to Doench et al. (2003): for the CXCR siRNA duplex, CXCR4 sense (5′-GAUUUCACUCCAGCUAACATT-3′) and CXCR4 antisense (5′-UGUUAGCUGGAGUGAAAUCTT-3′); for CXCR miRNA duplexes, CXCR4.bottom (5′-UGUUAGCUGGAGUGAAAACUU-3′) CXCR4.bulge1.top (5′-GUUUUCACAAAGCUAACACA-3′) and CXCR4.bulge2.top (5′-GUU AUUCACAAGGAGCUAACACA-3′). The oligos were mixed 1:1 and annealed (5 min at 80°C, then the temperature was decreased 0.01°C/sec to 50°C and held for 30 min at 50°C then decreased at 2°C/sec to 4°C). To construct mU6-expressed shRNA vectors, the following oligos were phosphorylated, annealed, and ligated into a BbsI- and XhoI-digested mU6 vector: for firefly luciferase, shRNA luc1 sense (5′-TTTGTGTCGCTCTGCCTCATAGAATTATTATTTCTATGAGGCAGAGCGACACTTTTTC-3′) and shRNA luc1 antisense (5′-TCGAGAAAAAGTGTCGCTCTGCCTCATAGAAATAATAATTCTATGAGGCAGAGCGACA-3′); for siRNA version of Has-mir-30a, 1148 (5′-TTTGCTGCAAACATCCGACTGGAAGTTATTAATCTTTCAGTCGGATGTTTGCAGCTTTTTC-3′) and 1149 (5′-TCGAGAAAAAGCTGCAAACATCCGACTGAAAGATTAATAACTTCCAGTCGGATGTTTGCAG-3′); for miRNA version of Has-mir-30a, 2264 (5′-TTTGCTGCAAACATCCTCGACTGGAAGTTATTAATCTTTCAGTCGGATGTTTGCAGCTTTTTC-3′) and 2265 (5′-TCGAGAAAAAGCTGCAAACATCCGACTGAAAGATTAATAACTTCCAGTCGAGGATGTTTGCAG-3′).
Protein analysis and immunostaining
Western blot analyses of several proteins expressed in ES cells and the detection of apoptotic cells by an active caspase-3 immunostaining (Cell Signaling) were performed as described previously (Wang et al. 1996).
Cell cycle analysis
FACScan analysis for cell cycle profiles using propidium iodide (PI) were performed as described previously (Wang et al. 1996) using a Beckman Coulter Epics XL-MCL at Northwestern's Flow Cytometry Core Facility.
RT–PCR and quantitative RT–PCR analysis
Total RNA was prepared using Trizol (Invitrogen), and reverse transcription was performed using Superscript III (Invitrogen) with an oligodT primer. For miRNA analysis, TaqMan microRNA assays for mir-19, mir-21, mir-130, mir-290, and sno135 were used according to the manufacturer's instructions (Applied Biosystems). Quantitative RT–PCR analysis was performed using a SYBR green kit (Applied Biosystems) on a 7500 Real-Time PCR System. RT–PCR and quantitative PCR primer sequences are provided in Supplemental Table 2.
ES cell transfection
For transfection, ES cells were passaged at least twice on gelatinized plates in ES cell media containing LIF to remove feeders. Feeder-free ES cells were then plated at 2 × 105 cells per well on a 24-well plate to transfect the following day. Cells were transfected using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). A ratio of 5:1 for Ff- and Rr-Luc was used (0.1 μg of Ff-Luc reporter and 0.02 μg of Rr-Luc reporter). For assays using DNA effectors, a concentration of 0.25 μg of effector was used. For assays using dsRNA, a final concentration of 2 nM was used. The transfected cells were harvested 48 h after transfection and analyzed with a dual-luciferase kit from Promega.
Antibodies
Antibodies were obtained from the following sources: Active Caspase-3 (Cell Signaling), Bim (Assay Designs), Bad (MBL), Bax (MBL), Ras (Millipore), PTEN (Millipore), E2F1 (Santa Cruz Biotechnologies), Nanog (Bethyl), Oct3/4 (Santa Cruz Biotechnologies), Sox2 (Abcam), β-tubulin (Developmental Studies Hybridoma Bank [DSHB]), Ago2 (Cell Signaling), and HA (DSHB).
Acknowledgments
We thank A. Bradley and G. Guo for NM5 ES cells, N.G. Copeland for reagents of BAC recombineering, the Welcome Trust Sanger Center for distributing 129Sv BAC clones, C. Cepko for CreERT2 plasmid, W. Sellers for myrHA-Akt1 plasmid, and N.H. Colburn for anti-PDCD4 antibody. This work was supported by grants from the NIH (5R21GM079528) and the Illinois Department of Public Health to X.W.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1749809.
Supplemental material is available at http://www.genesdev.org.
References
- Azuma-Mukai A., Oguri H., Mituyama T., Qian Z.R., Asai K., Siomi H., Siomi M.C. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Calabrese J.M., Seila A.C., Yeo G.W., Sharp P.A. RNA sequence analysis defines Dicer's role in mouse embryonic stem cells. Proc. Natl. Acad. Sci. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S., Jung S., Rothenberg S.M., Smolen G.A., Mlody B.G., Haber D.A. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc. Natl. Acad. Sci. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkers P.F., Birkenkamp K.U., Lam E.W., Thomas N.S., Lammers J.W., Koenderman L., Coffer P.J. FKHR-L1 can act as a critical effector of cell death induced by cytokine withdrawal: Protein kinase B-enhanced cell survival through maintenance of mitochondrial integrity. J. Cell Biol. 2002;156:531–542. doi: 10.1083/jcb.200108084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T., Zamore P.D. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Forstemann K., Horwich M.D., Wee L., Tomari Y., Zamore P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T., Nogami M., Yoshikawa M., Ikeno M., Okazaki T., Takami Y., Nakayama T., Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A.E., Conte D., Li N., Parrish S., Ha I., Baillie D.L., Fire A., Ruvkun G., Mello C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grishok A., Sinskey J.L., Sharp P.A. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes & Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Wang W., Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha–DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Zamore P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Villeneuve L.M., Morris K.V., Rossi J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Koralov S.B., Muljo S.A., Galler G.R., Krek A., Chakraborty T., Kanellopoulou C., Jensen K., Cobb B.S., Merkenschlager M., Rajewsky N., et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Lai E.C. miRNAs: Whys and wherefores of miRNA-mediated regulation. Curr. Biol. 2005;15:R458–R460. doi: 10.1016/j.cub.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Lee Y.S., Nakahara K., Pham J.W., Kim K., He Z., Sontheimer E.J., Carthew R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Liu P., Jenkins N.A., Copeland N.G. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lu Z., Liu M., Stribinskis V., Klinge C.M., Ramos K.S., Colburn N.H., Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.B., Yuan Y.R., Meister G., Pei Y., Tuschl T., Patel D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Mello C.C., Conte D., Jr Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C., et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C. Specificity of ARGONAUTE7–miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor L., Strasser A., O'Reilly L.A., Hausmann G., Adams J.M., Cory S., Huang D.C. Bim: A novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Okamura K., Lai E.C. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K., Ishizuka A., Siomi H., Siomi M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Artus C.G., Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X.J., Wildey G.M., Howe P.H. Evidence that Ser87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J. Biol. Chem. 2006;281:813–823. doi: 10.1074/jbc.M505546200. [DOI] [PubMed] [Google Scholar]
- Qi Y., He X., Wang X.J., Kohany O., Jurka J., Hannon G.J. Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature. 2006;443:1008–1012. doi: 10.1038/nature05198. [DOI] [PubMed] [Google Scholar]
- Rand T.A., Ginalski K., Grishin N.V., Wang X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri P.C., Rozenblatt-Rosen O., Caplen N.J., Wolfsberg T.G., Umayam L., Lee J.C., Hughes C.M., Shanmugam K.S., Bhattacharjee A., Meyerson M., et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H., Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Steiner F.A., Hoogstrate S.W., Okihara K.L., Thijssen K.L., Ketting R.F., Plasterk R.H., Sijen T. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tolia N.H., Joshua-Tor L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- Tomari Y., Du T., Zamore P.D. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Z., Lawson B., Brewer J.W., Zinszner H., Sanjay A., Mi L.J., Boorstein R., Kreibich G., Hendershot L.M., Ron D. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153) Mol. Cell. Biol. 1996;16:4273–4280. doi: 10.1128/mcb.16.8.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lin C., Lu D., Ning Z., Cox T., Melvin D., Wang X., Bradley A., Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sheng G., Juranek S., Tuschl T., Patel D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Belasco J.G. Let me count the ways: Mechanisms of gene regulation by miRNAs and siRNAs. Mol. Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Wu L., Fan J., Belasco J.G. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr. Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Srinivasan L., Calado D.P., Patterson H.C., Zhang B., Wang J., Henderson J.M., Kutok J.L., Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat. Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M., Irvine D.V., Martienssen R.A. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]