Abstract
RNA-directed DNA methylation (RdDM) is an RNAi-based mechanism for establishing transcriptional gene silencing in plants. The plant-specific RNA polymerases IV and V are required for the generation of 24-nucleotide (nt) siRNAs and for guiding sequence-specific DNA methylation by the siRNAs, respectively. However, unlike the extensively studied multisubunit Pol II, our current knowledge about Pol IV and Pol V is restricted to only the two largest subunits NRPD1a/NRPD1 and NRPD1b/NRPE1 and the one second-largest subunit NRPD2a. It is unclear whether other subunits may be required for the functioning of Pol IV and Pol V in RdDM. From a genetic screen for second-site suppressors of the DNA demethylase mutant ros1, we identified a new component (referred to as RDM2) as well as seven known components (NRPD1, NRPE1, NRPD2a, AGO4, HEN1, DRD1, and HDA6) of the RdDM pathway. The differential effects of the mutations on two mechanistically distinct transcriptional silencing reporters suggest that RDM2, NRPD1, NRPE1, NRPD2a, HEN1, and DRD1 function only in the siRNA-dependent pathway of transcriptional silencing, whereas HDA6 and AGO4 have roles in both siRNA-dependent and -independent pathways of transcriptional silencing. In the rdm2 mutants, DNA methylation and siRNA accumulation were reduced substantially at loci previously identified as endogenous targets of Pol IV and Pol V, including 5S rDNA, MEA-ISR, AtSN1, AtGP1, and AtMU1. The amino acid sequence of RDM2 is similar to that of RPB4 subunit of Pol II, but we show evidence that RDM2 has diverged significantly from RPB4 and cannot function in Pol II. An association of RDM2 with both NRPD1 and NRPE1 was observed by coimmunoprecipitation and coimmunolocalization assays. Our results show that RDM2/NRPD4/NRPE4 is a new component of the RdDM pathway in Arabidopsis and that it functions as part of Pol IV and Pol V.
Keywords: DNA methylation, transcriptional gene silencing, siRNAs, Pol IV, Pol V
DNA methylation is an epigenetic mark conserved in plants, mammals, and some fungi. It plays an important role in epigenetic processes such as the stable transcriptional silencing of transgenes and endogenous genes (Baulcombe 2004; Bender 2004; Chan et al. 2004; Matzke et al. 2004), paramutation (Chandler and Stam 2004), imprinting (Scott and Spielman 2004), and X inactivation (Heard 2004). In plants, 24-nucleotide (nt) siRNAs can direct sequence-specific DNA methylation (Mette et al. 2000). Piwi-interacting small RNAs (piRNAs) and certain endogenous siRNAs in mammalian cells can also direct DNA methylation (Aravin et al. 2007). In Schizosaccharomyces pombe, there is no DNA methylation, but siRNAs can still trigger transcriptional gene silencing (TGS) by directing heterochromatic histone modifications (Volpe et al. 2002; Ebert et al. 2004).
Plant heterochromatic siRNAs are ∼24 nt long and are produced in a pathway that depends on the RNA-dependent RNA polymerase 2 (RDR2) and Dicer-like 3 (DCL3) (Xie et al. 2004). The pathway also requires Pol IV, a plant-specific, presumed DNA-dependent RNA polymerase (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005). Interestingly, the functioning of the siRNAs depends on not only the effector proteins Argonaute 4 (AGO4) (Zilberman et al. 2003), chromatin remodeling protein DRD1 (Kanno et al. 2004), and de novo DNA methyltransferase DRM2 (Cao and Jacobsen 2002), but also on Pol IVb/Pol V, another plant-specific, presumed DNA-dependent RNA polymerase (Pontier et al. 2005; Wierzbicki et al. 2008). Pol IV has been hypothesized to transcribe methylated DNA to produce the initial transcripts that are the substrate of RDR2 (Li et al. 2006; Pontes et al. 2006). This activity, however, has not been demonstrated either in vitro or in vivo. Recently, Pol V and DRD1 were found to be required for production of uncapped and nonpolyadenylated RNAs that may serve as the scaffold for binding by the heterochromatic siRNAs, which are the sequence-specific guides of DNA methylation (Wierzbicki et al. 2008). It has been hypothesized that Pol IV transcripts move to a nucleolar body, where they may be processed into siRNAs by RDR2 and DCL3 (Li et al. 2006; Pontes et al. 2006). The siRNAs then may be loaded onto AGO4, which can bind to NRPE1 in the nucleolus. It is possible that AGO4 with bound siRNAs subsequently exits the nucleolus and finds target genomic regions by siRNA base-pairing with Pol V transcripts from the target loci (Wierzbicki et al. 2008). DRM2 presumably associates with the AGO4 complex to cause DNA methylation.
Thus far, forward genetic screens have identified only the largest subunits of Pol IV and Pol V (NRPD1a/NRPD1 and NRPD1b/NRPE1, respectively) and the second largest subunit (NRPD2a/NRPD2/NRPE2) that is shared by Pol IV and Pol V (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Pontier et al. 2005; Eamens et al. 2008). It is well known that RNA Pol II consists of many other subunits, in addition to its largest subunit RPB1 and second largest subunit RPB2 (Cramer 2002). It is unclear whether Pol IV and Pol V require other subunits for functioning and whether the other subunits are unique or shared with Pol II. In this study, we carried out a forward genetic screen and identified a new component (referred to as RDM2 for RNA-directed DNA methylation 2) as well as seven known components (NRPD1a, NRPD1b, NRPD2a, AGO4, HEN1, DRD1, and HDA6) of the RNA-directed DNA methylation (RdDM) pathway in Arabidopsis. The differential effects of the mutations on two mechanistically distinct transcriptional silencing reporters suggest that RDM2, NRPD1a, NRPD1b, NRPD2a, HEN1, and DRD1 function only in the siRNA-dependent pathway of transcriptional silencing, whereas HDA6 and AGO4 have roles in both siRNA-dependent and -independent pathways of transcriptional silencing. RDM2 is required for DNA methylation and transcriptional silencing at transposons and other repetitive sequences. It is also required for high-level accumulation of siRNAs from the affected loci. RDM2 shares sequence similarity with the fourth subunit of Pol II, RPB4. We show that RDM2 is not an RPB4 ortholog; rather, it is part of Pol IV and Pol V. Our results suggest that Pol IV and Pol V are multisubunit enzymes and that their NRPD4/NRPE4 subunit has evolved from the ancestral RPB4 subunit of Pol II but has diverged to assume functions specific to the RdDM pathway.
Results
Identification of components mediating TGS by screening for ros1 suppressors
We showed previously that in the wild-type Arabidopsis background, the low level of siRNAs produced from the RD29A promoter driving the LUCIFERASE (RD29A-LUC) transgene does not lead to promoter DNA hypermethylation and TGS because of the active DNA demethylation activity of ROS1 (Gong et al. 2002). In ros1 mutant plants, however, the siRNAs cause DNA hypermethylation and TGS of the RD29A-LUC transgene as well as of the homologous endogenous RD29A gene (Gong et al. 2002). The CaMV 35S promoter-driven NPTII transgene linked to the LUC transgene is also silenced in the ros1 mutant likely because of heterochromatic spreading from the adjacent RD29A-LUC locus (Kapoor et al. 2005).
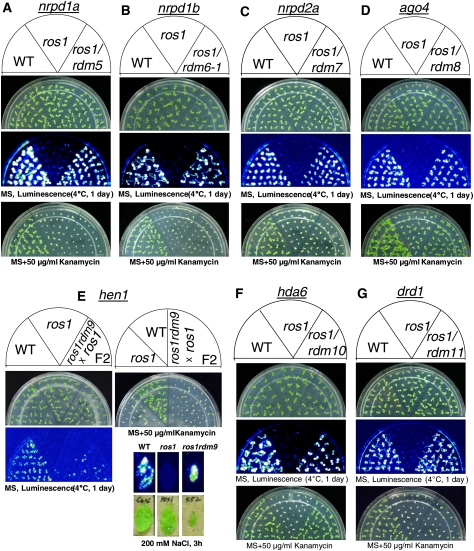
In this study, a T-DNA-mutagenized ros1 population was generated and systematically screened for suppressors of ros1 based on LUCIFERASE expression (i.e., luminescence) after cold treatment (1 d, 4°C). Thirteen independent mutants were identified in this study as suppressors of ros1. Through thermal asymmetric interlaced PCR (TAIL-PCR) or map-based cloning, the mutants of seven known genes (NRPD1a, NRPD1b, NRPD2a, AGO4, HEN1, DRD1, and HDA6) involved in TGS were identified as suppressors of ros1 (Fig. 1; Supplemental Fig. S1A–G). The results demonstrate that screening for suppressors of ros1 is a very effective strategy for identifying genes that are critical for TGS.
Figure 1.
Luminescence and kanamycin resistance phenotypes of ros1 suppressor mutations in known RdDM components. Wild type, ros1, and the double mutants ros1rdm5 (A), ros1rdm6-1 (B), ros1rdm7 (C), ros1rdm8 (D), ros1rdm9 (E), ros1rdm10 (F), and ros1rdm11 (G) were either grown on MS plates for 7–10 d and luminescence was imaged after cold treatment (1 d, 4°C), or were grown on MS plates supplemented with 50 μg/mL kanamycin and the pictures were taken after 1–2 wk. Because the ros1rdm9 mutant is sterile, the F2 progenies from a cross between ros1rdm9 and ros1, instead of ros1rdm9, were grown on plates for phenotype assay. The luminescence of ros1rdm9 was also assayed using NaCl-treated leaves from soil-grown plants. Because the rdm5, rdm6, rdm7, rdm8, rdm9, rdm10, and rdm11 mutants were identified as nrpd1a, nrpd1b, nrpd2a, ago4, hen1, hda6, and drd1, respectively, the names of these mutants are labeled on the top of each panel.
Differential effects of the nrpd1a, nrpd1b, nrpd2a, ago4, hen1, drd1, and hda6 mutations on RD29A-LUC and 35S-NPTII silencing
The release of the TGS phenotype of these known gene mutants was confirmed in their progenies. The results showed that all of these mutants are homozygous and dramatically suppressed the silencing of transgene RD29A-LUC and emitted strong luminescence after cold treatment (Fig. 1A–G). It is interesting that while all of the seven identified known genes have a substantial effect on silencing of the RD29A-LUC transgene, most of them (NRPD1a, NRPD1b, NRPD2a, HEN1, and DRD1) have no effect on the 35S-NPTII transgene, and mutations in these genes do not suppress kanamycin sensitivity of ros1 (Fig. 1A–C,E,G). However, mutation in AGO4 partially suppresses and mutation in HDA6 completely suppresses the kanamycin sensitivity of ros1 (Fig. 1D,F). For the hen1 mutant, the F2 progenies from a cross between ros1hen1 and ros1 were used for phenotype assay because the homozygous ros1hen1 mutant is sterile (Fig. 1E).
The molecular phenotypes of the known gene mutants ros1nrpd1b, ros1ago4, ros1hda6, and ros1drd1 were further investigated. RNA blot analysis detected transcripts from the endogenous RD29A as well as from the RD29A-LUC transgene in the suppressor mutants, suggesting that the silencing of the endogenous RD29A gene is suppressed by the known gene mutations (Supplemental Fig. S2A–D). Consistent with their kanamycin-response phenotypes, the nrpd1b and drd1 mutations had no effect on the transcript levels of the kanamycin-resistant gene NPTII, whereas ago4 partially rescued and hda6 completely rescued the expression of NPTII in the ros1 background (Supplemental Fig. S2A–D). Southern hybridization assays were used to test the effect of these mutations on DNA methylation. The results show that in ros1nrpd1b, ros1ago4, and ros1drd1, 5S rDNA methylation was reduced at all cytosine contexts (CG, CHG, and CHH), with especially dramatic reductions at the asymmetric CHH sites (Supplemental Fig. S3A). It is interesting that the hda6 mutation appeared to dramatically reduce CHG methylation but not CG and CHH methylation (Supplemental Fig. S3A). The effect of nrpd1b, ago4, and drd1 on DNA methylation was further confirmed by DNA methylation assay at the well-characterized retroelement AtSN1 (Zilberman et al. 2003; Xie et al. 2004; Onodera et al. 2005). After genomic DNA was digested with the DNA methylation-sensitive restriction enzyme HaeIII, the amplification of AtSN1 was blocked in ros1nrpd1b, ros1ago4, and ros1drd1, indicating decreased CHH methylation in these mutants (Supplemental Fig. S3B). In ros1hda6, however, the CHH methylation level at AtSN1 was the same as in the wild type (Supplemental Fig. S3B). In addition, small-RNA Northern analysis suggested that siRNA1003 accumulation was reduced in ros1nrpd1b, ros1ago4, and ros1drd1 but was drastically increased in ros1hda6 (Supplemental Fig. S3C). The transcript levels of the endogenous TGS targets AtGP1 and AtMU1 were assessed by RT–PCR. The results show that in ros1hda6, ros1ago4, and ros1drd1, the transcript levels of AtGP1 and AtMU1 increased substantially compared with those in ros1 and the wild type (Supplemental Fig. S3D).
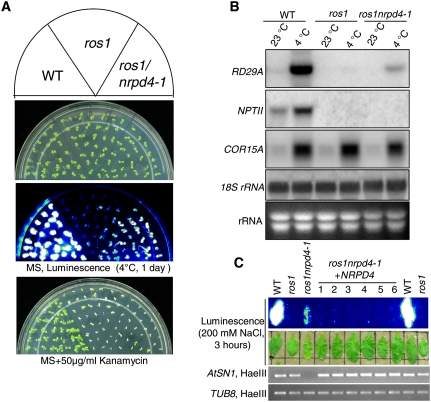
The nrpd4-1 mutation suppresses TGS in the ros1 mutant
Besides the seven known gene mutants, a new mutant, rdm2-1 (later redesignated as nrpd4-1 or nrpe4-1) was also identified as a suppressor of ros1. Figure 2A illustrates the luminescence phenotype of the wild type, ros1, and ros1nrpd4-1. The results show that after cold treatment, ros1nrpd4-1 emitted stronger luminescence than ros1, albeit the luminescence is still weaker than that from the wild type. The result suggests that the silencing of RD29A-LUC in ros1 is partially suppressed by the nrpd4 mutation. Interestingly, the kanamycin sensitivity of ros1nrpd4-1 was similar to that of ros1 (Fig. 2A), which indicates that the nrpd4 mutation has no effect on 35S-NPTII silencing in ros1.
Figure 2.
TGS is suppressed in the ros1nrpd4-1 mutant plants. (A) Luminescence and kanamycin resistance phenotypes. Wild type, ros1, and ros1nrpd4-1 were grown on MS plates and imaged after cold treatment (4°C, 24 h). Wild type, ros1, and ros1nrpd4-1 were grown on MS plates with kanamycin (50 μg/mL), and the pictures were taken after 1 wk. (B) The transcript levels of endogenous RD29A and NPTII transgene in wild type, ros1, and ros1nrpd4-1. Total RNA was extracted from 2-wk-old seedlings with or without cold treatment (24 h, 4°C). COR15A and 18S rRNA were used as cold treatment control and RNA loading control, respectively. (C) Assay for complementation of the ros1nrpd4-1 mutant. The plant expression vector PMDC164 harboring NRPD4 genomic sequence was transformed into the ros1nrpd4-1 mutant plants. The leaves from wild type, ros1, ros1nrpd4-1, and the six T1 individual transgenic lines were used for luminescence imaging after treatment with 200 mM NaCl for 3 h. After the genome DNA was digested with the methylation-sensitive enzyme HaeIII, the amplification of AtSN1 in the six transgenic lines was restored to the same level as that in wild type and ros1.
The ros1nrpd4-1 mutant was crossed to ros1, and the F1 plants were similar to ros1 plants in that they emitted little luminescence. F2 progenies from the selfed F1 plants segregated ∼3:1 for ros1: ros1nrpd4 luminescence phenotypes, suggesting that the nrpd4-1 mutation was recessive and in a nuclear gene (data not shown).
The luminescence phenotype of ros1nrpd4-1 plants suggested that TGS of the RD29A-LUC transgene was suppressed by the nrpd4-1 mutation. Northern blot assays revealed that the mRNA level of the endogenous RD29A was higher than that in ros1, although it was lower than that in the wild type (Fig. 2B). These results suggest that the nrpd4-1 mutation partially suppressed the silencing of both the RD29A-LUC transgene and the endogenous RD29A gene. Transcripts from the 35S-NPTII transgene were undetectable in the ros1nrpd4-1 mutant, as was the case in ros1 (Fig. 2B), which is in agreement with the kanamycin-sensitive phenotypes of ros1nrpd4-1 and ros1 plants. There was no difference among wild-type, ros1, and ros1nrpd4-1 plants in the 18S rRNA loading control or the stress-induced COR15A gene for cold treatment control (Fig. 2B). Taken together, these results demonstrate that the nrpd4-1 mutation partially suppresses TGS of the endogenous RD29A and the RD29A-LUC transgene but not the NPTII transgene in the ros1 mutant.
NRPD4 encodes a RPB4-like protein
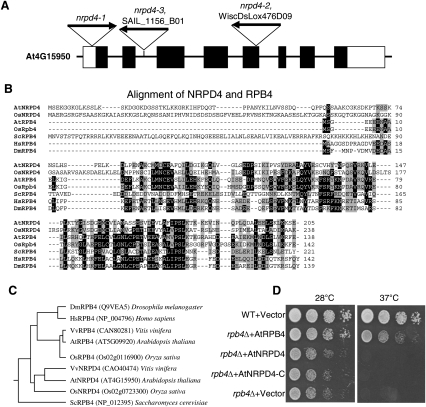
The T-DNA insertion site in ros1nrpd4-1 was determined by TAIL-PCR. A single T-DNA insertion was found in the 5′-UTR of AT4G15950. AT4G15950 encodes a protein with similarity to RPB4, a subunit of RNA polymerase II. Its function in the TGS pathway and its physical association with NRPD1a/NRPD1 and NRPD1b/NRPE1 (see below) suggest that it is a subunit of Pol IV and V, and thus was named “NRPD4/NRPE4” (nuclear RNA polymerase D/E 4). To confirm that NRPD4 is the correct gene, a construct harboring the full genomic sequence of NRPD4 was generated and transformed into the ros1nrpd4-1 mutant plants. The results show that like the ros1 plants, all six individual transgenic T1 plants emitted little luminescence as ros1 plants did after treatment with 200 mM NaCl for 3 h (Fig. 2C). The DNA methylation status of AtSN1 was tested in wild-type, ros1, ros1nrpd4-1, and the six transgenic T1 plants. After digestion with the methylation-sensitive enzyme HaeIII, the amplification of AtSN1 in all six transgenic plants was the same as in the wild type and ros1, while in ros1nrpd4-1, the amplification was completely blocked (Fig. 2C). The results suggest that the NRPD4 transgene complemented not only the luminescence phenotype but also the endogenous DNA methylation phenotype of ros1nrpd4-1. Furthermore, we analyzed two other nrpd4 mutant alleles from the Wisconsin T-DNA insertion lines (WiscDsLox476D09 and SAIL_1156_B1) named nrpd4-2 and nrpd4-3, respectively. The nrpd4-2 mutant has a T-DNA insertion in the fifth exon of AT4G15950, while nrpd4-3 has a T-DNA in the second intron. Like the ros1nrpd4-1 mutant, both the nrpd4-2 and nrpd4-3 mutants have a low level of DNA methylation at AtSN1 (Fig. 3). Therefore, we conclude that NRPD4 (At4G159050) is the gene that controls TGS and DNA methylation. We did not observe any developmental phenotypes on plants of any of the nrpd4 mutant alleles.
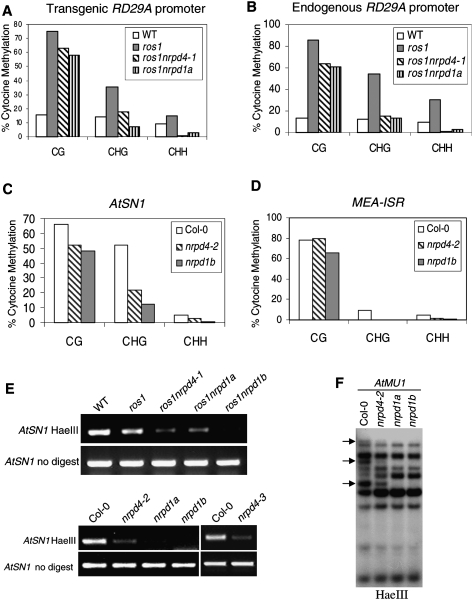
Figure 3.
Effect of nrpd4 mutations on DNA methylation. The percentage of cytosine methylation at transgene (A) and endogenous (B) RD29A promoters, and at AtSN1 (C) and MEA-ISR (D) were determined by bisulfite sequencing. The ros1nrpd1a or nrpd1b mutants were used as the mutant controls. (H) A, T, or C. The percentage of cytosine methylation on CG, CHG, and CHH sites is shown. (E) PCR assay of the effect of the nrdp4-1 mutation on DNA methylation of AtSN1. Amplification of AtSN1 was performed after the genomic DNA was digested with the methylation-sensitive restriction enzyme HaeIII. The undigested genomic DNA was amplified as a control. (F) Effect of the nrpd4 mutation on DNA methylation of AtMU1. Genomic DNA from Col-0, nrpd4-2, nrpd1a, and nrpd1b was digested with the methylation-sensitive restriction enzyme HaeIII, followed by Southern hybridization.
The nrpd4-1 mutation reduces DNA methylation at RdDM target loci
We examined the effect of nrpd4 mutations on DNA methylation at various loci (Fig. 3A–F). To test whether the suppression of TGS at RD29A-LUC transgene and endogenous RD29A in ros1nrpd4-1 mutant plants correlates with loss of DNA methylation, we analyzed the DNA methylation status at the RD29A promoter by bisulfite sequencing. The results show that, at a 361-base-pair (bp) region of the endogenous and transgenic RD29A promoter, heavy methylation occurred in all cytosine contexts (CG, CHG, and CHH) in the ros1 mutant but was reduced in ros1nrpd4-1 as well as in ros1nrpd1a plants (Fig. 3A,B). The reduced methylation was observed for both the endogenous and transgene RD29A promoters. Moreover, the reduction in cytosine methylation was most dramatic at asymmetric CHH sites, was modest at CHG sites, and was marginal at CG sites (Fig. 3A,B). For example, at the endogenous RD29A promoter, the CHH methylation is 30.2% in ros1, 0.7% in ros1nrpd4-1, and 2.4% in ros1nrpd1a, while the CG methylation is 86.2%, 63.9%, and 60.7% for the three genotypes, respectively (Fig. 3B). These results suggest that nrpd4-1, like nrpd1a, suppresses the TGS of the RD29A-LUC transgene and endogenous RD29A in ros1 by reducing DNA methylation at the RD29A promoter. It appears that, like NRPD1a and the other previously characterized genes including NRPD2a, NRPD1b, and DRD1 (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Pontier et al. 2005), NRPD4 is also required for de novo DNA methylation directed by the RdDM pathway.
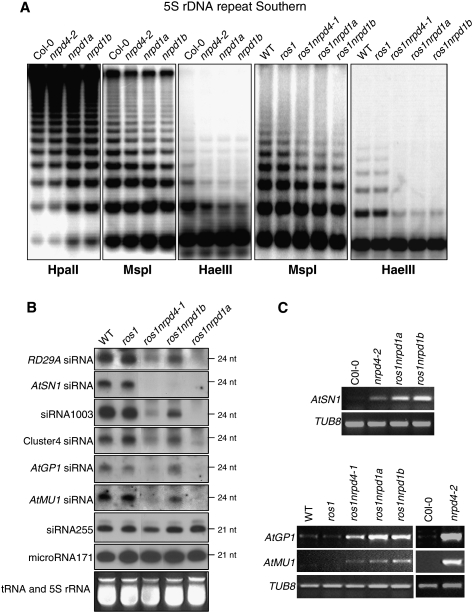
The DNA methylation status of the centromeric region was analyzed using restriction digestion by the methylation-sensitive restriction enzyme HaeIII and isoschizomers HpaII and MspI, followed by Southern hybridization. No difference in DNA methylation of the highly repetitive 180-bp centromeric repeat was detected among wild-type, nrpd4-2, nrpd1a, and nrpd1b mutant plants (Supplemental Fig. S4A). However, compared with the wild-type control, CHG and CHH methylation of 5S rDNA was reduced in nrpd4-2 as well as in nrpd1a and nrpd1b (Fig. 4A). Nevertheless, the CG methylation level of 5S rDNA in nrpd4-2 was similar to that in the wild type, although CG methylation in nrpd1a and nrpd1b was lower than that in the wild type (Fig. 4A), which is consistent with previous reports (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005). The suppressive effect of nrpd4 mutations on CHG and CHH methylation of 5S rDNA was further confirmed in ros1nrpd4-1 mutant plants (Fig. 4A, right two panels).
Figure 4.
Effect of nrpd4 mutations on DNA methylation, siRNA accumulation, and RNA transcript levels. (A) The nrpd4 mutations reduce DNA methylation at 5S rDNA repeats. Genomic DNA from the indicated genotypes was digested with HpaII (for CG and CHG methylation), MspI (for CHG methylation), and HaeIII (for CHH methylation), followed by Southern hybridization. (B) Detection of RD29A promoter siRNAs, AtSN1 siRNA, siRNA1003, Cluster4 siRNA, AtGP1 siRNA and AtMU1 siRNA, ta-siRNA255, and microRNA171 in the indicated genotypes. The ethidium bromide-stained gel corresponding to tRNA and 5S rRNA was used as a loading control. The positions of size markers are indicated (24 nt or 21 nt). (C) The nrpd4 mutations increase the expression of AtSN1, AtGP1, and AtMU1. The transcript level of the transposons was detected by RT–PCR. TUB8 was used as a control.
To assess the effect of nrpd4 mutations on methylation of genomic sequences located outside of centromeres and rDNA arrays, we examined the methylation status at two other genomic loci: AtSN1, a well-characterized retroelement (Zilberman et al. 2003; Xie et al. 2004; Onodera et al. 2005), and MEA-ISR, a subtelomeric repeat sequence present downstream from the MEA gene (Cao and Jacobsen 2002). The bisulfite sequencing results show that in wild-type plants, AtSN1 elements were heavily methylated, with 66.1%, 52.0%, and 5.2% cytosine methylation at CG, CHG, and CHH sites, respectively, but the methylation levels in nrpd4-2 plants were reduced to 52.3%, 22.1%, and 2.9%, respectively (Fig. 3C). Although the effect was a little less than that of nrpd1b, nrpd4-2 clearly reduced AtSN1 methylation. MEA-ISR is an ∼183-bp sequence present in seven direct repeats and located in subtelomeric regions (Cao and Jacobsen 2002). Our bisulfite sequencing results revealed a high level of DNA methylation at the MEA-ISR region in wild-type plants (Fig. 3D), which is consistent with a previous report (Cao and Jacobsen 2002). The CHG and CHH methylation but not the CG methylation at MEA-ISR was dramatically reduced in nrpd4-2 as well as in nrpd1b (Fig. 3D). These results show that nrpd4 mutations have strong effects on CHH and CHG methylation but have less influence on CG methylation, which is consistent with the effect of nrpd4-1 on methylation of the RD29A promoter.
The methylation status of AtSN1 was further confirmed with the methylation-sensitive enzyme HaeIII, followed by PCR. The result shows that in both the wild-type and ros1, AtSN1 elements were heavily methylated and resistant to HaeIII cleavage, but in ros1nrpd4-1, the methylation at AtSN1 was much reduced, and the reduction was similar to that in ros1nrpd1a but less than that in ros1nrpd1b (Fig. 3E). The reducing effect of nrpd4 mutations on AtSN1 methylation was further confirmed in another two nrpd4 mutant alleles, nrpd4-2 and nrpd4-3 (Fig. 3E). Southern hybridization was carried out to determine the methylation status of AtMU1 at CHH sites. Figure 3F shows that three HaeIII undigested bands present in the wild type were partially digested in nrpd4-2 and completely digested in nrpd1a and nrpd1b, suggesting that the nrpd4-2 mutation partially blocks CHH methylation at AtMU1. Taken together, our results demonstrate a strong effect of nrpd4 mutations in reducing CHH methylation, which suggests that RdDM is impaired in the mutants.
The nrpd4 mutations reduce 24-nt siRNA levels and increase expression of target loci
To determine whether the reduction of DNA methylation in the nrpd4 mutant plants is correlated with defects in the biogenesis of siRNAs that direct DNA methylation, small-RNA blot analysis was carried out. Figure 4B shows that accumulation of 24-nt siRNAs in ros1nrpd4-1 was reduced substantially compared with that in wild-type and ros1 plants. In ros1nrpd1a plants, the accumulation of 24-nt siRNAs was completely blocked, whereas in ros1nrpd1b plants, the reduction of siRNA accumulation only occurred for some of the tested siRNAs, including RD29A promoter siRNAs, AtSN1 siRNAs, siRNA1003, and AtMU1 siRNAs (Fig. 4B). The levels of Cluster4 siRNAs and AtGP1 siRNAs in ros1nrpd1b were the same as those in wild-type and ros1 plants, consistent with previous findings that NRPD1b has an siRNA-independent function in RdDM (Pontier et al. 2005; Mosher et al. 2008). From the small RNA blot analysis, we found that although the accumulation of 24-nt siRNAs in ros1nrpd4-1 was much reduced compared with the wild type and ros1, for most of the tested siRNAs, their levels were more than those in ros1nrpd1a but less than those in ros1nrpd1b (Fig. 4B). The 21-nt microRNA171 and 21-nt ta-siRNA255 were also tested in this study, and we found no effect of the nrpd4 mutation, or of the nrpd1a and nrpd1b mutations, on accumulation of these two small RNAs (Fig. 4B). The result suggests that the suppression of DNA methylation in ros1nrpd4-1 may be partially explained by the reduction of siRNA accumulation. The reduced levels of Cluster4 siRNAs, AtSN1 siRNAs, and siRNA1003 were also seen in the nrpd4-2 allele (Supplemental Fig. S4B). Interestingly, in nrpd4-2, the accumulation of AtGP1 siRNAs, AtMU1 siRNAs, and siRNA02 was virtually the same as that in Col-0 wild-type plants (Supplemental Fig. S4B). The stronger effect of nrpd4-1 relative to nrpd4-2 on the accumulation of 24-nt siRNAs may be because the nrpd4-1 mutation is in the ros1 mutant background and in the C24 ecotype. Considering the strong DNA methylation phenotypes of nrpd4 mutants, these small RNA results suggest that NRPD4 may have dual functions; i.e., it may associate with NRPD1 and function as part of Pol IV in siRNA biogenesis, but it may also associate with NRPD1b/NRPE1 and function as part of Pol V in DNA methylation independent of its role in siRNA biogenesis.
RT–PCR was carried out to determine the impact of nrpd4 mutations on the expression of the affected endogenous loci. The results show that AtSN1, AtGP1, and AtMU1 have higher transcript levels in nrpd4-2 or ros1nrpd4-1 than in their wild-type background controls. However, the expression levels of the affected loci appeared to be lower in the nrpd4 mutants than in the nrpd1a and nrpd1b mutants (Fig. 4C).
NRPD4 is a variant of RPB4 but is not functional in RNA polymerase II
The NRPD4 gene consists of eight exons and seven introns (Fig. 5A) and is predicted to encode a protein of 205 amino acids. With the exception of the N-terminal region, the main body of NRPD4 (amino acids 75–205) is highly similar to RPB4 (a subunit of RNA polymerase II) from diverse organisms including yeast, humans, and flies (Fig. 5B). There is a separate protein encoded by the Arabidopsis genome, AtRBP4, that is more similar to RPB4 from nonplant eukaryotes than NRPD4. The Fitch-Margoliash method was applied to analyze the protein sequences of NRPD4 and RPB4, and the sequences were then used to construct a distance tree. The analysis reveals that NRPD4 and RPB4 proteins in higher eukaryotes are grouped into one clade that is different from the RPB4 in budding yeast (Fig. 5C). Within the multicellular eukaryotic clade, AtRPB4 groups with its orthologs in rice, grape, and even humans, while the Arabidopsis NRPD4 and its orthologs in rice and grape form a plant-specific subclade (Fig. 5C). The result suggests that NRPD4 and RPB4 evolved from a common ancestor after this ancestor diverged from the yeast RPB4, and that NRPD4 belongs to a plant-specific branch that functions in TGS. Sequence analysis shows that the Arabidopsis NRPD4 has an extra N-terminal region that is absent in RPB4 from Arabidopsis, rice, Drosophila, and human (Fig. 5B). NRPD4 orthologs from other plants, including rice, grape, and Medicago, are also characterized by the N-terminal extension (Fig. 5B; data not shown). The N-terminal extension can only be found in plants and not in any other higher eukaryotic organisms, further supporting that NRPD4 may function in plant-specific polymerases.
Figure 5.
NRPD4 encodes a protein with sequence similarity to RPB4. (A) Diagram of the NRPD4 gene showing the positions of exons (solid boxes), introns, and sites of T-DNA insertions. (B) Sequence alignment of NRPD4 from Arabidopsis and rice and RPB4 from Arabidopsis, rice, yeast, Drosopila, and human. (C) Phylogenetic relationships among NRPD4 and RPB4 proteins. The NRPD4 proteins are from Arabidopsis, rice, and grape, while the RPB4 proteins are from the three plant species and budding yeast, Drosophila, and human. (D) Assay for complementation of the yeast rpb4Δ mutant. The cultures from each of the indicated strains were diluted and spotted onto YPD plates and incubated for 2 d at 28°C or 37°C.
To test whether NRPD4 can serve as an alternative member of RPB4 that functions in RNA polymerase II, we transformed the Arabidopsis RPB4 (AtRPB4), NRPD4, and an N-terminally truncated NRPD4 (75–205 amino acids) into yeast rpb4Δ cells (Woychik and Young 1989). NRPD4 or its N-terminally truncated version could not complement the high-temperature sensitivity phenotype of the yeast rpb4Δ mutant, whereas the Arabidopsis RPB4 partially complemented the yeast mutant (Fig. 5D). The result suggests that NRPD4 is not a functional subunit of RNA polymerase II. It is interesting to note that the budding yeast RPB4 (ScRPB4) also possesses an N-terminal extension (Fig. 5B), which may confer certain properties that are specific for the yeast protein. Nevertheless, AtRBP4, which does not have such an N-terminal extension, but not NRPD4 or its N-terminally truncated version, could partially substitute for ScRPB4 in yeast. This observation suggests that the functional conservation between AtRPB4 and ScRBP4 and the lack of functional conservation between NRPD4 and ScRPB4 are largely determined by their overall sequence similarities rather than the presence or absence of N-terminal extensions.
NRPD4 was used as a control in yeast two-hybrid assays to examine the interaction of another RdDM component, AGO4, with potential interacting partners. Interestingly, although no interaction was found between NRPD4 and AGO4 as expected, a strong self-activation was found for NRPD4 (Supplemental Fig. S5A). The result indicates that NRPD4 functions in transcriptional activation (Supplemental Fig. S5A).
To examine the tissue patterns of expression of NRPD4, the NRPD4 gene promoter was fused to the β-glucuronidase reporter gene (GUS). Analysis of the transgenic plants expressing NRPD4 promoter-GUS indicated that NRPD4 expression is strong in the shoot meristematic region and in root tips, is less strong but easily detectable in cotyledons and flowers, and is weak in young leaves (Supplemental Fig. S5B). To determine the subcellular localization of the NRPD4 protein, an YFP-NRPD4 translational fusion was constructed and introduced into Arabidopsis plants. Confocal imaging showed that the fusion protein was mainly localized in the nucleus, although weak signals were also present in the cytoplasm (Supplemental Fig. S5C).
Interphase subnuclear localization of NRPD4
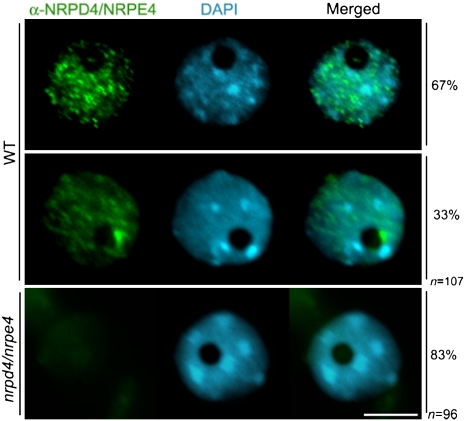
To determine the subnuclear organization of NRPD4, we performed immunolocalization with an anti-peptide antibody raised specifically to detect the native protein. Two distinct types of nuclear distribution were found for NRPD4 at interphase. In 67% of the cells, assigned as having type-1 interphase nuclei, NRPD4 was detected as small foci dispersed throughout the nucleoplasm (Fig. 6, top-row nucleus). Interestingly, NRPD4 was largely absent from the major heterochromatic DAPI-rich regions known as chromocenters (Soppe et al. 2002) and from the nucleolus region, which was easily identified as a dark hole after DAPI staining (Fig. 6, first-row nucleus). In contrast, in type-2 interphase nuclei, in addition to its diffuse nucleoplasmic distribution, NRPD4 was enriched in a round perinucleolar body or nucleolar dot (Fig. 6, middle-row nucleus). The failure to detect any NRPD4 signals in the nrpd4-1 loss-of-function mutant suggests that the antibody is specific for this protein (Fig. 6, bottom panel).
Figure 6.
Subnuclear distribution of NRPD4/NRPE4. Representative images of Arabidopsis interphase nuclei after immunolocalization of NRPD4/NRPE4 (in green) in the wild type (WT) and in the nrpd4/nrpe4 mutant. The frequency of nuclei displaying each interphase pattern is shown on the right. Nuclear DNA is stained with DAPI (blue). Bar, 5 μm.
NRPD4/NRPE4 is partially colocalized with both Pol IV and Pol V at interphase
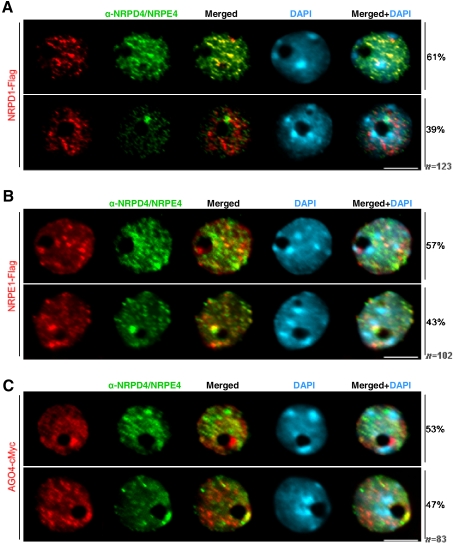
To determine whether the NRPD4/NRPE4 protein pools are substantially colocalizing with part of both Pol IV and Pol V complexes, we examined their relative localization in the nucleus by double immunostaining. For this purpose, we analyzed the localization of NRPD4/NRPE4 using the native antibody in transgenic lines expressing epitope-tagged NRPD1 and NRPE1. As shown in Figure 7A, NRPD1 is distributed in a heterogeneous subnuclear pattern, displaying discrete larger foci clustered near the heterochromatic regions and also more diffusely distributed signals throughout the nucleoplasm as described previously (Pontes et al. 2006). We observed that within NRPD4/NRPE4 type-1 nuclei showing small, dispersed foci in the nucleoplasm and no detectable signals in the nucleolus or nucleolar periphery, the larger NRPD1 foci were mostly located near heterochromatin, and NRPD4/NRPE4 were largely colocalizing (Fig. 7A). In contrast, no significant colocalization of immunostaining signals was found in NRPD4/NRPE4 type-2 nuclei that showed a prominent nucleolar dot (Fig. 7A).
Figure 7.
Colocalization of NRPD4/NRPE4 with Pol IV, Pol V, and AGO4. Immunofluorescence of NRPD4/NRPE4 (green) in transgenic lines expressing full-length epitope-tagged NRPD1 (A), NRPE1 (B), and AGO4 (C) (all in red). The merged images reveal bright yellow signals due to the overlap of red and green channels. The frequency of nuclei displaying each interphase pattern is shown to the right. Nuclear DNA is stained with DAPI (blue). Bar, 5 μm.
The largest subunit of Pol V, NRPE1, is clustered within heterochromatic regions and shows a nucleolar Cajal body-like structure near or within the nucleolus (Li et al. 2006, 2008; Pontes et al. 2006). In the type-1 NRPD4/NRPE4 nuclear distribution, five to seven spots were colocalizing with NRPE1 in the nucleoplasm near heterochromatin, while no colocalization within the nucleolar dot was observed (Fig. 7B). In contrast, in NRPD4/NRPE4 type-2 nuclei, a prominent bright-yellow signal was displayed within the nucleolar dot (Fig. 7B), suggesting that this nuclear region is the main site in the nucleus where this protein is colocalizing with NRPE1.
Previous studies have revealed that NRPE1 is colocalized with AGO4 in the nucleus and that these two proteins may be part of the same effector complex (Li et al. 2006). AGO4 was distributed in the nucleoplasm, excluded from major heterochromatic regions, and, like NRPE1, also showed a nucleolar dot (Fig. 7C). Our colocalization analyses revealed that NRPD4/NRPE4 was enriched at the AGO4 nucleolar dot and that the proteins colocalized to a lesser extent in the nucleoplasm (Fig. 7C).
NRPD4/NRPE4 interacts with NRPD1 and NRPE1
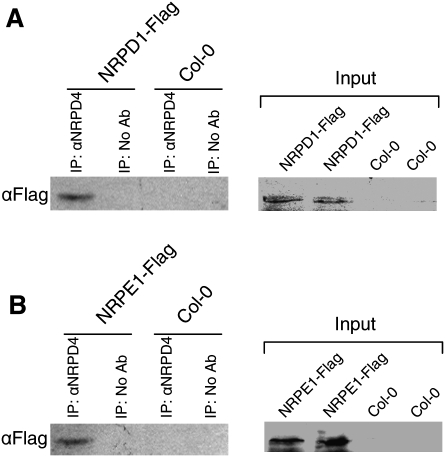
Because of the high similarity between NRPD4/NRPE4 and RPB4 and our findings that NRPD4/NRPE4 functions in the RdDM pathway, we asked whether NRPD4 might be a functional subunit of Pol IV and/or Pol V. Coimmunoprecipitation experiments were carried out to detect association between NRPD4/NRPE4 and the largest subunit of Pol IV or Pol V. Protein extracts from NRPD1-Flag transgenic plants, NRPE1-Flag transgenic plants, or wild-type plants without the transgenes were incubated with antibodies against NRPD4/NRPE4, followed by precipitation with protein A agarose beads. The bound protein complex was washed and then eluted for Western blot analysis using anti-Flag antibody to detect whether NRPD1a/NRPD1 or NRPD1b/NRPE1 was coimmunoprecipitated with NRPD4/NRPE4. The analysis revealed that both NRPD1-Flag (Fig. 8A) and NRPE1-Flag (Fig. 8B) could be coprecipitated by the NRPD4/NRPE4 antibodies. The results suggest that NRPD4/NRPE4 can interact with both NRPD1 and NRPE1 in vivo. Thus, NRPD4/NRPE4 might function as a subunit in both Pol IV and Pol V.
Figure 8.
NRPD4/NRPE4 interacts with NRPD1 and NRPE1. Coimmunoprecipitation analysis of the interaction between NRPD4 and NRPD1 (A) or NRPE1 (B). Anti-NRPD4 antibody was incubated with protein extracts from flowers of NRPD1-Flag plants, NRPE1-Flag plants, or wild-type plants without transgenes, followed by immunoprecipitation with protein A beads. The bound proteins were washed and eluted, followed by Western blotting with anti-Flag antibody.
Discussion
DNA methylation is an important epigenetic mark conserved in plants, mammals, and some fungi. Approximately one-third of methylated sequences in the Arabidopsis genome are associated with small RNAs (Zhu 2008). RdDM is a mechanism for achieving sequence-specific de novo DNA methylation. Several components have been identified for the RdDM pathway, mostly by forward genetic screens for suppressors of TGS in Arabidopsis. In our system, mutations in the DNA demethylase gene ROS1 caused TGS of the RD29A-LUC reporter gene, the endogenous RD29A gene, and the 35S-NPTII transgene. Our previous screens for ros1 suppressors, which were largely based on the kanamycin resistance phenotype, led to the identification of RPA2, a component of DNA replication and repair that is required for silencing of 35S-NPTII, possibly by functioning in the spreading of TGS from RD29A-LUC to the linked 35S-NPTII transgene or in maintaining the heterochromatin at 35S-NPTII (Kapoor et al. 2005). The screens also led to the identification of AGO6 (Zheng et al. 2007) as a component required for RdDM, and to the identification of SUP32/UBP26 (Sridhar et al. 2007), a ubiquitin protease. The sup32/ubp26 mutant revealed a necessary role of histone H2B deubiquitination in RdDM. In this study, we carried out a large-scale, systematic screen for ros1 suppressors. This effort led to the identification of seven known components for RdDM.
Mutations in NRPD1, NRPE1, NRPD2/NRPE2, HEN1, and DRD1 suppressed the TGS of RD29A-LUC but not of 35S-NPTII. These mutations also blocked or reduced the accumulation of 24-nt siRNAs and reduced DNA methylation, particularly CHH methylation of RdDM target loci (Figs. 3, 4; Supplemental Figs. S3, S4; data not shown). These results demonstrate that RD29A promoter siRNAs are the initial trigger of TGS in the ros1 mutant. The hda6 mutation, like sup32/ubp26 mutations (Sridhar et al. 2007), suppressed the TGS of both RD29A-LUC and 35S-NPTII. These observations indicate that histone deacetylation and deubiquitination are required for both siRNA-dependent (RD29A-LUC) and siRNA-independent (35S-NPTII) TGS. It is interesting that the ago4 mutation also partially suppressed the TGS of 35S-NPTII, suggesting that AGO4 may also have a siRNA-independent role in TGS.
Importantly, our effort led to the identification of several new components of RdDM. One of them, RDM2, is similar to the RPB4 subunit of RNA polymerase II. Our data show that another gene (AtRPB4), but not RDM2, can function as part of Pol II. This is consistent with the sequence divergence between RDM2 and RPB4 orthologs. In addition to divergence along the entire length of the AtRPB4 sequence, RDM2 has evolved an N-terminal extension specific to RDM2 orthologs in plants. We found that RDM2 is associated with both NRPD1 and NRPE1. Furthermore, RDM2 colocalizes, at least partly, with both NRPD1 and NRPE1 in subnuclear foci. These results suggest that RDM2 is part of Pol IV and Pol V and thus should be referred to as NRPD4/NRPE4. This conclusion is supported by the important genetic function of NRPD4/NRPE4 in RdDM. Mutations in this critical component result in reduced siRNA accumulation, defective DNA methylation, and reactivation of sequences normally silenced by RdDM.
Despite the important roles of Pol IV and Pol V in RdDM, neither their protein compositions nor their biochemical activities are presently known. It is possible that Pol IV and Pol V need only the largest and second largest subunits for their function in RdDM. However, our identification of a new subunit of Pol IV and Pol V in this study suggests that like Pol II (Cramer 2002), Pol IV and Pol V are multisubunit complexes. Consistent with this notion of multisubunit structure for both Pol IV and Pol V, proteomic analysis of NRPD1- or NRPE1-containing protein complexes from plants revealed several subunits shared by Pol IV, Pol V, and Pol II (Ream et al. 2008).
Research has clearly established that in Pol II, RPB4 and RPB7 form a dimer outside the core polymerase complex, and that in addition to transcriptional roles, RPB4 and RPB7 have functions independent of Pol II (Khazak et al. 1998; Larkin and Guilfoyle 1998; Choder 2004). Although coimmunoprecipitation experiments showed that NRPD4/NRPE4 can be associated with NRPD1 and NRPE1, immunolocalization studies show that NRPD4/NRPE4 does not always colocalize with either NRPD1 or NRPE1. Perhaps NRPD4/NRPE4 also has functions independent of Pol IV and Pol V. Indeed, some YFP-NRPD4 fusion proteins appear to be localized outside the nucleus (Supplemental Fig. S5C). Even within the interphase nucleus, it is unclear at the present time whether NRPD1/NRPD4 complexes and NRPE1/NRPE4 complexes are mutually exclusive or whether the disparate patterns of NRPD4/NRPE4 localization may be due to cell cycle regulation. It is also unclear whether NRPD4/NRPE4 may shuttle between Pol IV and Pol V and whether it is needed for only part of the functions of Pol IV and Pol V. NRPD4/NRPE4 may be needed for Pol IV function in initiating siRNA production from a subset of Pol IV-dependent loci. This scenario could explain why the apparently null allele (nrpd4-2) of NRPD4/NRPE4 substantially reduces siRNA accumulation from some but not all NRPD1-dependent loci in the Col-0 background (Supplemental Fig. S4B). On the other hand, the nrpd4/nrpe4 mutations have a strong effect on DNA methylation at all Pol IV- and Pol V-dependent loci examined. This observation suggests that NRPD4/NRPE4 is important for Pol V function even when not needed for Pol IV function. Nevertheless, it is clear that NRPD4/NRPE4 does not always associate with Pol V either. A portion of NRPE1 is always found in the nucleolar dot, whereas NRPD4/NRPE4 localizes in the nucleolar dot in only a subset of the cells examined. Although the nucleolar dot has been hypothesized to be a processing center for siRNAs because a portion of RDR2, DCL3, AGO4, NRPE1, and siRNAs are found there (Li et al. 2006; Pontes et al. 2006), the function of this structure are still unknown. Like its RPB4 counterpart in Pol II transcription and post-transcriptional RNA metabolism, NRPD4/NRPE4 also appears to be a dynamic regulator rather than a stable subunit of pol IV and pol V.
Materials and methods
Plant growth, mutant screening, and cloning
The wild-type C24 plants and the ros1 mutant plants carry the homozygous Rd29A-LUC transgene (Ishitani et al. 1997). A T-DNA-mutagenized population in the ros1 mutant background was generated as described previously (Kapoor et al. 2005). Plants were grown in a controlled room at 22°C with 16 h of light and 8 h of darkness. About 200 T2 seeds from each 25-line pool were sterilized and planted individually in agar plates containing Murashige-Skoog (MS) salts. RD29A-LUC expression was analyzed as described before (Ishitani et al. 1997). In brief, 7-d-old seedlings were sprayed with luciferin (Promega) for luminescence imaging under a CCD camera. The putative ros1 suppressors that emitted high luminescence in MS plates were transferred to grow in soil. After the seedlings grew for 1 mo, a young leaf from each seedling was cut and treated with 200 mM NaCl on filter paper for 3 h. The luminescence imaging was applied on the leaves to further eliminate false positives. The genomic sequence flanking the T-DNA insertion in the positive mutant plants was determined by using the TAIL-PCR (Liu et al. 1995). For untagged mutants, map-based cloning was used to identify the mutated genes.
The genomic sequence of NRPD4 was amplified and cloned into a Gateway vector PMDC164 for complementation assay. The cDNA clone containing the full-length NRPD4 ORF was obtained from ABRC (clone no. U67510) and was cloned into the Gateway vector PEG104 for YFP-NRPD4 expression in transgenic plants. The RPB4 full-length cDNA, NRPD4 full-length cDNA, and its truncated version were cloned into the yeast expression vector pYPGE15 for yeast complementation assay. The NRPD4 and AGO4 cDNAs were cloned into pDEST22 and pDEST33 for yeast two-hybrid assay.
RNA blot analysis
For Northern blot analysis, Arabidopsis seedlings were grown in MS plates for 2 wk and harvested after cold treatment (4°C, 1 d) or no treatment. Total RNA was extracted using Trizol (Invitrogen) and dissolved in DEPC-treated H2O. Twenty micrograms of total RNA was separated on 1.2% denaturing agarose gels containing 10% formaldehyde and was transferred to Hybond-N+ membranes (Amersham) for Northern hybridization. The small RNA used in this study was extracted from flowers. In the total RNA of flowers, the small-molecular-weight RNA was separated from high-molecular-weight RNA by adding an equal volume of PEG 8000 solution (20% PEG, 1 M NaCl). Therefore, high-molecular-weight RNA was precipitated after centrifugation, and small-molecular-weight RNA in the supernatant was precipitated by adding an equal volume of isopropanol overnight at −20°C. Sixty micrograms of small-molecular-weight RNA was separated on a 15% polyacrylamide gel at 200 V for 3 h. The gel was electroblotted to Hybond-N+ membranes (Amersham). Membranes were cross-linked and hybridized overnight at 38°C with [γ-32P]ATP-labeled DNA oligonucleotides or α-32PdCTP-labeled amplified DNA in PerfectHyb buffer (Sigma). Northern blots were washed twice with 2× SSC, 0.1% SDS and then once with 1× SSC, 0.1% SDS. Washed blots were exposed to X-ray films. The DNA oligo probes and primers for probe amplification are listed in Supplemental Table S1.
Yeast strains and yeast complementation
The yeast strains used for complementation assay of RPB4 and NRPD4 were the wild-type strain (YSB1140) and its isogenic derivative rpb4Δ (YSB1755) (Runner et al. 2008). The yeast two-hybrid assay was performed in the yeast strain MaV203 (Invitrogen). Yeast transformation was performed using the yeast transformation buffer (0.1 M LiAc, 40% PEG3350 in 1× TE). Transformants were plated and selected on synthetic complete medium (SC-aa) that lacked the specified amino acid. The positive colonies were incubated and used for the growth assay. For the rpb4Δ complementation assay, the culture was diluted and spotted onto YPD plates. The plates were incubated for 2 d at 28°C and 37°C, respectively. For the yeast two-hybrid assay, the culture was diluted and spotted onto SC-aa medium plates lacking leucine, tryptophan, and histine and containing 50 mM 3-amino-1,2,4-triazole. The plates were incubated for 2–4 d at 28°C for interaction analysis.
DNA methylation assays
For Chop-PCR, genomic DNA (500 ng) was digested with the methylation-sensitive restriction enzyme HaeIII overnight. After digestion, ∼10% of digestion reaction DNA was used for amplification of AtSN1 using gene-specific primers (Supplemental Table S1). PCR conditions were 5 min at 94°C followed by 35 cycles of 30 sec at 94°C, 30 sec at 56°C, and 1 min at 72°C. PCR products were then subjected to electrophoresis. For the Southern hybridization assay, 5 μg of genomic DNA was digested with HpaII, MspI, or HaeIII. The digested DNA was loaded onto a 1.2% agrose gel and transferred to Hybond-N+ membranes. The 180-bp DNA repeat and 5S rDNA repeat were labeled with α-32PdCTP for Southern hybridization to determine their DNA methylation status. For bisulfite sequencing, 4 μg of genomic DNA was digested overnight with EcoRI and HindIII, and the digested DNA was denatured and then used for bisulfite treatment in a DNA sodium bisulfite treatment mixture including 15 μL of DNA, 102 μL of fresh 40.5% sodium bisulfite (pH 5.0; Sigma), and 3 μL of fresh 20 mM hydroquinone (Sigma). Bisulfite treatment was performed in a PCR machine for 16 h at 55°C with a jolt to 95°C for 5 min every 3 h. The sodium bisulfite-treated DNA was purified with the Wizard DNA Clean-up system (Promega) and dissolved in 200 μL of H2O. The purified DNA was incubated for 20 min at 37°C after 13 μL of 5 N NaOH was added, followed by precipitation with 3 volumes of ethanol overnight at −20°C. The collected DNA was used for amplification of endogenous and transgenic RD29A promoters, AtSN1, and MEA-ISR elements. The PCR product was cloned into the pGEM-T easy vector (Promega), and at least 10 individual clones were sequenced for each sample. The primer information for RD29A promoter, AtSN1, and MEA-ISR was obtained from Kapoor et al. (2005), Xie et al. (2004), and Cao and Jacobsen (2002).
RT–PCR analysis
Total RNA was extracted with Trizol reagent (Sigma) from flowers. Contaminating DNA was removed with DNase (Promega). For semiquantitative RT–PCR analysis, 5 μg of total RNA was used for the first-strand cDNA synthesis with the SuperScript III RT–PCR System (Invitrogen). The cDNA reaction mixture was diluted and then used as template for PCR. The PCR conditions were preincubation for 5 min at 95°C, followed by 28–35 cycles of denaturation for 30 sec at 95°C, annealing for 30 sec at 56°C, and extension for 60 sec at 72°C. TUB8 was used as the internal control. All primers used in RT–PCR analysis are listed in Supplemental Table S1.
Coimmunoprecipitation assays and immunoblotting
The immunoprecipitation assay was carried out as described previously (Li et al. 2006).
One gram of seedlings from NRPD1a-Flag plants, NRPD1b-Flag plants, or wild-type plants without transgenes was used for preparation of cell lysates with 2 mL of protein extraction buffer. The crude protein extracts were precleared with protein A agarose beads (Pierce), followed by incubation with anti-NRPD4 antibody. The protein complex was captured and precipitated by protein A agarose beads, washed five times with extraction buffer, and eluted by boiling the beads in SDS sample buffer. The eluted samples were resolved on an 8% SDS-PAGE gel for Western blotting by anti-Flag antibody. Rabbit anti-NRPD4 antibodies were generated against the synthetic peptide CGKSDKPTKSSKNSL-amide and affinity-purified using the synthetic peptide.
Immunostaining
Interphase nuclei were isolated as described previously (Jasencakova et al. 2000). Upon 4% paraformaldehyde post-fixation, the nuclei were incubated overnight at 4°C with primary antibodies for NRPD4/NRPE4 (1:100), anti-Flag (1:200; Sigma), and anti-cmyc (1:200; Chemicon). Secondary antibodies, anti-rabbit Alexa 488 (Invitrogen) and anti-mouse Alexa 594, were diluted at 1:500 in PBS and incubated for 3 h at 37°C. DNA was counterstained with 1 μg/mL DAPI in Prolong Gold mounting medium (Invitrogen). The preparations were inspected with a Nikon Eclipse E800i epifluorescence microscope equipped with a Photometrics Coolsnap ES Mono digital camera. Images were acquired by the Phylum software and pseudocolored and merged in Adobe Photoshop.
Acknowledgments
We thank R. Stevenson for technical assistance, S. Buratowski for providing yeast strains, and M. Zheng for use of a fluorescence microscope. This work was supported by National Institutes of Health Grants R01GM070795 and R01GM059138 to J.-K.Z. O.P. was supported by an Edward Mallinckrodt Foundation Award.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1765209.
Supplemental material is available at http://www.genesdev.org.
References
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Bender J. DNA methylation and epigenetics. Annu. Rev. Plant Biol. 2004;55:41–68. doi: 10.1146/annurev.arplant.55.031903.141641. [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. 2002;99:16491–16498. doi: 10.1073/pnas.162371599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Chandler V.L., Stam M. Chromatin conversations: Mechanism and implications of paramutation. Nat. Rev. Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- Choder M. Rpb4 and Rpb7: Subunits of RNA polymerase II and beyond. Trends Biochem. Sci. 2004;29:674–681. doi: 10.1016/j.tibs.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Cramer P. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- Eamens A., Vaistij F.E., Jones L. NRPD1a and NRPD1b are required to maintain post-transcriptional RNA silencing and RNA-directed DNA methylation in Arabidopsis. Plant J. 2008;55:596–606. doi: 10.1111/j.1365-313X.2008.03525.x. [DOI] [PubMed] [Google Scholar]
- Ebert A., Schotta G., Lein S., Kubicek S., Krauss V., Jenuwein T., Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes & Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldan-Arjona T., David L., Zhu J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Heard E. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Stevenson B., Zhu J.K. Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell. 1997;9:1935–1949. doi: 10.1105/tpc.9.11.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z., Meister A., Walter J., Turner B.M., Schubert I. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell. 2000;12:2087–2100. doi: 10.1105/tpc.12.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T., Mette M.F., Kreil D.P., Aufsatz W., Matzke M., Matzke A.J. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Kanno T., Huettel B., Mette M.F., Aufsatz W., Jaligot E., Daxinger L., Kreil D.P., Matzke M., Matzke A.J. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat. Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- Kapoor A., Agarwal M., Andreucci A., Zheng X., Gong Z., Hasegawa P.M., Bressan R.A., Zhu J.K. Loss-of-function mutations in a conserved replication protein suppress transcriptional gene silencing in a DNA methylation-independent manner in Arabidopsis. Curr. Biol. 2005;579:5889–5898. doi: 10.1016/j.cub.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Khazak V., Estojak J., Cho H., Majors J., Sonoda G., Testa J.R., Golemis E.A. Analysis of the interaction of the novel RNA polymerase II (pol II) subunit hsRPB4 with its partner hsRPB7 and with pol II. Mol. Cell. Biol. 1998;18:1935–1945. doi: 10.1128/mcb.18.4.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin R.M., Guilfoyle T.J. Two small subunits in Arabidopsis RNA polymerase II are related to yeast RPB4 and RPB7 and interact with one another. J. Biol. Chem. 1998;273:5631–5637. doi: 10.1074/jbc.273.10.5631. [DOI] [PubMed] [Google Scholar]
- Li C.F., Pontes O., El-Shami M., Henderson I.R., Bernatavichute Y.V., Chan S.W., Lagrange T., Pikaard C.S., Jacobsen S.E. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Li C.F., Henderson I.R., Song L., Fedoroff N., Lagrange T., Jacobsen S.E. Dynamic regulation of ARGONAUTE4 within multiple nuclear bodies in Arabidopsis thaliana. PLoS Genet. 2008;4:e27. doi: 10.1371/journal.pgen.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995;8:457–463. doi: 10.1046/j.1365-313x.1995.08030457.x. [DOI] [PubMed] [Google Scholar]
- Matzke M., Aufsatz W., Kanno T., Daxinger L., Papp I., Mette M.F., Matzke A.J. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim. Biophys. Acta. 2004;1677:129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Mette M.F., Aufsatz W., van der Winden J., Matzke M.A., Matzke A.J.M. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher R.A., Schwach F., Studholme D., Baulcombe D.C. Pol IVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc. Natl. Acad. Sci. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Nunes P.C., Pontes O., Pikaard C.S. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Pontes O., Li C.F., Nunes P.C., Haag J., Ream T., Vitins A., Jacobsen S.E., Pikaard C.S. The Arabidopsis chromatin-modifying nuclear siRNA pathway involves a nucleolar RNA processing center. Cell. 2006;126:79–92. doi: 10.1016/j.cell.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Pontier D., Yahubyan G., Vega D., Bulski A., Saez-Vasquez J., Hakimi M.A., Lerbs-Mache S., Colot V., Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & Dev. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ream T.S., Haag J.R., Wierzbicki A.T., Nicora C.D., Norbeck A., Zhu J.K., Hagen G., Guilfoyle T.J., Pasa-Tolic L., Pikaard C.S. Subunit composition of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell. 2008 doi: 10.1016/j.molcel.2008.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runner V.M., Podolny V., Buratowski S. The Rpb4 subunit of of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol. Cell. Biol. 2008;28:1883–1891. doi: 10.1128/MCB.01714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R., Spielman M. Epigenetics: Imprinting in plants and mammals—The same but different? Curr. Biol. 2004;14:R201–R203. doi: 10.1016/j.cub.2004.02.022. [DOI] [PubMed] [Google Scholar]
- Soppe W.J., Jasencakova Z., Houben A., Kakutani T., Meister A., Huang M.S., Jacobsen S.E., Schubert I., Fransz P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wierzbicki A.T., Haag J.R., Pikaard C.S. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woychik N.A., Young R.A. RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol. 1989;9:2854–2859. doi: 10.1128/mcb.9.7.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L., Gustafson A., Kasschau K., Lellis A., Zilberman D., Jacobsen S., Carrington J. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Zhu J., Kapoor A., Zhu J.K. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. Epigenome sequencing comes of age. Cell. 2008;133:395–397. doi: 10.1016/j.cell.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Jacobsen S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]