Abstract
Calicivirus structure proteins are expressed from a subgenomic mRNA with two overlapping cistrons. The first ORF of this RNA codes for the viral major capsid protein VP1, and the second for the minor capsid protein VP2. Translation of VP2 is mediated by a termination/reinitiation mechanism, which depends on an upstream sequence element of ∼70 nucleotides denoted “termination upstream ribosomal binding site” (TURBS). Two short sequence motifs within the TURBS were found to be essential for reinitiation. By a whole set of single site mutations and reciprocal base exchanges we demonstrate here for the first time conclusive evidence for the necessity of mRNA/18S rRNA hybridization for translation reinitiation in an eukaryotic system. Moreover, we show that motif 2 exhibits intramolecular hybridization with a complementary region upstream of motif 1, thus forming a secondary structure that positions post-termination ribosomes in an optimal distance to the VP2 start codon. Analysis of the essential elements of the TURBS led to a better understanding of the requirements for translation termination/reinitiation in eukaryotes.
Keywords: Translation initiation, termination, reinitiation, TURBS, mammalian mRNA translation
Translation of proteins from mRNAs represents one of the basic processes in living organisms that allows us to differentiate them from nonliving replicating molecular systems like viruses (Sachs et al. 1997; Kapp and Lorsch 2004). In contrast to prokaryotes, the overwhelming amount of mRNAs in eukaryotes is monocistronic. Accordingly, the eukaryotic mRNAs are usually organized in a way that the translational start site is located rather close to the 5′ end, so that protein synthesis can be initiated by the so-called scanning process (Kozak 1987, 1989). This process implies assembly of a preinitiation complex at the mRNAs 5′-terminal cap structure and movement of this complex in 3′ direction to reach the initiation site. There are only rare cases in eukaryotic translation that deviate from this rule and start protein synthesis at internal sites of a mRNA (Sonenberg and Hinnebusch 2007). These processes are either promoted by an internal ribosomal entry site (IRES) (Hellen and Sarnow 2001; Pestova et al. 2001; Sarnow et al. 2005), discontinuous movement of the ribosome in a process named “ribosomal shunting” (Latorre et al. 1998; Cuesta et al. 2001; Park et al. 2001; Ryabova et al. 2002; Xi et al. 2005), or reinitiation of protein synthesis after termination of translation of an upstream cistron (Kozak 1987, 2002; Meyers 2003; Gould and Easton 2005, 2007; Ryabova et al. 2006; Luttermann and Meyers 2007; Pöyry et al. 2007).
The so far best studied case of translation reinitiation, occurring in higher eukaryotic cells, is found in caliciviruses (Meyers 2003, 2007; Luttermann and Meyers 2007; Pöyry et al. 2007), a group of important pathogens that cause gastrointestinal diseases in humans and a variety of sometimes fatal syndromes in animals (Green et al. 2000). Characteristic features of the family Caliciviridae are the formation of nonenveloped virions of typical morphology and the nonsegmented ssRNA genomes of positive polarity. The calicivirus genome is 7–8 kb long and contains two or three functional ORFs (Fig. 1; Green et al. 2000). ORF1 codes for the nonstructural proteins, which are translated as a polyprotein that is processed post-translationally by a virus-encoded protease. A 3′-terminal small ORF codes for the minor capsid protein VP2 with a size that varies among the caliciviruses from 8 to 20 kDa. The major capsid protein VP1 (∼60 kDa) is encoded by a separate ORF2 in members of the genera Vesivirus and Norovirus or is part of the ORF1-encoded polyprotein in viruses belonging to the genera Lagovirus and Sapovirus. As a unique feature of the genus Vesivirus the VP1 protein is translated as a precursor protein (Fig. 1), which is cleaved into the leader protein and the mature capsid protein by a viral protease (Sosnovtsev et al. 1998). VP1 represents the major protein component of the virion, the basic structure of which is made up by 180 copies of this protein. In contrast, the viral particle contains only a few molecules of VP2 (Prasad et al. 1994). The function of VP2 is unknown but it was found to be essential for virus infectivity (Sosnovtsev et al. 2005).
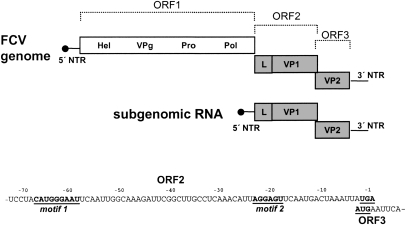
Figure 1.
Schematic map of FCV genomic and subgenomic RNA and location of the essential elements of the TURBS in the 3′-terminal sequence of ORF2. In the scheme, the basic structure and organization of the FCV genomic and subgenomic RNAs are illustrated (not drawn to scale). Gray bars represent ORF2 and ORF3 coding for the capsid proteins. The white bar symbolizes the nonstructural protein coding region (ORF1). Below, the 3′-terminal 72 nt of the ORF2 sequence are shown. Motif 1 and motif 2, determined by previous analyses (Luttermann and Meyers 2007), and the start/stop codons in the overlap region of ORF2 and ORF3 are highlighted. VPg (virus protein, genome-linked) present at the 5′ ends of both the genomic and subgenomic RNAs is symbolized by a black circle at the end of a line representing the 5′ nontranslated regions (NTR) of the RNAs. The encoded proteins are indicated by abbreviations. (Hel) Putative helicase domain; (VPg) virus protein, genome linked; (Pro) 3C-like protease; (Pol) RNA-dependent RNA polymerase; (L) leader; (VP1) major capsid protein; (VP2) minor capsid protein.
The genomic RNA serves as a template for the synthesis of the nonstructural polyprotein while a 3′ coterminal subgenomic mRNA is used for translation of the major capsid protein VP1 and the minor capsid protein VP2 (Herbert et al. 1996; Luttermann and Meyers 2007). As a special feature of the calicivirus genome organization the frames coding for the two capsid proteins overlap by 1–8 nucleotides (nt). Both the full-length and the subgenomic RNAs do not have a 5′ cap, but carry a covalently linked viral protein (VPg). VPg serves as a primer for genome replication (Rohayem et al. 2006) and presumably functions as a cap substitute promoting translation of the viral RNAs. To achieve translation initiation, VPg interacts with initiation factor 4E and/or eukaryotic initiation factor 3 (eIF3) (Daughenbaugh et al. 2003; Goodfellow et al. 2005; Chaudhry et al. 2006).
Translation initiation close to the RNA's 5′ end is mediated by the bound VPg protein and leads to expression of the 5′-terminal ORFs of both the genomic and subgenomic RNAs. An alternative mechanism is required to allow access of ribosomes to the downstream frame and translation of VP2 from the bicistronic subgenomic mRNA. Previous studies on FCV (Vesivirus) and RHDV (Lagovirus) showed that translation of VP2 is achieved by a termination/reinitiation mechanism (Meyers 2003, 2007; Luttermann and Meyers 2007; Pöyry et al. 2007). Since eukaryotic translation is usually initiated close to the 5′ end and not designed for promoting a restart after termination, caliciviruses use a special mechanism to promote reinitiation. This mechanism depends on an upstream sequence named termination upstream ribosomal binding site (TURBS; ∼70 nt), which is thought to bind the post-termination ribosome and therefore increase the chance of reinitiation (Meyers 2003, 2007; Luttermann and Meyers 2007; Pöyry et al. 2007). The TURBS region contains two short sequence motifs essential for VP2 translation as determined by deletion mapping (Fig. 1; Luttermann and Meyers 2007; Meyers 2007). Motif 1 is found to be conserved among caliciviruses and is located at similar positions in the mRNAs of the different caliciviruses, upstream of the 3′-terminal ORFs. This motif is the core of a region, which is complementary to the loop region of helix 26 within 18S rRNA. Hybridization of motif 1 to 18S rRNA could tether the ribosome to the viral RNA. Published data indicate interaction of motif 1 with initiation factor eIF3 (Pöyry et al. 2007), a process that could assist the hypothetical hybridization-mediated contact or could represent an alternative mechanism for tethering of the post-termination ribosome. In contrast to motif 1, the sequence of motif 2 is not conserved. This sequence was supposed to play a role in positioning of the ribosome relative to the start site of the 3′-terminal ORF (Luttermann and Meyers 2007; Meyers 2007).
In this study, experiments are described that allow us to evaluate the importance of the proposed hybridization of motif 1 to 18S rRNA and give for the first time more detailed insight into the function of motif 2.
Results
Characterization of motif 1 by single base substitutions
A major determinant for VP2 expression is represented by TURBS motif 1 that was proposed to establish intermolecular base pairing to a sequence in helix 26 of ribosomal 18S rRNA (Luttermann and Meyers 2007; Meyers 2007). The pentameric core of motif 1 (UGGGA) is conserved among caliciviruses. In the earlier analyses a limited number of single base exchanges in this sequence were tested for their effects on VP2 expression. The results supported the predicted interaction of motif 1 with the complementary region of 18S rRNA (Luttermann and Meyers 2007; Meyers 2007). To obtain further support for the hypothesis we introduced additional mutations in the motif of the FCV RNA in order to investigate the importance of every single nucleotide (Fig. 2). At two positions (nucleotides −66 and −61) two exchanges were tested, the substitution of G for A, which preserves the putative binding to the 18S rRNA by a G–U pairing, and an exchange C for A, which prevents the binding at this position. As described before (Luttermann and Meyers 2007) analysis of VP2 expression was done via transient expression, quantitative immunoprecipitation, and phosphoimager determination of the incorporated radiolabel in the proteins detected after SDS-PAGE. Expression of VP1 from the preceding ORF in the same plasmid served as an internal control for normalization of transfection efficiency and the general expression level. The VP2 translation efficiency from mutated RNAs was compared with the expression level of a wild-type construct and showed for most substitutions an influence on VP2 synthesis.
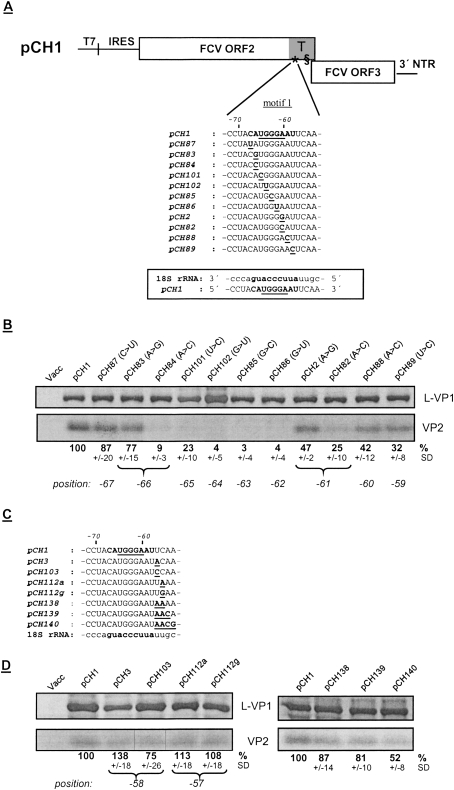
Figure 2.
Influence of single base substitutions in motif 1 on VP2 expression. (A) On top, a schematic representation of the basic structure of the pCH1 construct is given (not drawn to scale). The TURBS region at the 3′-terminal end of ORF2 is highlighted in gray (T) and the essential motif 1 (*) and motif 2 (§) within the TURBS region are indicated. The sequences below and in C represent part of FCV ORF2 containing motif 1, written in bold. The pentanucleotide conserved among calicivirus sequences is underlined. Below the wild-type sequence, the sequences of the mutants with single substitutions within motif 1 (A) and mutations downstream from the region showing complementarity to the 18S rRNA (C) are shown with the exchanged residues highlighted and underlined. (A) In the box below, the sequence of the mouse 18S rRNA (nucleotides 1105–1121) is given in 3′–5′ orientation and the 9 nt complementary to motif 1 of the FCV RNA are marked in bold. The wild-type FCV mRNA sequence (5′–3′orientation) is aligned to the 18S rRNA sequence and motif 1 is marked in bold. (B,D) Gels with the proteins precipitated after transient expression of the indicated constructs. (B) The results for the exchanges within motif 1; (D) the substitutions downstream from the region showing complementarity to the 18S rRNA in the wild-type FCV mRNA. The calculated VP2 expression efficiencies in percent and the standard deviations (SD) are given below the autoradiographs (relative to wild-type pCH1, normalized to the expression of L-VP1). Braces connect the different mutations located at the same position of the FCV ORF2 sequence. Below the braces the positions of the mutations are given relative to the ORF2 3′ end. Cells infected with vaccinia virus MVA-T7 served as a negative control (Vacc). Please note that Northern blots conducted during earlier experiments with pCH1 and mutants thereof gave no indication for the transcription of smaller RNAs coding for VP2 as could be expected if a cryptic T7 promoter was located in the TURBS region (Luttermann and Meyers 2007), so that VP2 is only expressed from the bicistronic RNA.
All substitutions disturbing the putative interaction between motif 1 and the 18S rRNA showed reduced VP2 synthesis (Fig. 2B). The strongest reduction was observed for the mutants affecting the three Gs at position −62 to −64, showing nearly a complete loss of VP2 expression (pCH102, pCH85, pCH86) (Fig. 2B). The substitution of C for A at position −66 (pCH84) also led to a strong reduction of VP2 translation efficiency (9% of wild-type level). At positions −65 (pCH101) and −61 (pCH82) the exchange of C for U or A, respectively, resulted in reduction of VP2 expression to ∼25%. There was also a reduction of VP2 synthesis by mutations at positions −60 (A to C, pCH88) or −59 (U to C, pCH89) to 42% or 32% of the wild-type level, respectively. The substitution at position −67 (C to U, pCH87) showed just a mild reduction of VP2 translation to 87%. This mutation would preserve the binding to the 18S rRNA via a U–G pairing. At this position a mutation preventing base-pairing was not possible, because such a change would have generated a stop codon within the ORF2 sequence. Exchanges at positions −66 (A to G, pCH83) and −61 (A to G, pCH2) that preserved the putative hybridization via a G–U pair exhibited higher levels of VP2 translation than substitutions at the same positions that would prevent hybridization with the 18S rRNA. At position −61 mutations preventing the binding (A to C, pCH82) reduced VP2 translation (25% of wild-type level) much more than mutations that still allowed pairing (A to G, pCH2, 47% of wild-type level). At position −66 this difference was even bigger with an approximately eightfold higher VP2 translation efficiency for the G for A mutant, allowing a G–U pair, compared with the exchange C for A (VP2 expression of 77% and 9% for mutants pCH83 and pCH84, respectively).
Taken together, these results demonstrate the importance of the complete motif 1 sequence for VP2 translation. Moreover, the data strongly support the hypothesis that motif 1 hybridizes to 18S rRNA.
All the tested substitutions within motif 1 (nucleotides −67 to −59) have a clear negative influence on VP2 expression, unless they preserve the proposed interaction with the 18S rRNA via a G–U hybridization. To analyze if sequences flanking the putative binding region are important for VP2 expression and if VP2 translation efficiency can be enhanced by elongation of the stretch of hybridizing nucleotides of the FCV and 18S rRNA, substitutions of nucleotides downstream from the motif 1 sequence were made (Fig. 2C). At position −58 exchange of A for U (pCH3) could lead to an A–U pairing between FCV RNA and 18S rRNA whereas C for U (pCH103) has no influence on the proposed interaction. The latter exchange (pCH103) had a mild repressive effect on VP2 translation with 75% of wild-type level, whereas the substitution that would allow an additional base pairing (pCH3) raised VP2 translation to 138% of wild-type activity (Fig. 2D). At position −57, two exchanges were introduced (A or G for C, pCH112a or pCH112g) that allowed an additional pairing (A–U or G–U) that was separated from the rest of the heteroduplex by one nonpaired residue. These mutations showed translation efficiencies near the wild-type level with 113% or 108%, respectively. A continuous elongation of the stretch of hybridizing nucleotides of the FCV and 18S rRNA by 2, 3, or 4 nt (pCH138–pCH140) led to a reduction of VP2 translation efficiency to 87%, 81%, or 52%, respectively. Thus, only the lengthening of the complementary region by one base pairing is able to enhance translation of the second frame, whereas further elongation of the hybridizing sequence has a negative influence on the reinitiation efficiency. In consequence a stretch of 10 18S rRNA complementary residues gives the highest level of VP2 translation. Taken together the results show that the flanking nucleotides tested here have only a minor effect on VP2 synthesis.
Verification of the hybridization model in a yeast system
Direct evidence that motif 1 interacts with the helix 26 of 18S rRNA requires the ability to manipulate ribosomal 18S rRNA in order to generate reciprocal mutations. To this end we used a yeast system that enabled us to alter rRNA sequences. The Saccharomyces cerevisiae strain NOY908 lacks all chromosomal copies of the 35S rRNA gene and expresses the rRNA precursor from the episomally maintained plasmid pNOY373 (Wai et al. 2000). Transfection of a plasmid encoding a further copy of the rRNA precursor allows the provision of mutated rRNAs when sequence alterations are introduced in this plasmid prior to transfection. In order to test translation reinitiation templated by a FCV mRNA-like RNA in this system, NOY908 cells were transformed with two plasmids, pNOY353, a yeast 18S rRNA plasmid, and the FCV test plasmid pYF21A. The latter contains a cDNA fragment with two frames arranged in the same way as in FCV subgenomic mRNA. The first frame in the construct is equivalent to part of FCV ORF2, codes for VP1, and contains the TURBS together with the start/stop region. The second frame in the construct overlaps the preceding ORF as in the FCV mRNA. It starts with the first 8 nt of FCV ORF3, the rest of which was replaced by a sequence coding for GFP fused with a 3′-terminal c-Myc tag (GFPmyc) (Fig. 3A).
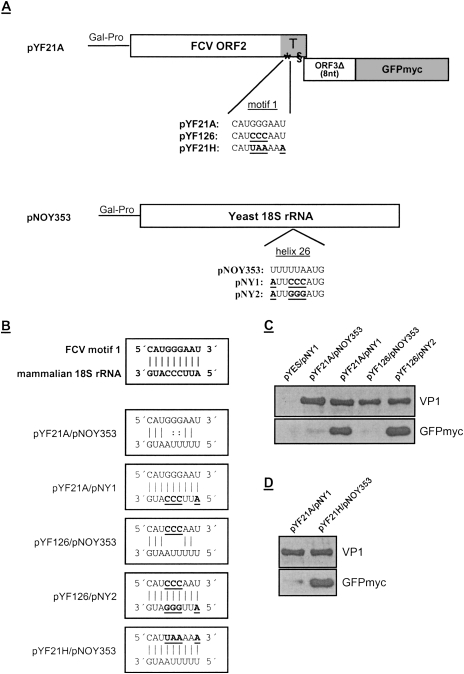
Figure 3.
Interaction of motif 1 of the FCV TURBS with helix 26 of 18S rRNA. (A) Schematic drawing of the plasmids introduced into the yeast strain NOY908. On top, pYF21A is indicated, containing two frames arranged the same way as in FCV subgenomic mRNA. The first frame corresponds to FCV ORF2, codes for VP1, and contains the TURBS (T, highlighted in gray) with the two essential motifs indicated. The second frame starts with the first 8 nt of the VP2 coding sequence that is fused with the sequence coding for GFP with a 3′-terminal c-Myc tag (GFPmyc). Below the schematic representation of pYF21A, the motif 1 sequence is shown for FCV wild type and for substitution mutants pYF21H and pYF126, with the exchanges within motif 1 highlighted. Below, the pNOY353 construct is shown, coding for the 18S rRNA of yeast, and the sequence of helix 26 (nucleotides 1056–1164 in yeast 18S rRNA) is given. Mutations introduced into this region in the constructs pNY1 and pNY2 are indicated. (B) On top, the putative base pairing between the FCV TURBS motif 1 and the mammalian 18S rRNA sequence is indicated by vertical lines. Below, the possible hybridizing residues of the motif 1 regions of the indicated pYF constructs and the helix 26 sequences of the 18S rRNA constructs are illustrated for the plasmid combinations introduced into the yeast strain NOY908. (C,D) The proteins translated in the yeast cells, transfected with the plasmid combinations given above the gels, are shown in a Western blot analysis with antisera against VP1 (top panel) or GFP (bottom panel). Please note that in comparison with C the blot with the anti GFP serum in D was exposed for a much shorter time to be able to demonstrate the increase in GFPmyc expression in the right lane. The ratio between VP1 and GFPmyc for the construct combination pYF21A/pNY1 can therefore not be compared between the blots C and D. (Gal-Pro) Yeast galactose-promoter.
To analyze whether a reinitiation event leading to synthesis of GFPmyc occurred and, if so, which requirements have to be fulfilled for reinitiation, different derivates of these plasmids were established and introduced into the cells. These constructs contained mutations affecting the complementary regions of TURBS motif 1 and 18S rRNA helix 26 (Fig. 3A,B). The proteins expressed from plasmid pYF21A in the yeast system were analyzed by SDS-PAGE and Western blotting.
First we investigated the efficiency of GFPmyc synthesis in yeast cells transfected with a combination of plasmids pYF21A (FCV/GFPmyc) and pNOY353 (yeast 18S rRNA) that display the respective wild-type sequences. The yeast 18S rRNA helix 26 differs in the region complementary to TURBS motif 1 by 4 nt from the mammalian sequence. Interaction between yeast 18S rRNA and motif 1 would nevertheless be possible via 7 nt, because two of the differing nucleotides would lead to G–U pairing. Analysis of the proteins translated from pYF21A revealed a weak expression of the second frame coding for GFPmyc (Fig. 3C). This could be interpreted in a way that interaction via the seven complementary nucleotides between yeast 18S rRNA and FCV motif 1 was sufficient to mediate some reinitiation events. To improve the hybridization proposed to be essential for VP2 translation in mammalian cells, plasmid pNY1 was established. In pNY1 the helix 26 sequence of yeast 18S rRNA was changed at four positions, generating the sequence found in mammalian 18S rRNA at the proposed binding site, thus allowing the interaction via all 9 nt as proposed for the mammalian system. When pNY1 was transfected together with pYF21A, a GFPmyc expression was observed that was much stronger than the signals detected for the combination of pYF21A and the yeast pNOY353 plasmid (Fig. 3C).
To analyze if less than seven complementary bases led to a loss of GFPmyc synthesis, pYF21A was changed by replacing the three Gs in the core of motif 1 with C residues resulting in plasmid pYF126. This mutated motif 1 could interact with the yeast 18S rRNA only via five complementary nucleotides (Fig. 3B). Nevertheless, transfection of plasmids pYF126 and pNOY353 still led to a low level of GFPmyc synthesis (Fig. 3C). However, adaptation of the corresponding region in 18S rRNA (pNY2) to the changes in motif 1 introduced in pYF126 enhanced the translation of GFPmyc remarkably (Fig. 3C, right lane). Thus, reciprocal substitutions in the putative hybridizing regions of FCV RNA and 18S rRNA restored the efficient translation of the second frame.
In the above-described analysis only a minor part of the ribosomes contained the mutated 18S rRNA encoded by plasmids pNY1 or pNY2, whereas the majority of the ribosomes should carry the wild-type yeast rRNA transcribed from the pNOY373 plasmid (Dresios et al. 2006), present in the cells to provide ribosomes for translation of yeast proteins. To enhance the expression of the second frame and to confirm the interaction between the mRNA and the rRNA, FCV TURBS motif 1 was mutated at four positions in a way that the sequence was complementary to the 18S rRNA sequence of yeast (pYF21H) (Fig. 3A,B). In this setting all ribosomes of the transfected yeast cells would be able to interact with the mutated FCV mRNA sequence. Analysis of protein synthesis resulting from the plasmid combination pYF21H/pNOY353 showed a remarkably high level of GFPmyc expression that exceeded the level observed for pYF21A/pNY1 by far (Fig. 3D).
These experiments definitely show that motif 1 promotes translation of the downstream ORF via hybridization with 18S rRNA.
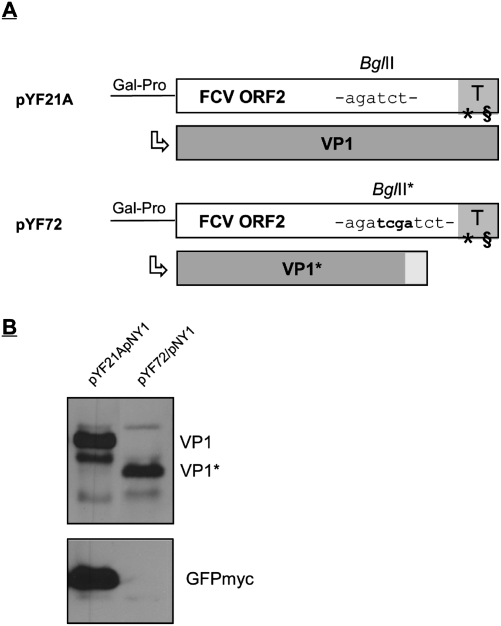
We also had to prove that VP2 expression in yeast is due to reinitiation of translation since tethering of ribosomes to a mRNA could also result in a functional IRES. In the mammalian cells we showed that translation of the second frame is not mediated via an IRES (Luttermann and Meyers 2007), but in yeast cells the mechanism could be different. The finding that translation of the downstream frame occurred even when only 5 nt were able to hybridize could indicate mechanistic differences in the yeast cells since such a weak interaction was not able to promote reinitiation in mammalian cells. It was therefore necessary to show that expression of GFP in yeast resulted from reinitiation of translation after termination at the end of the upstream ORF. To this end, construct pYF72 was generated that is based on pYF21A but contains an insertion of 4 nt at the BglII site of the upstream ORF leading to a frameshift and therefore to termination of VP1 translation ∼200 nt upstream of the start/stop site (Fig. 4A). It has been shown before that such a construct is no longer able to promote downstream ORF translation in the mammalian system (Luttermann and Meyers 2007). As expected, transfection of the plasmid combination pYF72/pNY1 led to translation of a smaller VP1 protein (VP1*) (Fig. 4B). Importantly, GFPmyc translation was not observed for this combination (Fig. 4B). Thus, also in the yeast system initiation of translation of the second frame depends on translation of the first frame down to the start/stop site just as was shown before in mammalian cells.
Figure 4.
Translation of the second frame in pYF constructs in yeast depends on translation of the first frame. (A) Schematic drawing of the upstream frame of plasmid pYF72 compared with pYF21A (not drawn to scale). The TURBS (T) is highlighted in gray and the two essential motifs are indicated. Insertion of four extra nucleotides (written in bold) at a BglII site (BglII*) 275 nt upstream of the ORF2 stop codon results in a frameshift and premature termination of ORF2 translation. The gray bars below indicate the proteins encoded by ORF2. The VP1* expressed from the frameshift mutant pCH72 is truncated by 70 amino acids, and the C-terminal 18 amino acids of the protein represent non-FCV residues, resulting from the introduced frameshift (light-gray area). Note that ORF3 is not shown in the scheme. (B) The proteins translated in the yeast system are shown in a Western blot analysis with antisera against VP1 (top panel) or GFP (bottom panel). (Gal-Pro) Yeast galactose-promoter.
Characterization of motif 2 by single substitutions
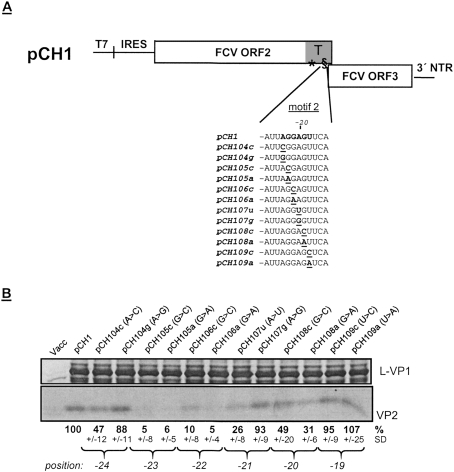
In comparison with the situation with motif 1, hypotheses with regard to the role of TURBS motif 2 in translation reinitiation are much less defined. Previous analyses indicated that this motif might be engaged in positioning the ribosome relative to the VP2 start codon (Luttermann and Meyers 2007; Meyers 2007). To further elucidate the function of motif 2 in the reinitiation mechanism we wanted to determine the specific importance of individual nucleotides within the sequence of this motif. For this purpose single base substitutions were introduced at each position of the crucial hexamer sequence (Fig. 5A). Two different exchanges were made for each nucleotide of the original sequence and the resulting mutants were tested in the mammalian cell system.
Figure 5.
Influence of single substitutions in motif 2. (A) On top, a schematic representation of the basic structure of the pCH1 construct is given (not drawn to scale). The TURBS region at the 3′-terminal end of ORF2 is highlighted in gray (T) and the two essential motifs within the TURBS region are indicated. The sequences below represent part of the FCV ORF2 sequence containing motif 2, written in bold. Mutants thereof with substitutions in motif 2 are shown below with the exchanged residues highlighted and underlined. (B) Gels with the proteins precipitated after transient expression of the indicated constructs. The calculated VP2 expression efficiencies in percent and the standard deviations (SD) are given below the autoradiographs (wild-type pCH1 set to 100%; values were normalized to the expression of L-VP1). Braces mark the different mutations affecting the same position of the FCV ORF2 sequence. Below the braces the positions of the mutated residues are given relative to the ORF2 3′ end. Cells infected with vaccinia virus MVA-T7 served as a negative control (Vacc). (T7) Phage T7 promoter; (IRES) internal ribosome-binding site from Encephalo myocarditis virus.
The strongest effect was seen for the exchange of A or C for G at the positions −23 and −22 of the wild-type sequence with an expression level of 5%–10% (pCH105, pCH106) (Fig. 5B). Exchange of C or A for G at position −20 (pCH108) resulted in a milder but still pronounced reduction to 49% or 31%, respectively. Substitution of the last nucleotide of the hexamer at position −19 (U to C or A, pCH109) led to VP2 translation at about wild-type efficiency. The substitutions at positions −24 (pCH104) and −21 (pCH107) showed different rates of VP2 translation dependent on the inserted nucleotide. The exchange of G for A at these two positions had only minor effects on VP2 expression (88% or 93% of wild-type level, respectively), whereas mutation of A to C (nucleotide −24) or A to U (nucleotide −21) led to reduction of VP2 expression to 47% or 26%, respectively. These results could be indicative of interaction of motif 2 with another nucleic acid sequence, as the substitution of G for A would enable hybridization via a G–U instead of A–U pairing, whereas the substitutions of C or U for A would prevent hybridization.
Motif 2 interacts with a cis-element
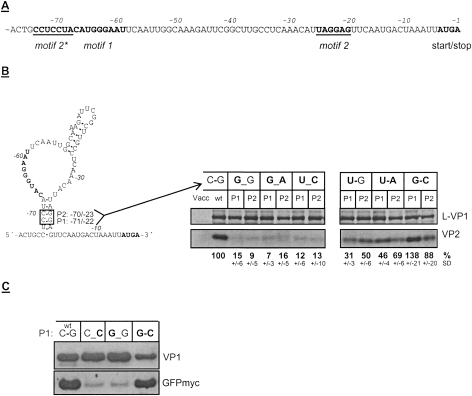
In contrast to motif 1, the primary sequence of motif 2 is not conserved among caliciviruses. Nevertheless, the results of the single substitutions described above indicate an interaction with a nucleic acid. Hybridization of a variable viral sequence with a probably rather conserved cellular counterpart seems not very likely and we therefore searched for a region within the FCV RNA complementary to motif 2. We found a hexamer (nucleotides −73 to −68) directly upstream of motif 1 (Fig. 6A), which is complementary to a sequence located at nucleotides −25 to −20, which is almost identical to the region −24 to −19 mapped before as motif 2. The secondary structure resulting from the putative hybridization of these regions shown in Figure 6B is similar to the structure proposed by Pöyry et al. (2007). To analyze the importance of the reverse complementary region, now designated motif 2*, mutants were generated with single substitutions within this sequence. The C residues at positions −71 (P1) and −70 (P2) in the stem region of the predicted structure were selected for mutation (Fig. 6B). These residues are located at positions corresponding to the motif 2 residues that showed the highest reduction of VP2 translation upon mutation (nucleotides −22 and −23, pCH105, pCH106) (see Fig. 5B). The different mutations in motif 2* were designed to disrupt the putative binding to motif 2 (G_G) or to preserve this hybridization by a U–G pairing. Therefore, substitutions of U or G for the C were introduced at both of these positions. Furthermore, the same substitutions were made in the context of the above-described mutants with changes in motif 2 (A or C for G at positions −23 and −22). This led in some cases to reciprocal exchanges restoring complementarity (e.g., mutation of C to A at position −71 and mutation G to U at position −22 would allow an A–U pairing). Thus the different exchanges disrupt (C–G to U_C or G_A) or restore (C–G to U–A or G–C) the putative binding between these two motifs.
Figure 6.
Interaction between motif 2 and a complementary sequence upstream of motif 1. (A) The 3′-terminal 78 nt of FCV ORF2 are given. The essential elements start/stop site, motif 1, motif 2, and the region complementary to motif 2 (designated motif 2*) are highlighted. (B) On the left side, the putative secondary structure of the sequence shown above, based on the interaction between motif 2 and motif 2*, is indicated. Two positions (P1 and P2) within the stem formed by motif 2* and motif 2 are marked. At these positions substitutions in the two motifs were introduced and analyzed. The autoradiograph on the right side shows the proteins L-VP1 and VP2 translated from the different mutants. In all cases the same combination of nucleotides was tested at position P1 (nucleotides −71 and −22) and at position P2 (nucleotides −70 and −23) as indicated just above the lanes. The calculated VP2 expression efficiencies in percent and the standard deviations (SD) are given below the autoradiographs (wild-type pCH1 set to 100%; values were normalized to the expression of L-VP1). (C) The proteins translated in yeast cells after transfection of the plasmid pNY1 and a pYF plasmid with mutations in motif 2 and/or motif 2* at position P1 (nucleotides −71 and −22, given above the gel) are shown in a Western blot analysis with antisera against VP1 (top panel) or GFP (bottom panel).
Mutations that prevented the putative binding between motif 2* and motif 2 by substitution of one (G_G) or two of the complementary nucleotides (G_ A, U_ C) resulted in a clear reduction of ORF3 expression to 7%–16% compared with the wild-type construct with the putative C–G binding (Fig. 6B, left gel). The substitution of U for C in motif 2*, preserving the interaction via a U–G binding, resulted in a milder reduction to 31% (P1: nucleotide −71) or 50% (P2: nucleotide −70) (Fig. 6B, right gel). When reciprocal exchanges were made in both regions, permitting a binding by U–A or G–C pairing, the translation efficiency of VP2 was much higher than for the corresponding single mutations. U–A interaction resulted in VP2 translation levels of 46% (P1: nucleotide −71/−22) or 69% (P2: nucleotide −70/−23). The G–C pair at position −71/−22 showed an even higher VP2 translation efficiency than the wild type (138%), whereas the G–C pair at position −70/−23 resulted in an expression level near the wild type (88%) (Fig. 6B).
To confirm the importance of the interaction between motifs 2 and 2* the mutations at position P1 (nucleotide −71/−22) were tested again in the yeast system. Mutations that prevented the putative binding between motif 2* and motif 2 by substitution of one nucleotide (G_G, C_C) resulted in a dramatic reduction of GFPmyc translation compared with the wild-type construct with the putative C–G binding (Fig. 6C). In contrast, the reciprocal substitution of the residues at position −71/−22 resulting in a G–C pair led to a GFPmyc translation level comparable with that of the wild-type construct. These results clearly argue in favor of a hybridization of motif 2 and motif 2*, probably leading to a secondary structure similar to the one shown in Figure 6B.
Mutations confirm the secondary structure loop located between motif 2*/1 and motif 2
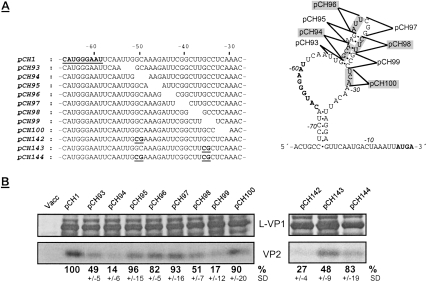
As shown in previous analyses, large deletions between motif 1 and motif 2 are tolerated without significant loss of VP2 expression (Luttermann and Meyers 2007), so that it can be concluded that neither the sequence itself connecting the two motifs nor the distance between the two crucial elements is essential. We wanted to analyze whether, nevertheless, sequences in this spacer region are important for the efficiency of VP2 translation; e.g., via the formation of the secondary structure proposed above. For this purpose we generated a set of constructs with 3-mer deletions between motif 1 and motif 2 (Fig. 7A). Most of the deletions showed no effect on VP2 translation since the expression levels were comparable with the wild type (Fig. 7B). Two 3-mer deletions (pCH93: nucleotides −54 to −52; pCH98: nucleotides −39 to −37) showed a reduction of VP2 translation to half of the wild-type level. Importantly, a prominent reduction of VP2 synthesis to 14% (pCH94: nucleotides −51 to −49) or 17% (pCH99: nucleotides −36 to −34) was observed when the 3 nt directly downstream from the above-mentioned trimers were deleted. The four 3-mer deletions leading to reduced VP2 expression all affected the predicted stem composed of nucleotides −52 to −46 and −34 to 39. It can be speculated that the deletions present in pCH94 and pCH99 reduced the putative hybridization stability much more than the other two deletions as they affect the basis of the stem consisting of three G–C base pairs.
Figure 7.
Effect of mutations disturbing the secondary structure of the TURBS region. (A) On the left side, sequences of the 3′-terminal region of FCV ORF2 are shown, with motif 1 marked in bold and underlined in the wild-type pCH1 construct. The positions of the 3-mer deletions and the mutations within the stem built by nucleotides −52 to −46 and −34 to −39 are given below the wild-type sequence. On the right side, the predicted secondary structure based on the interaction between motif 2* and motif 2 is shown and the location of the 3-mer deletions is indicated. (B) Analysis of VP2 expression efficiencies for the indicated mutants. The calculated VP2 expression efficiencies in percent and the standard deviations (SD) are given below the autoradiographs (relative to wild-type pCH1, normalized to the expression of L-VP1). Cells infected with vaccinia virus MVA-T7 served as a negative control (Vacc).
To verify the predicted hybridization described above, double substitutions were introduced into the sequences forming the stem structure to prevent the proposed interaction. GC at positions −51/−50 or the corresponding GC at positions −36/−35 were replaced by CG in constructs pCH142 or pCH143, respectively. Both mutations led to a clear reduction of VP2 translation efficiency to 27% or 48%, respectively (Fig. 7B). In contrast, a mutant combining both double exchanges and thus restoring the stem structure (pCH144) showed almost wild-type level of VP2 expression (83%). These results support the importance of the predicted stem–loop generated by interaction between nucleotides −52 to −46 and −34 to −39.
Discussion
Expression of different proteins from one di- or polycistronic mRNA is very common in prokaryotes and relies on restart of translation after termination of expression of the preceding ORF. In contrast, the overwhelming amount of eukaryotic mRNAs is monocistronic. The reason for this discrepancy can in part be explained by differences in the mechanisms leading to the identification of translational start sites in prokaryotic and eukaryotic cells. Prokaryotes use a rather simple system guided by sequence complementarity between the mRNA and ribosomal 16S rRNA for binding and positioning of ribosomes at the initiation site (Shine and Dalgarno 1975; Jacob et al. 1987). In eukaryotes, a multistep process relying on a large number of initiation factors and other components leads to the selection of the proper start site for translation, and since this process employs the mRNAs 5′-terminal cap structure as a major determinant for ribosome binding, eukaryotic translation initiation usually occurs close to the 5′ end. There are, however, some cases of translational restart templated by eukaryotic bicistronic mRNAs. In order to better understand the differences and parallels between eukaryotic and prokaryotic translation it is interesting to investigate the mechanism underlying these eukaryotic reinitiation events.
In the present study we investigated the mechanism of translation reinitiation on a natural bicistronic mRNA that is used by caliciviruses for expression of their capsid proteins VP1 and VP2 from two overlapping ORFs. Reinitiation was found to be dependent on a sequence element denoted TURBS that is located in the 3′ region of the VP1 coding cistron and contains three essential sequence motifs. Motif 1 encompasses a pentameric sequence (“UGGGA”) conserved among the caliciviruses. In FCV this pentamer is located in the center of a 9-nt sequence that is complementary to a part of helix 26 of mammalian 18S rRNA. Single substitutions in this region of motif 1 showed the importance of the complete complementary sequence. Final proof for the hypothesis of an interaction between the FCV mRNA and 18S rRNA was obtained with the help of a yeast system, in which mutations could be introduced into both RNAs, motif 1 of FCV, and helix 26 of 18S rRNA. In this system only a basal expression of the downstream frame was observed when the two partners were able to hybridize via 5 or 7 base pairs. However, reciprocal mutations in the 18S rRNA allowing hybridization via 9 nt led to high levels of translation of the downstream frame. When the FCV motif 1 sequence was adapted to the yeast 18S rRNA sequence to allow all ribosomes to hybridize with the TURBS via a stretch of 9 nt, a dramatic increase in translation of the downstream frame was observed. The results from these experiments confirmed once again the hypothesis that motif 1 interacts with 18S rRNA as the effect of mutations in one of these RNAs could be compensated by corresponding mutations in the other RNA, and the available amount of ribosomes with the fitting 18S rRNA sequence determined the level of VP2/GFPmyc expression from the downstream ORF.
VP2/GFPmyc translation in the yeast system used here is due to reinitiation and not to IRES mediated de novo start since termination of translation of the first frame far upstream of the start codon of the second frame led to loss of VP2/GFPmyc expression. This result demonstrates that the ribosome has to approach the start/stop site via translation as was also demonstrated in mammalian cells before (Luttermann and Meyers 2007). Moreover, the analysis of motif 2/2* mutants in yeast demonstrated that the same basic mechanism underlies VP2 expression in yeast and mammalian cells.
The second essential motif in the FCV TURBS is located at position −19 to −24 relative to the 3′ end of ORF2 (Luttermann and Meyers 2007). As shown here by analyses with (reciprocal) mutations in mammalian cells and yeast, motif 2 functions via hybridization to another piece of viral RNA, denoted motif 2*, that was found directly upstream of motif 1. Motif 2* was not detected as an essential sequence in earlier studies because the tested deletions could by chance be compensated by neighboring sequences, leading to only slight reductions in VP2 expression.
The sequences of motifs 2 and 2* are not conserved among caliciviruses, but mutation analyses conducted with RHDV sequences revealed the functional importance of these motifs, which are located at equivalent positions in the TURBS of RHDV (Meyers 2007) and FCV. Similarly, sequence elements able to form a motif 2/2* hybridization structure were also found in other caliciviruses (Fig. 8). Importantly, the putative motif 2* sequences are all found directly upstream of the conserved motif 1 and the hypothetical motif 2 is located in a very similar distance upstream of the VP2 start site (distance of 13 to 20 nt) (Fig. 8).
Figure 8.
Comparison of the putative TURBS sequence of different caliciviruses. An exemplary set of TURBS sequences from different caliciviruses, encompassing members of all four genera, is shown. The essential motifs are indicated for the FCV sequence. In the sequences below, the start and stop codons are marked in bold, motif 1 is highlighted in capital letters, and the putative motifs 2 and 2* are underlined. The distance between the 3′ end of motif 2 and the start codon of the VP2 coding ORF is indicated on the right side of the sequences. (FCV) Feline calicivirus; (SMSV) San Miguel sealion virus; (BCV) bovine calicivirus; (RV) rabbit vesivirus; (RHDV) rabbit hemorrhagic disease virus; (EBHSV) European brown hare syndrome virus; (NV) Norwalk virus; (BEC) bovine enteric calicivirus; (SV) sapovirus; (PESV) porcine enteric sapovirus; (WCV) walrus calicivirus; (CCV) canine calicivirus.
The intramolecular interaction between motif 2 and motif 2* probably leads to a secondary structure as shown in Figures 6B and 7A. The results from the structure probing mentioned by Pöyry at al. (2007) are in agreement with this putative secondary structure. The model-structure predicts the formation of a stem–loop built by interaction between nucleotides −52 to −46 and −34 to −39 (see Fig. 7A). Indeed, 3-mer deletions that affected the formation of this stem–loop reduced the VP2 translation level by ∼85%, when the deletions prevented base pairing in the basis of the stem. Analyses with (reciprocal) mutations introduced into the sequences forming the basis of the stem confirmed this interaction. The conclusion that these results show the importance of the predicted secondary structure and are not due to loss of a primary sequence element is based on earlier data showing that nearly the complete spacer between motif 1 and motif 2 (deletion of nucleotides –57 to −28) can be deleted without affecting the VP2 translation level (Luttermann and Meyers 2007). Thus, neither the spacer sequence itself nor the distance between motif 2*/1 and motif 2 are essential for the reinitiation process. Interestingly, different shorter deletions between motif 1 and motif 2 had considerable effects on translation of VP2 (Luttermann and Meyers 2007), which now can be correlated with changes in the predicted secondary structure, since deletions leading to a strong reduction of VP2 translation exhibit less stable secondary structures. Taken together, our results indicate that the TURBS has to adopt a compact structure to allow efficient reinitiation by the ribosome. Small deletions disturbing secondary structure formation would prevent proper packaging of the TURBS whereas deletion of major parts of the spacer between motif 2*/motif 1 and motif 2 could have similar effects as the formation of the predicted structure via hybridization and reduce the size of the TURBS below a critical level. Such deletions are probably not occurring naturally since the respective sequence is not only important for VP2 translation but also codes for a crucial part of the VP1 protein.
The termination/reinitiation mechanism leading to translation of the minor capsid protein VP2 is dependent on intra- and intermolecular base pairing. In our model motif 2 would interact with motif 2* and build the proposed secondary structure. This structure could help to present motif 1 in a way that allows binding and positioning of the ribosome for the reinitiation process. As part of this structure, motif 1 is able to tether a certain proportion of the terminating ribosomes via hybridization with the helix 26 sequence of the 18S rRNA that is part of the 40S subunit of the ribosome. Thereby, time would be available to assemble an initiation complex containing the necessary factors. The tethering of this complex at the translation start site would bypass the requirement for a good initiation codon context that is observed in scanning-dependent initiation, thereby allowing initiation on noncanonical start codons, as shown before (Luttermann and Meyers 2007). This model is in accordance with the recent finding that a residue of helix 26 (nucleotide 1107 of mammalian 18S rRNA) interacts in initiation events with the mRNA. This interaction occurs at position −17 of the mRNA as shown by cross-linking experiments (Pisarev et al. 2008). This position corresponds to the 3′ end of motif 2 of the FCV TURBS that is located at position −16 relative to the ORF3 start codon (nucleotide −20 with regard to the ORF2 stop codon). Thus, the position of the ribosome bound to the RNA for reinitiation seems to parallel the arrangement in standard initiation complexes.
Cross-linking analyses have shown that the TURBS region of FCV RNA is able to bind the multisubunit eIF3 (Pöyry et al. 2007). eIF3 is a crucial factor in nearly all translation initiation events. It is engaged in different steps of the initiation process but one major function of this factor is to provide kind of a scaffold for the assembly of the initiation complex. It was shown only recently that eIF3 also plays a major role in translation termination and ribosome recycling (Pisarev et al. 2007). Following peptide chain release initiation factors eIF1, eIF1A, and eIF3 are sufficient and necessary to achieve complete disassembly of the post-termination ribosome and its dissociation from the mRNA. eIF3 promotes dissociation of the 60S subunit, leaving eIF3/40S still bound to the mRNA with a deacetylated tRNA in the P-site. Ejection of this deacetylated tRNA is promoted by eIF1, and, finally, the loosely associated eIF3j subunit promotes release of the eIF3/40S complex from the mRNA. The tethering of the 40S subunit via 18S rRNA hybridization together with the binding of eIF3 to the TURBS demonstrated by Pöyry et al. (2007) could help to prevent dissociation of the RNA/eIF3/40S subunit complex. This would give some extra time to recruit the necessary factors for reinitiation and eIF3 could help to speed up this process. It has been reported that binding of eIF3 to the TURBS region was prevented by substitutions of the three Gs in motif 1, suggesting motif 1 as an eIF3-binding site (Pöyry et al. 2007). In the yeast system, the same mutation had no negative influence on translation of the second frame when the ribosomal RNA was changed reciprocally to allow hybridization of the mutated sequence. If, as should be expected, eIF3 is also necessary for VP2 expression in yeast, the primary sequence of motif 1 cannot be crucial for factor binding since the introduced changes can be tolerated as shown here. Further work is necessary to define the details of the interaction between TURBS and eIF3. This might also help to explain the results obtained for the mutant at position −61 (pCH2), which showed a clearly reduced translation efficiency (47%) despite the possible interaction via a G–U pairing. This result cannot be explained by the influence of the exchanges on nucleic acid hybridization alone, and should therefore reflect effects on other factors, like, e.g., the interaction of the RNA with eIF3.
Interestingly, also reinitiation in the yeast GCN4 system was recently found to be dependent on a sequence element located upstream of the start/stop site that is interacting with eIF3 (Szamecz et al. 2008). This system relies on translation of very short uORFs and thus differs considerably from the calicivirus system analyzed here. Besides other differences, eIF3 is believed to stay bound to the 40S subunit during translation of a short uORF (Park et al. 2001; Pöyry et al. 2004; Szamecz et al. 2008). After translation of a long uORF as in FCV, eIF3 has to be recruited de novo. Nevertheless, it will be interesting to learn more on the detailed function of the respective upstream element in the GCN4 mRNA and compare the data with those obtained for FCV.
The mechanism proposed here for translation of calicivirus VP2 is somewhat reminiscent of termination/reinitiation events in the prokaryotic system. A requirement for prokaryotic translation reinitiation is a vestigial Shine-Dalgarno (SD) motif present at the reinitiation site of the downstream frame that is too weak to recruit ribosomes from the free pool. The post-terminating ribosome is believed to undergo a bidirectional diffusion for a very limited period of time (and therefore a limited distance) (Adhin and van Duin 1990) and thus a certain proportion of the ribosomes reach the weak SD motif, bind to the complementary sequence, and restart translation. In the eukaryotic system a TURBS-like region is supposed to have an equivalent function in tethering of the ribosome to allow reinitiation.
Interaction of a mRNA sequence with 18S rRNA was described before as a possible way to trigger translation initiation (Tranque et al. 1998; Yueh and Schneider 2000; Chappell et al. 2006; Dresios et al. 2006). In one case, the involvement of such an interaction in enhancement of translation initiation at the 5′ end of a mRNA was further proven by experiments in a yeast system (Dresios et al. 2006). We provide here for the first time conclusive evidence for the necessity of such a hybridization in a eukaryotic termination/reinitiation system. This principle of translation initiation is obviously used by all caliciviruses, especially as the results we obtained for RHDV (Meyers 2007) fit nicely with the model postulated for FCV in this study. Interestingly, reinitiation of translation depending on a motif 1-like sequence has been described very recently for VP1 expression from the genomic RNA of bovine norovirus, a member of the genus Norovirus of the Caliciviridae (McCormick et al. 2008). In this case, the mechanism is used to express the major capsid protein by an alternative way that is independent of the subgenomic viral mRNA. The biological reason why caliciviruses use this way of translation initiation for expression of VP2 and in some cases VP1 is still obscure. The fact that termination/reinitiation of translation has been described for other viruses, including important human pathogens like influenza viruses (Horvath et al. 1990) and respiratory syncytial viruses (Gould and Easton 2005), indicates that this mechanism has some special advantages, and it will most likely be only a question of time until similar strategies are discovered in nonviral systems. The recent work on calicivirus gene expression has elucidated how reinitiation of translation functions in eukaryotes and has also added significantly to our understanding of translation initiation in general.
Materials and methods
Cells and viruses
BHK-21 cells (kindly provided by T. Rümenapf) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and nonessential amino acids.
Vaccinia virus MVA-T7 (Wyatt et al. 1995) was kindly provided by B. Moss (NIH, Bethesda, MD) and the FCV vaccine strain 2024 by K. Danner (Hoechst Roussel Vet GmbH).
The yeast strain NOY908 (kindly provided by M. Nomura) was grown in YEPD medium (1% yeast extract, 2% BactoPeptone [Difco Laboratories], 2% glucose).
Construction of recombinant plasmids
Restriction and subcloning were done according to standard procedures. Restriction and modifying enzymes were purchased from New England Biolabs and Fermentas GmbH.
pCH constructs were established on the basis of plasmid pCH1 (Luttermann and Meyers 2007).
The yeast expression plasmid pYES (Invitrogen) containing a yeast galactose-promoter was used to generate the plasmids pYF21A by insertion of FCV ORF2 (without the region coding for the leader protein) as the first frame. The second overlapping frame started with the first 8 nt of FCV ORF3 fused to a GFP coding region with a c-Myc tag fused to the 3′ end. pYF126 and pYF21H are derivatives of pYF21A. pNOY353 (kindly provided by M. Nomura) served as starting material for the establishment of pNY1 and pNY2.
Point mutations and deletions were introduced by standard PCR-based site-directed mutagenesis methods using thermostable Pfu polymerase (Promega) and synthetic primers purchased from Invitrogen or Metabion. The cloned PCR products were all verified by nucleotide sequencing with the BigDye Terminator Cycle Sequencing Kit (PE Applied Biosystems). Sequence analysis and alignments were done with Genetics Computer Group software (Devereux et al. 1984). Details of the cloning procedure and the sequences of the primers are available on request.
Expression, detection, and quantification of proteins in the mammalian cell system
Transient expression of plasmids in BHK-21 cells using vaccinia virus MVA-T7, metabolic labeling with [35S]methionine (ICN), preparation of cell extracts, and recovery of immunoprecipitates with double precipitation were done as described previously (Luttermann and Meyers 2007). Briefly, VP2 expression efficiency was quantified after SDS-PAGE separation of VP1 and VP2 precipitated with antisera V1 (Thumfart and Meyers 2002) and V2 (raised against bacterially expressed VP2), respectively. Double precipitation was used to ensure quantitative recovery of the proteins as tested before (Luttermann and Meyers 2007). The precipitates were combined, and aliquots thereof were separated by 10% PAGE. The gels were analyzed with a FujiFilm BAS-1500 PhosphorImager, and the intensities of the signals were determined with TINA 2.0 software (Raytest). The molar ratio of VP1 and VP2 was calculated based on the number of labeled residues within the proteins and the measured radioactivity. For comparison of expression efficiencies of different constructs, the VP2 expression level of the wild-type construct pCH1 was defined as 100%. The amounts of VP2 expression of the other constructs were normalized to the values determined for VP1 as an internal standard. The normalized value for VP2 was then used for calculation of the expression efficiency, given as a percentage of the wild-type value. The data presented here represent the average of at least three independent experiments.
Generation of yeast cells
Haploid yeast strain NOY908 lacks all chromosomal rDNA genes and grows via a single rDNA gene expressed from plasmid pNOY373 (Wai et al. 2000). This strain was transformed via standard LiAc procedure with plasmid pNOY353 (Wai et al. 2000) or pNY1 or pNY2 and plasmid pYF21A or pYF126 or pYF21H, plated on selective media containing 2% galactose, and incubated for 2–5 d at 30°C. Yeast cells were selected for their ability to grow in the absence of tryptophan (indicating the presence of pNOY353), uracil (indicating the presence of pYF21A), and histidine (indicating maintenance of the deletion of the chromosomal rDNA locus).
Analysis of protein expression in the yeast cell system
The selected yeast cells were cultured at 30°C on synthetical-defined medium lacking tryptophan, uracil, and histidine. The medium contained galactose to induce transcription of the engineered 35S rRNA and the VP1/GFPmyc mRNA. Cells were grown, harvested, and lysed, and the expression of the proteins was encoded by pYF21A detected in Western blots.
For Western blot analysis, the proteins were separated by 10% SDS-PAGE and blotted onto a nitrocellulose membrane (Schleicher & Schuell Bioscience GmbH) at 100 V for 1 h. The membrane was blocked with Blotto buffer (PBS buffer, 5% milk powder, 0.05% Tween 20). The blots were washed four times with PBS-Tween (0.05% Tween 20). The membranes were incubated with the appropriate first antibody (V1 [Thumfart and Meyers 2002], GFP polyclonal antibody sc-8334 [Santa Cruz Biotechnologies, Inc.]) in PBS-Tween overnight at 4°C. The blots were washed four times with PBS-Tween, and the corresponding secondary antibody (α-rabbit peroxidase-conjugated antibody from Dianova) was added in PBS-Tween for 2 h at room temperature. The membranes were washed four times with PBS-Tween and then incubated with SuperSignal West Pico chemoluminescent substrate (Pierce). The signal was detected with Kodak Biomax films.
Acknowledgments
We thank Maren Ziegler, Petra Wulle, and Janett Wieseler for excellent technical assistance. We are very grateful to M. Nomura at the University of California at Irvine, Irvine, CA, for providing yeast strain NOY908 and plasmids pNOY353 and pNOY373. We are also grateful to J.O. Thumfart and H. Schirrmeier for the preparation of the VP2 antiserum. This work was supported by grant Me 1367/3 from the Deutsche Forschungsgemeinschaft.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.507609.
References
- Adhin M.R., van Duin J. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 1990;213:811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- Chappell S.A., Dresios J., Edelman G.M., Mauro V.P. Ribosomal shunting mediated by a translational enhancer element that base pairs to 18S rRNA. Proc. Natl. Acad. Sci. 2006;103:9488–9493. doi: 10.1073/pnas.0603597103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Y., Nayak A., Bordeleau M.E., Tanaka J., Pelletier J., Belsham G.J., Roberts L.O., Goodfellow I.G. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 2006;281:25315–25325. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- Cuesta R., Xi Q., Schneider R.J. Preferential translation of adenovirus mRNAs in infected cells. Cold Spring Harb. Symp. Quant. Biol. 2001;66:259–267. doi: 10.1101/sqb.2001.66.259. [DOI] [PubMed] [Google Scholar]
- Daughenbaugh K.F., Fraser C.S., Hershey J.W., Hardy M.E. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J. 2003;22:2852–2859. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresios J., Chappell S.A., Zhou W., Mauro V.P. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2006;13:30–34. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- Goodfellow I., Chaudhry Y., Gioldasi I., Gerondopoulos A., Natoni A., Labrie L., Laliberte J.F., Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6:968–972. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P.S., Easton A.J. Coupled translation of the respiratory syncytial virus M2 open reading frames requires upstream sequences. J. Biol. Chem. 2005;280:21972–21980. doi: 10.1074/jbc.M502276200. [DOI] [PubMed] [Google Scholar]
- Gould P.S., Easton A.J. Coupled translation of the second open reading frame of M2 mRNA is sequence dependent and differs significantly within the subfamily Pneumovirinae . J. Virol. 2007;81:8488–8496. doi: 10.1128/JVI.00457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.Y., Ando T., Balayan M.S., Berke T., Clarke I.N., Estes M.K., Matson D.O., Nakata S., Neill J.D., Studdert M.J., et al. Family Caliciviridae. In: Regenmortel M.H.V., et al., editors. Virus taxonomy. Academic Press; New York: 2000. pp. 725–734. [Google Scholar]
- Hellen C.U., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Herbert T.P., Brierley I., Brown T.D. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 1996;77:123–127. doi: 10.1099/0022-1317-77-1-123. [DOI] [PubMed] [Google Scholar]
- Horvath C.M., Williams M.A., Lamb R.A. Eukaryotic coupled translation of tandem cistrons: Identification of the influenza B virus BM2 polypeptide. EMBO J. 1990;9:2639–2647. doi: 10.1002/j.1460-2075.1990.tb07446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob W.F., Santer M., Dahlberg A.E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc. Natl. Acad. Sci. 1987;84:4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp L.D., Lorsch J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol. Cell. Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: An update. J. Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre P., Kolakofsky D., Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol. Cell. Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C., Meyers G. A bipartite sequence motif induces translation reinitiation in feline calicivirus RNA. J. Biol. Chem. 2007;282:7056–7065. doi: 10.1074/jbc.M608948200. [DOI] [PubMed] [Google Scholar]
- McCormick C.J., Salim O., Lambden P.R., Clarke I.N. Translation termination reinitiation between open reading frame 1 (ORF1) and ORF2 enables capsid expression in a bovine norovirus without the need for production of viral subgenomic RNA. J. Virol. 2008;82:8917–8921. doi: 10.1128/JVI.02362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers G. Translation of the minor capsid protein of a calicivirus is initiated by a novel termination-dependent reinitiation mechanism. J. Biol. Chem. 2003;278:34051–34060. doi: 10.1074/jbc.M304874200. [DOI] [PubMed] [Google Scholar]
- Meyers G. Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus. J. Virol. 2007;81:9623–9632. doi: 10.1128/JVI.00771-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.S., Himmelbach A., Browning K.S., Hohn T., Ryabova L.A. A plant viral “reinitiation” factor interacts with the host translational machinery. Cell. 2001;106:723–733. doi: 10.1016/s0092-8674(01)00487-1. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Hellen C.U., Pestova T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Kolupaeva V.G., Yusupov M.M., Hellen C.U., Pestova T.V. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T.A., Kaminski A., Jackson R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T.A., Kaminski A., Connell E.J., Fraser C.S., Jackson R.J. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes & Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.V., Matson D.O., Smith A.W. Three-dimensional structure of calicivirus. J. Mol. Biol. 1994;240:256–264. doi: 10.1006/jmbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- Rohayem J., Robel I., Jager K., Scheffler U., Rudolph W. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 2006;80:7060–7069. doi: 10.1128/JVI.02195-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova L.A., Pooggin M.M., Hohn T. Viral strategies of translation initiation: Ribosomal shunt and reinitiation. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:1–39. doi: 10.1016/S0079-6603(02)72066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova L.A., Pooggin M.M., Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006;119:52–62. doi: 10.1016/j.virusres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Sachs A.B., Sarnow P., Hentze M.W. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Sarnow P., Cevallos R.C., Jan E. Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 2005;33:1479–1482. doi: 10.1042/BST0331479. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A.G. New modes of translational control in development, behavior, and disease. Mol. Cell. 2007;28:721–729. doi: 10.1016/j.molcel.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Sosnovtseva S.A., Green K.Y. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 1998;72:3051–3059. doi: 10.1128/jvi.72.4.3051-3059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S.V., Belliot G., Chang K.O., Onwudiwe O., Green K.Y. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 2005;79:4012–4024. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B., Rutkai E., Cuchalova L., Munzarova V., Herrmannova A., Nielsen K.H., Burela L., Hinnebusch A.G., Valasek L. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes & Dev. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumfart J.O., Meyers G. Feline calicivirus: Recovery of wild-type and recombinant viruses after transfection of cRNA or cDNA constructs. J. Virol. 2002;76:6398–6407. doi: 10.1128/JVI.76.12.6398-6407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranque P., Hu M.C., Edelman G.M., Mauro V.P. rRNA complementarity within mRNAs: A possible basis for mRNA-ribosome interactions and translational control. Proc. Natl. Acad. Sci. 1998;95:12238–12243. doi: 10.1073/pnas.95.21.12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai H.H., Vu L., Oakes M., Nomura M. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: Use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt L.S., Moss B., Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- Xi Q., Cuesta R., Schneider R.J. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J. Virol. 2005;79:5676–5683. doi: 10.1128/JVI.79.9.5676-5683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A., Schneider R.J. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes & Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]