Abstract
At discrete points in development, transient signals are transformed into long-lasting cell fates. For example, the asymmetric identities of two Caenorhabditis elegans olfactory neurons called AWCON and AWCOFF are specified by an embryonic signaling pathway, but maintained throughout the life of an animal. Here we show that the DNA-binding protein NSY-7 acts to convert a transient, partially differentiated state into a stable AWCON identity. Expression of an AWCON marker is initiated in nsy-7 loss-of-function mutants, but subsequently lost, so that most adult animals have two AWCOFF neurons and no AWCON neurons. nsy-7 encodes a protein with distant similarity to a homeodomain. It is expressed in AWCON, and is an early transcriptional target of the embryonic signaling pathway that specifies AWCON and AWCOFF; its expression anticipates future AWC asymmetry. The NSY-7 protein binds a specific optimal DNA sequence that was identified through a complete biochemical survey of 8-mer DNA sequences. This sequence is present in the promoter of an AWCOFF marker and essential for its asymmetric expression. An 11-base-pair (bp) sequence required for AWCOFF expression has two activities: One region activates expression in both AWCs, and the overlapping NSY-7-binding site inhibits expression in AWCON. Our results suggest that NSY-7 responds to transient embryonic signaling by repressing AWCOFF genes in AWCON, thus acting as a transcriptional selector for a randomly specified neuronal identity.
Keywords: Homeodomain, olfactory development, transcriptional maintenance, transcriptional repression, activity-dependent gene expression, cGMP-dependent protein kinase
Most neurons are born in embryogenesis but maintain their morphologies, physiological properties, and patterns of gene expression throughout life. The genetic pathways required to maintain neural identity can be distinct from those required for its establishment, suggesting that maintenance is an active genetic process. For example, MeCP2, a transcriptional regulator defective in the autism-like disorder Rett syndrome, acts in mature neurons to maintain dendritic complexity and synaptic function. Because of this late action of MeCP2, children with Rett syndrome are normal at birth and become symptomatic only months or years later, often losing language and motor skills that they acquired early in life (Dragich et al. 2000; Chen et al. 2001; Kishi and Macklis 2004; Guy et al. 2007).
In the nematode Caenorhabditis elegans, distinct genetic mechanisms control the initial specification and subsequent maintenance of a pair of olfactory neuron identities called AWCON and AWCOFF. AWCON and AWCOFF neurons have similar morphologies but different functions and patterns of gene expression (Troemel et al. 1999). AWCON senses the odor butanone, expresses the G protein-coupled receptor gene str-2, and can promote either attractive or repulsive behaviors based on modulatory inputs from a guanylate cyclase (Troemel et al. 1999; Wes and Bargmann 2001; Tsunozaki et al. 2008). The contralateral neuron, AWCOFF, senses the odor 2,3-pentanedione, expresses the G protein-coupled receptor gene srsx-3, and has only been observed to mediate attractive behaviors (Wes and Bargmann 2001; Bauer Huang et al. 2007). Each animal has one AWCON and one AWCOFF neuron, but these identities are randomly assumed by the right or left AWC neuron in each individual (Troemel et al. 1999). This element of randomness is unusual in C. elegans, which is otherwise notable for the predictability of its cell fate decisions (Sulston and Horvitz 1977). The initial AWCON/AWCOFF decision depends on a signaling pathway in which a claudin-like transmembrane protein, NSY-4, and an embryonic gap junction network generated by the innexin NSY-5 induce one AWC to exit the AWCOFF default identity and become AWCON (Vanhoven et al. 2006; Chuang et al. 2007). The induced AWCON neuron then provides feedback to the contralateral neuron to stabilize the AWCOFF identity. Genetic analysis indicates that NSY-5/NSY-4 signaling acts through the calcium channel UNC-2/UNC-36 and the membrane protein OLRN-1, and regulates a kinase cascade that includes UNC-43 (CaMKII), NSY-1 (ASK1/MAPKKK), SEK-1 (SEK/MAPKK), and TIR-1 (an adaptor protein). High activity of the calcium/kinase cascade is associated with the future AWCOFF neuron, and low activity with the future AWCON (Sagasti et al. 2001; Tanaka-Hino et al. 2002; Chuang and Bargmann 2005; Bauer Huang et al. 2007).
The NSY-5 gap junction network observed in electron micrographs disappears after hatching, and the downstream signaling pathway apparently becomes inactive as well (Chuang and Bargmann 2005; Chuang et al. 2007). AWC left–right asymmetry is stable throughout the lifetime of the animal, so these transient signaling events must be captured and stabilized by molecules that act after embryogenesis. Indeed, post-embryonic expression of AWC markers requires a different set of genes than those that act in the embryo. Several genes that affect olfactory signal transduction are required to maintain the AWCON marker str-2∷GFP after the first larval stage (Troemel et al. 1999). These include odr-1 and daf-11, which encode receptor guanylate cyclase homologs that are localized to the sensory cilia and thought to produce cGMP during chemosensation (Birnby et al. 2000; L'Etoile and Bargmann 2000). Mutations in genes encoding olfactory Gα proteins also boost str-2 expression during larval stages (Lans et al. 2004; Lans and Jansen 2006). The genetic requirement for olfactory signal transduction molecules suggests that ongoing sensory activity maintains AWC gene expression.
The contrast between embryonic signaling mutants and post-embryonic maintenance mutants defines a period of interest near hatching when the AWC identity is crystallized. Here we show that nsy-7, which encodes a divergent transcriptional regulator, coordinates the transition from establishment to maintenance of the AWCON identity. nsy-7 mutants exhibit evidence of left–right asymmetry during the first larval stage, but lose this asymmetry and markers of AWCON function by adulthood. We find that nsy-7 expression is an early target of the developmental signaling pathway that induces AWCON, and provide evidence that it directly represses AWCOFF genes to ensure a stable AWCON identity.
Results
Left–right AWC asymmetry emerges in the L1 larval stage
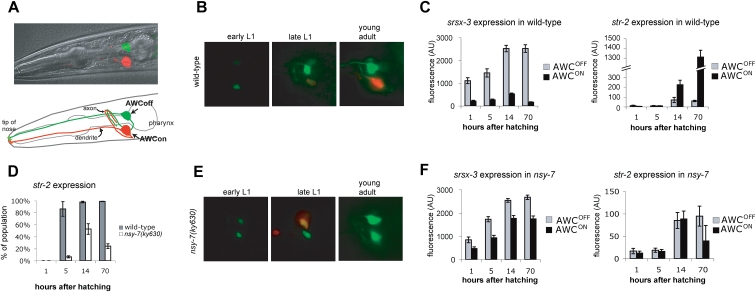
To visualize the AWCON and AWCOFF neurons simultaneously, we generated a transgenic strain, kyIs408, with integrated str-2∷dsRed2 (AWCON) and srsx-3∷GFP (AWCOFF) reporter genes. In adult animals, srsx-3∷GFP and str-2∷dsRed2 were expressed in a mutually exclusive pattern in contralateral AWC neurons (Fig. 1A). We used this strain to characterize the appearance of left–right asymmetry during development.
Figure 1.
nsy-7(ky630) mutants are defective for maintenance of asymmetric identity in AWC. (A) Adult animal with the integrated array kyIs408. A fluorescence image of AWCON, expressing str-2∷dsRed2, and AWCOFF, expressing srsx-3∷GFP, is overlaid on a DIC image. (B) AWC neurons in wild-type animals in early L1 (1 h after hatching), late L1 (14 h after hatching), or in the young adult. (Red fluorescence) str-2∷dsRed2; (green fluorescence) srsx-3∷GFP. (C) Quantitation of srsx-3∷GFP and str-2∷dsRed fluorescence (n = 25–38 animals). In each animal, the cell with higher srsx-3∷GFP expression was defined as AWCOFF. (D) Fraction of nsy-7(ky630) animals expressing str-2∷dsRed2 at 1, 5, 14, and 70 h after hatching; 1–14 h are L1 stage, and 70 h is adult. (E) AWC neurons in nsy-7(ky630) animals, as in B. (F) Quantitation of srsx-3∷GFP and str-2∷dsRed fluorescence in nsy-7(ky630) mutants, as in C (n = 33–45 animals).
In newly hatched animals, srsx-3∷GFP was expressed in both AWCs (Fig. 1B). The two AWCs differed approximately fivefold in their level of srsx-3 expression, suggesting that left–right asymmetry had been initiated by this time (Fig. 1C). Over the course of the first larval stage (14 h), expression of srsx-3 increased twofold to threefold in both AWC neurons. Between the L2 larval stage and the adult stage, srsx-3 expression fell in AWCON (defined as the AWC with low srsx-3 expression), and remained high in AWCOFF (the AWC with high srsx-3 expression).
As previously observed with the marker str-2∷GFP (Troemel et al. 1999), str-2∷dsRed was not expressed in the AWCs at hatching. Beginning ∼5 h after hatching (mid-L1), faint str-2 expression could be detected in one AWC neuron, which was invariably the AWC with weaker expression of srsx-3. At the L1/L2 transition, >95% of animals expressed str-2, always in the AWC with weaker srsx-3 expression (Fig. 1B–D). If either AWCON or AWCOFF is killed at this stage, the surviving AWC does not change its pattern of gene expression, suggesting that the AWCON and AWCOFF identities have been irreversibly determined (Troemel et al. 1999). Between the L2 larval stage and the adult stage, str-2 expression increased in the AWCON neuron.
These results establish the timing of three events in AWC differentiation: repression of srsx-3 in AWCON, which begins by hatching; induction of str-2 in AWCON during the mid- to late L1 stage; and the increased expression of srsx-3 and str-2 in the appropriate AWCs between L1 and adult. As expected from previous studies (Troemel et al. 1999), AWCL and AWCR were equally likely to enter the str-2+srsx-3− AWCON state or the str-2−srsx-3+ AWCOFF state.
nsy-7(ky630) initiates but fails to maintain AWCON asymmetry
nsy-7(ky630) was isolated from a screen for mutants that fail to express str-2 in either AWC neuron (Vanhoven et al. 2006). As adults, these mutants had a typical 2-AWCOFF phenotype in which both AWC neurons expressed srsx-3. Like olfactory transduction mutants, but unlike most other mutants from this screen, nsy-7(ky630) larvae exhibited evidence of AWC left–right asymmetry during the L1 stage, when many animals expressed str-2 in one of the two AWC neurons (Fig. 1D). The fraction of animals expressing str-2 decreased later in development (Fig. 1D).
To define the differences between wild type and nsy-7 more precisely, the levels of str-2∷dsRed and srsx-3∷GFP expression were compared quantitatively at different developmental stages. Throughout development, both AWCs in nsy-7 mutants expressed srsx-3 at similar levels resembling the levels in normal AWCOFF neurons (Fig. 1E,F). At the L1/L2 transition, half of the nsy-7 animals also expressed str-2 in one AWC neuron (Fig. 1D,E), the signature of an AWCON-like identity. str-2 expression was always restricted to one of the two AWCs; this cell was equally likely to be the cell with slightly higher or slightly lower srsx-3∷GFP expression (Fig. 1F). The fraction of animals expressing str-2∷dsRed fell after the L1 stage, as did the level of str-2∷dsRed expression within AWC (Fig. 1F). Some adult animals expressed str-2∷dsRed at low levels, but because of the low levels, the fraction scored as positive varied between experiments. All AWC neurons in adults expressed srsx-3. These results suggest that nsy-7(ky630) mutants initiate elements of an asymmetric AWCON-like identity, but cannot repress the AWCOFF marker or maintain the AWCON marker after the L1 stage.
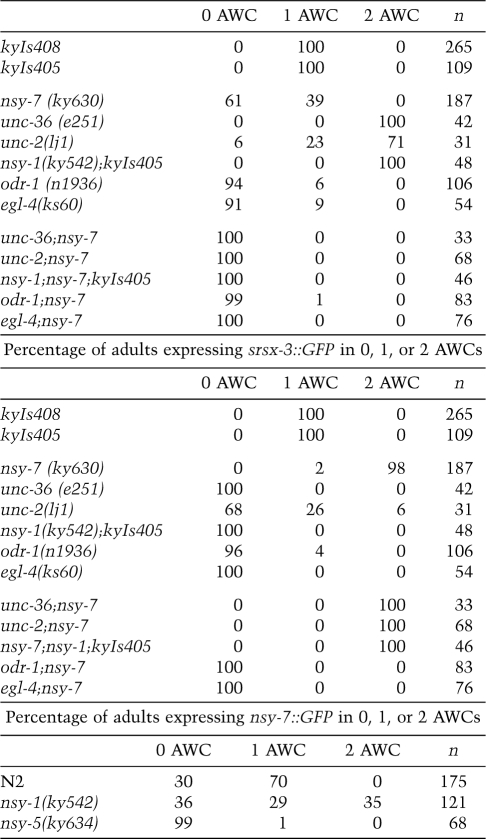
If nsy-7 mutants cannot stabilize the post-embryonic AWCON identity, AWCON markers should be lost regardless of the activity of the upstream signaling pathway. This prediction was tested by making double mutants between nsy-7 and genes in the signaling pathway for AWC asymmetry. Null mutants in the calcium channel genes unc-2 and unc-36 or in the downstream MAPKKK gene nsy-1 have a 2-AWCON phenotype due to disruptions in signaling between the embryonic AWC neurons (Troemel et al. 1999). nsy-7;unc-2 mutants, nsy-7;unc-36 mutants, and nsy-7;nsy-1 mutants all resembled nsy-7: In the double mutants, both AWCs expressed srsx-3 and neither expressed str-2, a 2-AWCOFF phenotype (Table 1). These results are consistent with an essential function for nsy-7 downstream from unc-2, unc-36, and nsy-1, and suggest that nsy-7 may maintain the AWCON identity and suppress the AWCOFF identity.
Table 1.
AWC gene expression in single and double mutants
Olfactory cGMP transduction pathways affect both AWCON and AWCOFF
The time course of str-2 expression in nsy-7 mutants resembles that of odr-1 olfactory transduction mutants, which initiate but fail to maintain asymmetric str-2 expression in AWCON (Troemel et al. 1999). odr-1 encodes a guanylate cyclase homolog involved in olfactory transduction, whose late role may represent an activity-dependent input into AWC gene expression (L'Etoile and Bargmann 2000). Unlike nsy-7 mutants, however, we found that odr-1 mutants were also defective in maintenance of the AWCOFF marker srsx-3, so that srsx-3 expression was present in young odr-1 larvae but absent in odr-1 adults (Table 1; data not shown). These results suggest defects in both AWCON and AWCOFF in olfactory transduction mutants.
A possible target of cGMP produced by ODR-1 is EGL-4, a cGMP-dependent kinase homolog that is required for normal olfaction and olfactory plasticity in AWC, and for expression of some chemoreceptor genes in AWB neurons (Daniels et al. 2000; L'Etoile et al. 2002; van der Linden et al. 2008). Like odr-1 mutants, egl-4-null mutants expressed str-2 and srsx-3 in the AWC neurons of L1 animals, but not in adults (Table 1; data not shown). Thus, odr-1 and egl-4 are required to maintain and increase expression of both AWCON and AWCOFF genes between the L1 stage and the adult stage, in contrast with nsy-7, which is required specifically for appropriate gene induction and repression in AWCON. nsy-7;odr-1 and nsy-7;egl-4 double mutants resembled the odr-1 or egl-4 single mutants: Adults expressed neither str-2 nor srsx-3 (Table 1).
nsy-7 encodes a homeodomain-like protein that acts in AWC
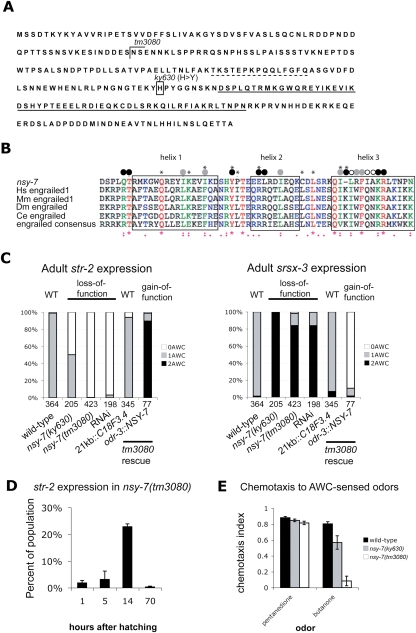
nsy-7(ky630) was mapped using standard methods to the predicted ORF C18F3.4, which encodes an uncharacterized protein (Fig. 2A). Although it had no significant homologs outside nematodes by BLAST searches, low-stringency searches and motif searches revealed a region with distant similarity to a homeodomain (Fig. 2B). The ky630 mutation was associated with a C → T transition in the third exon of the predicted gene, resulting in a missense H → Y mutation immediately before the homeodomain-like region (Fig. 2A). nsy-7(ky630) is recessive and RNAi against C18F3.4 generated a nsy-7-like phenotype, with two srsx-3-expressing AWCOFF cells and no str-2-expressing AWCON cells (Fig. 2C). These results identify nsy-7(ky630) as a likely reduction-of-function mutation in C18F3.4.
Figure 2.
nsy-7 encodes a protein with distant similarity to homeodomains. (A) Sequence of the long isoform of NSY-7. The ky630 and tm3080 mutations are marked, and the predicted homeodomain-like region is underlined. Dotted line indicates residues absent in the short NSY-7 isoform. (B) Alignment of the NSY-7 homeodomain-like region with engrailed-family homeodomains. Engrailed residues that bind specific bases (open circles) or the DNA backbone (black circles) and residues that form the hydrophobic core (gray circles) are marked. Asterisks above the sequences mark residues important for homeodomain function (Gehring et al. 1994; Draganescu and Tullius 1998; Fraenkel et al. 1998; Sato et al. 2004; Chi 2005). (C) Expression of srsx-3∷GFP and str-2∷dsRed in AWC neurons of wild type, nsy-7 mutants, C18F3.4-RNAi animals, and nsy-7(tm3080) rescued with a genomic clone or an odr-3∷nsy-7 cDNA clone. Numbers of animals scored are indicated. (D) Percentage of nsy-7(tm3080) animals expressing str-2∷dsRed2 at 1, 5, 14, and 70 h after hatching; 1–14 h are L1 stage, and 70 h is young adult. (E) Chemotaxis to the AWCON-specific odor 2-butanone and the AWCOFF-specific odor 2,3-pentanedione.
A deletion mutation in C18F3.4, tm3080, was kindly provided by the National Bioresource Project in Japan. tm3080 deletes parts of the second and third exons and then generates a frameshift that leads to early termination. Therefore, tm3080 probably represents a null mutation in C18F3.4. The nsy-7(tm3080) mutant phenotype was similar to but stronger than the nsy-7(ky630) phenotype: Almost all adult animals had two srsx-3-expressing AWCOFF cells and no str-2-expressing AWCON cells (Fig. 2C). A fraction of L1 larvae expressed str-2 in one AWC neuron, suggesting initiation of the AWCON identity, but this expression was unstable (Fig. 2D). nsy-7(ky630) and nsy-7(tm3080) mutants had no obvious defects in growth rate, brood size, morphology, or coordination.
The functions of AWCON and AWCOFF were examined by testing chemotaxis to butanone and 2,3-pentanedione, odors sensed by AWCON and AWCOFF, respectively (Wes and Bargmann 2001). nsy-7(ky630) animals had a mild defect and nsy-7(tm3080) had a severe defect in butanone chemotaxis, and both mutants were normal for chemotaxis to 2,3-pentanedione (Fig. 2E). The milder butanone chemotaxis defect in nsy-7(ky630) correlated with its weaker effect on adult str-2 expression (Fig. 2C). Thus, nsy-7 is required for multiple properties of AWCON, including sensation of butanone, expression of str-2, and repression of srsx-3.
When expressed in both AWC neurons of nsy-7(tm3080) mutants under the AWC-selective odr-3 promoter, C18F3.4 cDNAs caused an overexpression phenotype opposite to the nsy-7 mutant phenotype. Both AWCs in these transgenic animals expressed str-2 and not srsx-3, exhibiting the properties of AWCON cells (Fig. 2C). These results suggest that C18F3.4 functions in AWC to regulate expression of str-2 and srsx-3 and confer the AWCON identity.
The AWC asymmetry pathway regulates nsy-7 transcription
Regulatory sequences for nsy-7 were identified by characterizing the genomic regions required for rescue of nsy-7(tm3080) mutants. Partial rescue was obtained with the C18F3.4 genomic coding region and 9 kb of upstream sequence, and rescue was improved by increasing the amount of upstream sequence (Supplemental Fig. S1). Near-complete rescue was achieved using the genomic coding region with 21 kb upstream of the start site (Fig. 2C).
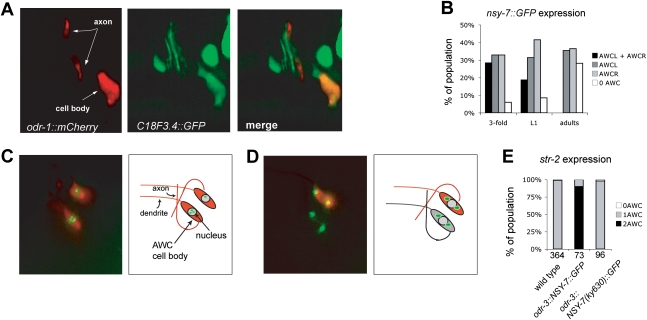
When fused to a GFP reporter, the 21-kb region upstream of nsy-7 drove expression in numerous cell types including gut, the amphid sheath glial cells, and head and tail neurons including AWC, ASE, and ASH (Fig. 3A; data not shown). This broad expression pattern suggests that nsy-7 may have uncharacterized functions that are unrelated to AWC asymmetry. In adult animals, nsy-7∷GFP was asymmetrically expressed in one AWC neuron (Fig. 3A). The nsy-7-expressing neuron was identified by crossing a line expressing nsy-7∷GFP with a line expressing an srsx-3∷mCherry (AWCOFF) reporter or a str-2∷dsRed2 (AWCON) reporter. nsy-7 was consistently expressed in the same cell as str-2 (145/158 animals), and contralateral to the cell expressing srsx-3 (99/99 animals). Therefore, nsy-7 appears to be expressed in AWCON but not AWCOFF.
Figure 3.
Expression of nsy-7∷GFP and nuclear localization of NSY-7. (A) Expression of a 21-kb nsy-7∷GFP promoter fusion (green) and the AWC marker odr-1∷mCherry (red). The AWC cell body and AWC axons in the nerve ring are marked. (B) nsy-7∷GFP expression across development. (C) Nuclear localization of a NSY-7∷GFP protein expressed in both AWCs under the odr-3 promoter. str-2∷dsRed marks AWCON; note gain-of function 2-AWCON phenotype. (D) Cytoplasmic localization of NSY-7rGFP with the ky630 mutation (odr-3 promoter, str-2∷dsRed marker); note wild-type asymmetric expression of str-2∷dsRed. (E) Quantification of str-2∷dsRed expression in wild type, or wild type overexpressing odr-3∷nsy-7∷GFP with or without the ky630 mutation. Numbers of animals scored are indicated.
The developmental time course of nsy-7∷GFP expression was examined in a transgenic strain expressing nsy-7∷GFP and a marker for both AWC neurons, odr-1∷mCherry. At the threefold embryonic stage, when the signaling pathway initiates AWC asymmetry, most animals expressed nsy-7∷GFP asymmetrically in one AWC neuron, which was equally likely to be AWCL or AWCR (Fig. 3B). At this stage, a subset of embryos expressed nsy-7∷GFP bilaterally in AWC; bilateral AWC expression fell beginning at the L1 larval stage, and was not observed in adults. These results suggest that nsy-7∷GFP expression is an early marker for AWC asymmetry. To determine whether the apparent asymmetry was regulated by the embryonic signaling pathway, nsy-7∷GFP expression was examined in signaling mutants with 2-AWCOFF or 2-AWCON phenotypes. In 2-AWCOFF nsy-5(ky634) mutants, adult nsy-7∷GFP expression was lost (Table 1). In 2-AWCON nsy-1 mutants, 35% of adults expressed nsy-7∷GFP in both AWC neurons (Table 1). The regulation of nsy-7∷GFP by the embryonic AWC asymmetry genes suggests that the nsy-7 promoter is a transcriptional target of the AWC signaling pathway.
nsy-7∷GFP was expressed bilaterally in the ASE neurons, another pair of chemosensory neurons that exhibits asymmetry. The asymmetric identities of the ASE neurons can be followed using the reporters gcy-5∷GFP, expressed in ASER, and gcy-7∷GFP, expressed in ASEL (Yu et al. 1997). Both gcy-5∷GFP and gcy-7∷GFP were appropriately and asymmetrically expressed in nsy-7 mutants, indicating that ASE asymmetry does not require nsy-7 (data not shown).
NSY-7 is a nuclear protein that binds the consensus DNA sequence CCTTAAC
NSY-7 has distant similarity to homeodomain proteins, suggesting that it might act as a transcription factor. Consistent with this possibility, a C-terminally tagged NSY-7∷GFP fusion protein was localized to the nucleus of AWC (Fig. 3C). The fusion protein was biologically active, as it induced a gain-of-function 2-AWCON phenotype when expressed in both AWC neurons in a wild-type background (Fig. 3E). A NSY-7∷GFP fusion protein carrying the missense mutation H179Y encoded by the ky630 mutation failed to localize to the nucleus (Fig. 3D), and also failed to induce a 2-AWCON phenotype when expressed in both AWCs (Fig. 3E). These results suggest that NSY-7 nuclear localization may be required for its activity.
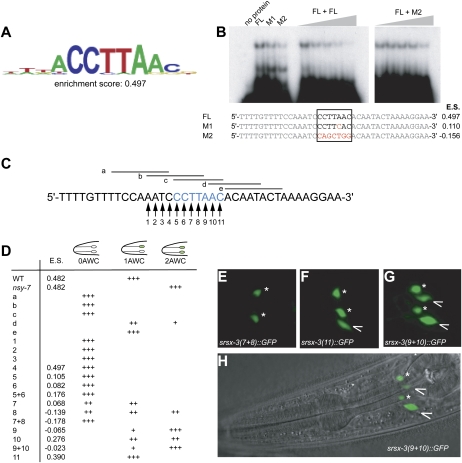
To ask whether NSY-7 can bind to DNA and to identify potential target sequences, we made use of universal protein-binding microarrays (Berger et al. 2006, 2008). The DNA microarrays consisted of 41,944 60-mer oligonucleotide features that collectively represent all possible 8-mer DNA sequences, with each 8-mer represented ∼32 times in the array. Full-length GST-tagged NSY-7 protein was produced in Escherichia coli, purified, and applied to the DNA microarray; NSY-7 binding at each DNA spot was detected and quantified using fluorescence-conjugated anti-GST antibody. NSY-7-binding preferences over all 8-mers were calculated from the 60-mer probe data and used to generate an “enrichment score” between −0.5 and 0.5, reflecting the preference of the protein for any given eight-nucleotide sequence. NSY-7 bound the specific recognition site ACCTTAAC with an enrichment score of 0.497 (Fig. 4A; Supplemental Table S1; these data are freely available on the UniPROBE database; Newburger and Bulyk 2009); an enrichment score >0.45 is indicative of nonrandom binding (Berger et al. 2008). These results demonstrate that NSY-7 is a sequence-specific DNA-binding protein.
Figure 4.
NSY-7 binds to the sequence CCTTAAC. (A) Consensus NSY-7-binding sequence determined by protein-binding microarray experiments. The enrichment score correlates with the binding affinity of NSY-7 for the sequence, and is measured on a scale of −0.5 to 0.5. (B) In vitro binding of 6His-tagged NSY-7 to the predicted consensus sequence. FL, full-length consensus sequence in a 39-nucleotide context from the srsx-3 promoter. Consensus binding site is boxed. M1 and M2, mutations predicted to decrease the enrichment score (E.S.), shown to the right of the sequences. All lanes except the first contain 500 ng of 6His:NSY-7. Lanes 5–9 contain a labeled FL probe and increasing amounts of cold FL competitor; lanes 10–14 show labeled FL probe + cold M2. The bottom band in lanes 2–4 probably represents a degradation product of the recombinant protein. (C,D) Mutations made in the srsx-3 promoter (C), and resulting expression patterns (D). Enrichment scores are shown for each point mutation. (+++) 80%–100% of population; (++) 30%–80%; (+) 10%–30%; (blank) <1%. (E) GFP expression under an srsx-3 promoter containing mutations 7 and 8 (0 AWC). (F) GFP expression under an srsx-3 promoter containing mutation 11 (1 AWC). (G) GFP expression under an srsx-3 promoter containing mutations 9 and 10 (2 AWC). (H) DIC image of worm shown in G. In E–H, asterisks indicate AWB and arrowheads indicate AWC.
The promoter regions of genes that are expressed in AWC neurons were examined for the presence of the ACCTTAAC-binding site identified in the protein-binding microarrays. None of these promoters contained the complete 8-mer, but a perfect match for the sequence CCTTAAC was found in the srsx-3 promoter, 425 base pairs (bp) upstream of the predicted start site. Binding of NSY-7 to the preferred sequence from the srsx-3 promoter was verified by electromobility shift assays (EMSAs) using a 32P-labeled double-stranded oligonucleotide with the sequence CCTTAAC flanked by 16 nucleotides on each side. A full-length, bacterially-produced His-tagged NSY-7 protein bound the labeled probe, and this interaction was competed away by an unlabeled probe (Fig. 4B; Supplemental Fig. S2). Mutations in the predicted binding sequence reduced binding of NSY-7 to a labeled probe and reduced the ability of unlabeled probe to compete NSY-7 protein away from a wild-type labeled probe (M1 and M2) (Fig. 4B). NSY-7 also weakly bound the M2 mutant probe, suggesting some sequence nonspecific binding activity. We conclude that NSY-7 protein can selectively bind the sequence CCTTAAC.
The NSY-7-binding sequence CCTTAAC is required for asymmetric srsx-3 expression
The importance of the putative NSY-7-binding sequence in the srsx-3 promoter was assessed by mutating an srsx-3∷GFP reporter gene. Deletion of the CCTTAAC sequence eliminated all expression of srsx-3∷GFP in AWC (Fig. 4C [b,c], D [b,c]). Further mutational analysis of this element and surrounding sequences in the srsx-3 promoter was performed by deleting short sequence blocks in the binding site and surrounding sequence (Fig. 4C, a–e) and by mutating individual nucleotides to residues that produced the lowest enrichment score in the protein-binding microarray experiment (Fig. 4C, 1–11). Expression of these mutated elements in transgenic C. elegans demonstrated that the CCTTAAC sequence was embedded in a larger site with the ability to either repress or activate AWC srsx-3 expression, AATCCCTTAAC (Fig. 4C–H). Mutation or deletion of the four bases preceding the NSY-7-binding site, or the first three bases of the site itself (deletions a–c, point mutations 1–7), resulted in loss of all srsx-3 expression in AWC. Mutation of the following three bases of the NSY-7-binding site produced a phenotype resembling that of the nsy-7 mutant, in which both AWCs expressed srsx-3 (point mutations 8–10). Mutation of the last base of the site or bases beyond it had no effect. Thus, the region near the NSY-7-binding site is a compound element associated with both activating and repressing activities. The simplest explanation for these results is that the compound element binds NSY-7, which represses expression in AWCON, and also binds a second activating factor that is present in both AWCOFF and AWCON (see the Discussion).
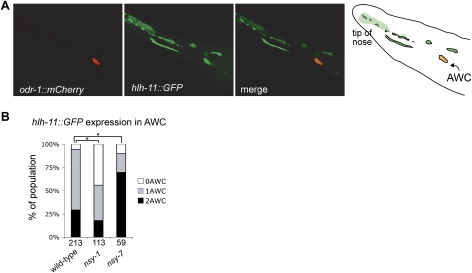
The extended activator/repressor sequence defined by mutagenesis of the srsx-3 promoter, AATCCCTTAAC, occurs only 22 times in the C. elegans genome. To ask whether this sequence was associated with asymmetric AWC expression in other cases, we examined one additional gene in which this sequence appears 1.5 kb upstream of the transcriptional start site, hlh-11. hlh-11 encodes one of ∼35 helix–loop–helix transcription factors in the C. elegans genome (Okkema and Krause 2005); it was examined purely as a reporter gene, as the deletion mutant hlh-11(ok2944) has no effect on str-2 or srsx-3 expression in AWC (n = 82). An hlh-11∷GFP reporter with 3 kb of upstream sequence was expressed in one AWC neuron in 65% of animals, and in both AWC neurons in 30% of animals (Fig. 5A,B); it was also expressed in a variety of additional cell types. The AWC neuron expressing hlh-11 was significantly more likely to be AWCOFF than AWCON (74% coexpression of hlh-11∷GFP with srsx-3∷mCherry, n = 54). Expression of hlh-11∷GFP in AWC was less frequent in nsy-1 (2-AWCON) mutants, and more frequent in nsy-7 mutants, suggesting that nsy-7 represses hlh-11∷GFP in AWCON (Fig. 5B). These results suggest that the 11-nucleotide motif can predict AWCOFF-biased expression in AWC neurons.
Figure 5.
hlh-11 is regulated by nsy-7. (A) Expression of an hlh-11∷GFP reporter with 3 kb of sequence upstream of the start site, showing colocalization with the AWC marker odr-1∷mCherry. (B) hlh-11∷GFP expression in wild-type, nsy-1(ky542), and nsy-7(tm3080) mutant backgrounds. Asterisk indicates results different at P < 0.001 (χ2 test). Numbers of animals scored are indicated.
Discussion
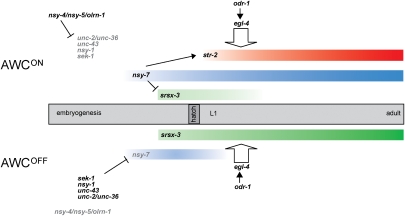
The AWC neurons acquire asymmetric identities early in development, but require three sets of genes to maintain these properties into adulthood: (1) nsy-7, which is specifically required to maintain AWCON properties; (2) odr-1 and egl-4, which are required to maintain markers for both AWCON and AWCOFF; and (3) odr-3 and other G proteins, which increase str-2 expression during larval stages (Troemel et al. 1999; Lans et al. 2004; Lans and Jansen 2006). nsy-7 mutants initiate str-2 expression but do not maintain AWCON identity, as the adult animals do not express str-2 or sense butanone. A nsy-7∷GFP reporter is an early marker of the AWCON neuron, suggesting that asymmetric expression of nsy-7 is a transcriptional readout of the embryonic signaling pathway. The sequence-specific DNA-binding properties of NSY-7 suggest that it binds directly to promoters of genes expressed in AWCOFF to repress their expression in AWCON.
A complex signaling pathway involving a multicellular gap junction network, claudins, calcium channels, and protein kinases acts in embryogenesis to distinguish the AWCON and AWCOFF cells (Troemel et al. 1999; Sagasti et al. 2001; Tanaka-Hino et al. 2002; Chuang and Bargmann 2005; Vanhoven et al. 2006; Bauer Huang et al. 2007; Chuang et al. 2007). Previous genetic studies and cell ablations suggest that the AWCOFF identity is a default, or ground state, and AWCON is an induced state (Troemel et al. 1999). The developmental analysis of asymmetric marker genes presented here provides a dynamic view of AWCON induction (Fig. 6). The bilateral early expression of the AWCOFF marker srsx-3 is consistent with an AWCOFF-like ground state. The first detectable marker of AWCON identity is nsy-7∷GFP, whose asymmetric expression beginning in the embryo overlaps with and anticipates the repression of the AWCOFF markers srsx-3 and hlh-11 in the L1 stage. Repression of srsx-3 is followed by asymmetric expression of the AWCON marker str-2 in late L1 larvae.
Figure 6.
Developmental stabilization of stochastic AWC asymmetry. Gene expression patterns in AWCON and AWCOFF across development are shown, along with inferred roles of the initial signaling cascade, the nsy-7 transcriptional regulator, and the odr-1/egl-4 activity-dependent pathway.
The induction of str-2 expression in AWCON is not fully explained by the action of nsy-7, implying the existence of additional transcriptional regulators in AWC. str-2 expression is undetectable in null mutants for signaling genes such as nsy-5, but is detectable in L1 larvae of nsy-7-null mutants, suggesting that another target of the signaling pathway initiates str-2 expression in AWCON. nsy-7; odr-1 double mutants also express str-2 in the L1 stage, so the olfactory transduction pathway involving odr-1 and egl-4 is unlikely to represent this missing target. Moreover, nsy-7 acts genetically to maintain str-2 expression in AWCON, but there is no predicted NSY-7-binding site in the str-2 promoter. As str-2 activation in AWCON is delayed compared with srsx-3 repression, we speculate that nsy-7 activates AWCON markers indirectly; for example, by repressing a transcriptional repressor. Further genetic screens may identify additional transcriptional effectors of AWC asymmetry.
NSY-7 binds to a specific sequence, CCTTAAC, that is present in the promoter region of the srsx-3 and hlh-11 target genes that are repressed in AWCON. Therefore, it is likely to act directly as a repressor of AWCOFF markers. This DNA-binding specificity was determined by protein-binding microarrays, which established the DNA-binding ability of this nonconserved protein, confirmed its sequence specificity, and identified an optimal site in a single experiment. The DNA-binding specificities of 168 mammalian homeodomain proteins have been established using this technique, and the NSY-7-binding site was distinct from any known site (Berger et al. 2006, 2008). This result is not unexpected, given the divergence of the potential DNA-binding domain of NSY-7 from canonical homeodomains. The I/V47, Q/K50, and N51 residues that recognize the core TAAT in the homeodomain-binding motif are not present in NSY-7. However, the hydrophobic residues that stabilize the homeodomain protein core are conserved or retain hydrophobic character in NSY-7, and several residues that interact with the DNA backbone are present (Draganescu and Tullius 1998; Fraenkel et al. 1998; Chi 2005). The NSY-7 sequence is therefore consistent with a homeodomain-like structure that recognizes a noncanonical DNA sequence.
The NSY-7-binding motif falls within a larger element responsible for asymmetric srsx-3 expression in AWCOFF, AATCCCTTAAC. This 11-bp element includes sequences that promote symmetric AWC expression, which overlap partly but not completely with the NSY-7-binding site. The separation of activator and repressor functions by point mutations suggests that the motif binds at least two different proteins. We propose that the compound element binds the asymmetric repressor protein NSY-7 in AWCON, and an activator protein that is present in both AWC neurons. A candidate for the activator protein is CEH-36, a homeodomain protein that is expressed symmetrically in AWC neurons and required for AWC fates (Lanjuin et al. 2003; Koga and Ohshima 2004). ceh-36 mutants fail to express both str-2 or srsx-3 in AWC, and CEH-36 and other Otx-class K50 homeodomains bind the sequence TAATCC, which is similar to the AATCCC sequence that activates AWC expression of the srsx-3 promoter (Affolter et al. 1990; Schier and Gehring 1993; Lanjuin et al. 2003; Chaney et al. 2005; Berger et al. 2008; Etchberger et al. 2009). NSY-7 might compete with CEH-36 or another activator for binding to the compound 11-bp element, specifically repressing expression in AWCON. This proposed mechanism resembles the function of the Drosophila Brinker protein in DPP signaling. DPP receptors regulate the activity of MAD/SMAD transcriptional activators, and regulate the expression of Brinker, a poorly conserved repressor protein with a divergent homeodomain that competes with MAD proteins at MAD-binding DNA elements in target genes (Campbell and Tomlinson 1999; Jazwinska et al. 1999; Rushlow et al. 2001; Saller and Bienz 2001). An alternative model is that NSY-7 and the proposed activator protein can bind the compound element simultaneously, but that NSY-7 recruits transcriptional corepressors and converts activation into repression in AWCON.
Maintenance of AWC cell identity involves both asymmetric transcriptional regulation by nsy-7, which is controlled by a developmental signaling pathway, and symmetric transcriptional regulation by activity-dependent odr-1/egl-4 cGMP signaling. The cGMP-dependent kinase EGL-4 appears to function in the nucleus, where it might interact with NSY-7 and other transcriptional regulators (L'Etoile et al. 2002). The convergence of developmental and activity-dependent regulators of neuronal function is of potential neurological and psychiatric interest. Disorders such as autism, schizophrenia, affective disorders, and epilepsy have genetic and developmental components, but are also influenced by experience, and manifest themselves in childhood, adolescence, or even adulthood (Charney and Nestler 2004; Keshavan et al. 2004; Chubb et al. 2008; Costa e Silva 2008; Wigle and Eisenstat 2008). Analysis of neuronal maintenance pathways like those used in AWCON may provide insight into some of these late-onset disruptions of neuronal function.
Materials and methods
Genetics and strains
C. elegans strains were cultured using standard methods (Brenner 1974). All strains were grown at 20°C. Strains used were as follows: wild-type Bristol (N2), CB4856 (Hawaiian), CX7894 kyIs408 [srsx-3∷GFP;str-2∷dsRed2;elt-2∷GFP] II, CX7893 kyIs405 [srsx-3∷GFP;str-2∷dsRed2;elt-2∷GFP] V, CX3695 kyIs140 [str-2∷GFP] I, CX6336 nsy-7(ky630) IV, CX8096 kyIs408 II; nsy-7(ky630) IV, CX5737 kyIs140 I; nsy-7(ky630) IV, CX10232 nsy-7(tm3080) IV, CX10231 kyIs408 II; nsy-7(tm3080) IV, CX9975 kyIs408 II; eri-1(mg366) IV; lin-15B(n744) X, CX2519 unc-5(e53) dpy-20(e1282ts) IV, RW1350 unc-44(e362) unc-82(e1323)/stDf7 IV, CB3824 eDf19/unc-24(e138) dpy-20(e1282) IV, BC352 let-51(s41) unc-22(s7)/sDf2 IV, CX8147 kyIs140 I; nsy-7(ky630) dpy-20(e1282ts) IV, CX8801 kyIs408 II; unc-36(e251) III, CX8800 kyIs408 II; unc-36(e251) III; nsy-7(ky630) IV, CX8985 kyIs408 II; unc-2(lj1) X, CX8799 kyIs408 II; nsy-7(ky630) IV; unc-2(lj1) X, CX8666 nsy-1(ky542) II; kyIs405 V, CX8782 nsy-1(ky542) II; nsy-7(ky630) IV; kyIs405 V, CX8089 kyIs408 II; odr-1(n1936) X, CX8547 kyIs408 II; nsy-7(ky630) IV; odr-1(n1936) X, CX8342 kyIs408 II; egl-4(ks60) IV, CX8546 kyIs408 II; egl-4(ks60) nsy-7(ky630) IV, CX10939 kyEx2843 [nsy-7(21kb)∷GFP; ofm-1∷dsRed2]; kyEx2424 [odr-1∷mCherry; ofm-1∷dsRed2], CX9709 kyIs408 II; nsy-7(ky630) IV; kyEx2125 [odr-3∷NSY-7∷GFP; ofm-1∷dsRed2], CX10533 kyEx2592 [nsy-7(13.4kb)∷NSY-7∷GFP; ofm-1∷dsRed2]; kyEx2424, CX8156 kyEx1253 [str-2∷dsRed2; elt-2rGFP], CX9431 kyEx1253; kyEx1984 [odr-3∷NSY-7; ofm-1∷dsRed2], CX9818 kyEx1253; kyEx2167 [odr-3∷NSY-7(ky630)∷GFP; ofm-1∷dsRed2], CX9810 kyEx1253; kyEx2163 [odr-3∷GFP:NSY-7; ofm-1∷dsRed2], CX11203 kyEx2982 [srsx-3(mut9 + 10)∷GFP; ofm-1∷dsRed2], CX11199 kyEx2978 [srsx(mut9)∷GFP; ofm-1∷dsRed2], CX11061 kyEx2899 [odr-1∷NLS-mCherry; elt-2∷GFP]; kyEx2900 [hlh-11∷GFP; ofm-1∷dsRed2], CX11046 nsy-7(tm3080) IV; kyEx2899; kyEx2900, CX11049 nsy-1(ky542) II; kyEx2899; kyEx2900, CX10098 kyIs408 II; nsy-7(ky630) IV; kyEx2290 [odr-3∷6His-NSY-7; ofm-1∷dsRed2], CX11050 kyEx2424; kyEx2902 [nsy-7(21kb)∷NSY-7∷GFP; ofm-1∷dsRed2], CX11059 nsy-1(ky542) II; kyEx2902; kyEx2424, CX11070 nsy-5(ky634) I; kyEx2902; kyEx2424.
Genetic mapping
nsy-7(ky630) was isolated from a screen for 2-AWCOFF mutants (Vanhoven et al. 2006). In three-factor mapping experiments, ky630 mapped to chromosome IV between unc-5(e53) (IV:1.78) and dpy-20(e1282ts) (IV:5.22). Trans-heterozygotes of ky630 with the deletions stDf7, eDf19, and sDf2 each displayed a wild-type str-2 expression phenotype, suggesting that the mutation fell in the interval IV:1.78–2.43 or IV:3.50–3.60. We confirmed that the mutation fell in the latter region by SNP mapping: The strain CX8147 nsy-7(ky630) dpy-20(e1282ts) IV; kyIs140 I was crossed to the polymorphic strain CB4856, and lines displaying either Nsy non-Dpy or Dpy non-Nsy phenotypes were evaluated at known polymorphic loci. The nsy-7 mutation fell between the SNPs CE4-150 (IV:3.59) and C07G1:1158 (IV:3.65). Sequencing of the ORF C18F3.4 in this interval revealed a G → A mutation at position 30,769 of cosmid C18F3.
RNAi
RNAi was performed by injection of dsRNA into the sensitized strain CX9975 kyIs408 II; eri-1(mg366) IV;lin-15B(n744) X (Sieburth et al. 2005). dsDNA template corresponding to the second exon of the C18F3.4 ORF was amplified from N2 lysate with a T7 sequence added at the 5′ ends using the following primers (T7 sequence underlined): primer1, TAATACGACTCACTATAGGGAGAGTTGCGAAAGGATATTCAGATG; primer2, TAATACGACTCACTATAGGGAGACTTAGCAAACAAGTTGGTGAGT.
Transcription was performed using the T7 RiboMAX Express RNAi System (Promega, P1700) according to instructions. Eight microliters of unpurified PCR reaction were used in a reaction volume of 20 μL. The transcription reaction was diluted 4× in RNAse-free water and 2 μL of each reaction was run on an agarose gel for quantification. The unpurified reaction mix was injected into the body cavity, gut, or gonad of adult hermaphrodites. Injected animals were transferred to fresh plates after 24 h of recovery and transferred again after 48 h; only F1s from the second and third sets of plates were scored. F1 progeny were scored after 3–4 d for str-2 and srsx-3 expression.
Molecular biology
Standard molecular biology techniques were used. The 21 kb nsy-7 promoter was PCR-amplified from wild-type (N2) lysate and cloned into the pSM-GFP expression vector between the NotI and AscI sites in MCS1 (primers, 5′-TTAAGAACAACACACATGACCTAC-3′ and 5′-TCTGAAAAAAAGTTTAGATGTTTGA-3′). For rescue, the same promoter was inserted into MCS1 of the pSM expression vector, and the nsy-7 genomic sequence was inserted into MCS2 between NheI and KpnI sites (primers, 5′-TTCCGAATCAAACATCTAAACTT-3′ and 5′-CGAAGAAACCAAACACTTTTTCA-3′).
NSY-7 cDNAs were obtained by PCR from a C. elegans cDNA library using primers at the beginning and end of the predicted ORF (5′-ATGTCTTCGGATACAAAATACAAA-3′ and 5′-TCAAGCCGTCGTCTCTTGCA-3′). Sequencing of cDNA fragments made from a circularized C. elegans RNA library revealed that some NSY-7 sequences included an SL1 spliced leader sequence at the predicted 5′ end, confirming that the predicted 5′ exon is (at least in some cases) the true starting exon. Two classes of cDNAs for C18F3.4 were identified from RT–PCR analysis, differing in the presence or absence of 15 amino acids at the beginning of the third exon. Both cDNAs resulted in a 2-AWCON phenotype when expressed in both AWC neurons under the odr-3 promoter (Fig. 2C; data not shown); only the longer isoform was examined in the remainder of this study.
The NSY-7∷GFP fusion plasmid was constructed by insertion of the NSY-7 cDNA into pSM-GFP between the NheI and KpnI restriction sites in MCS2, in frame with the GFP coding sequence. The GFP∷NSY-7 fusion plasmid was created by amplification of a GFP:NSY-7 fragment by PCR using overlap extension, with pSM-GFP and the NSY-7 cDNA as starting templates. This fragment was cloned into MCS2 of pSM between the NheI and KpnI restriction sites.
To create the GST∷NSY-7 bacterial expression vector for protein production, the full-length NSY-7 cDNA was cloned into pGEX 4T-3 using SalI and NotI sites. For the 6His:NSY-7 bacterial expression vector, the same NSY-7 cDNA was cloned into pSV271 (Pokala and Handel 2004) using NarI and XhoI sites. For expression in C. elegans, the GST:NSY-7 sequence was cloned into MCS2 of a modified pSM vector (containing a NotI site in MCS2 instead of MCS1) between the NheI and NotI sites, with a SalI site between the GST and NSY-7 sequences. 6His:NSY-7 was PCR-amplified from the bacterial expression vector to include NheI and KpnI sites, and inserted into MCS2 of pSM using these sites.
Mutation of the srsx-3∷GFP plasmid (Bauer Huang et al. 2007), was carried out by site-directed mutagenesis using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, 200524).
The hlh-11 promoter sequence was PCR-amplified from N2 lysate (primers, 5′-TCGTTTCTCTCGTTTCCTCC-3′ and 5′-TCTGGATTACCTGAAACTTTACAA-3′) and cloned into MCS1 of pSM-GFP between the FseI and AscI sites.
Sequence alignment
The nsy-7 homeodomain sequence was aligned with engrailed homeodomain sequences using ClustalW at PBIL (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_clustalw.html). GenBank accession numbers are Homo sapiens engrailed1, NM_001426; Mus musculus engrailed1, NM_010133; Drosophila melanogaster engrailed isoform A, NM_078976; and C. elegans engrailed (ceh-16), NM_066497. The engrailed consensus sequence is from Gehring et al. (1994).
Microscopy and fluorescence quantification
Fluorescence microscopy was carried out on a Zeiss Axioplan2 imaging system with a Hamamatsu Photonics C2400 CCD camera or a Zeiss Axio Imager.Z1 with ApoTome with a Zeiss AxioCam MRm CCD camera. Most animals were scored under a 20× or 40× Plan-Neofluar objective, and photographs were taken under a 40× Plan-Neofluar or 63× Plan-Apochromat objective. For quantification, images were collected on the Zeiss Axioplan2 system under standardized detector settings using Metamorph software. A region within the cell body of the AWC neuron was selected and average fluorescence intensity was calculated for the same region in red and green channels. A region of similar size was selected for the contralateral cell of the same animal, and the same data was collected. Data shown represents average values for at least 25 animals of each genotype at each time point.
Developmental analysis
To evaluate marker expression at precise stages during post-embryonic development, larvae were staged by hatch-off. Late embryos were picked to an NGM plate seeded with the E. coli strain OP50. After 30 min, just-hatched L1s were transferred to a fresh plate and grown for the specified time at 20°C. Individuals scored at any given time point were discarded; therefore, a separate set of staged worms was scored at each time point.
Behavioral assays
Chemotaxis assays were performed as described (Bargmann et al. 1993). Odors were diluted in ethanol and tested at standard concentrations (1:1000 butanone and 1:10,000 2,3-pentanedione). Three independent assays of each strain were conducted for each odor.
Protein production and purification
GST:NSY-7 and 6His:NSY-7 were produced by expression and purification in E. coli strain BL21(DE3). Transformed cells were grown overnight in LB medium + 50 μg/mL carbenicillin (for GST∷NSY-7) or 50 μg/mL kanamycin (for 6His∷NSY-7). Overnight cultures were diluted 1:100 in fresh LB + antibiotic; diluted cultures were grown at 37°C to OD600 ∼0.5, induced with IPTG at a final concentration of 1 mM, grown for an additional 4 h, and pelleted by spinning at ∼2500g for 15 min at 4°C. Pellets were washed and resuspended in ice-cold PBS, and then lysed by sonication. For 6His:NSY-7, the pellet was incubated on ice for 30 min in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole at pH 8.0) with 1 mg/mL lysozyme before sonication. Lysates were centrifuged for 10–30 min at 4°C. The GST fusion protein was purified from lysate supernatant using a Microspin GST Purification Module (Amersham, 27-4570-03) according to instructions, and His-tagged protein was purified using an Ni-NTA Spin Kit (Qiagen, 31314) according to instructions for native conditions. Presence of intact protein in the cell pellet before lysis, in lysate supernatant, and after purification was verified by SDS-PAGE followed by staining with Coomassie Blue. Protein concentration was estimated by spectrophotometry of the purified sample.
Protein-binding microarrays
Protein-binding microarray experiments were performed as described previously (Berger and Bulyk 2009; Berger et al. 2006). Briefly, protein-binding microarrays were performed using a custom-designed microarray from Agilent Technologies consisting of ∼44,000 features of 60-nt oligonucleotides, encompassing all possible combinations of 10-mers (Berger et al. 2006). Primer extension was performed on the single-stranded slides utilizing a common 24-nt region to generate a dsDNA microarray. GST-tagged NSY-7 was diluted to a final concentration of 500 nM in PBS, 2% (wt/vol) milk, 51.3 ng/μL salmon testes DNA (Sigma), 0.2 μg/μL BSA (New England Biolabs), and incubated for 1 h at 20°C. After washing, arrays were incubated with an Alexa488-conjugated anti-GST antibody (Invitrogen) for 1 h at 20°C. After subsequent washing, DNA-protein interaction was visualized using a Perkin Elmer ScanArray 5000 scanner to detect Alexa488 fluorescence. Arrays were normalized as previously described (Berger et al. 2006), and contiguous and gapped 8-mer enrichment scores as well as position weight matrices were generated using the “Seed-and-Wobble” algorithm (Berger et al. 2006).
EMSAs
Annealing and labeling
Double-stranded oligonucleotides were produced by annealing ssDNA fragments, synthesized by Integrated DNA Technologies (IDT), corresponding to the FL, M1, and M2 sequences. Sequences used were FL, 5′-TTTTGTTTTCCAAATCCCTTAACACAATACTAAAAGGAA-3′; FLrc, 5′-TTCCTTTTAGTATTGTGTTAAGGGATTTGGAAAACAAAA-3′; M1, 5′-TTTTGTTTTCCAAATCCCTTCACACAATACTAAAAGGAA-3′; M1rc, 5′-TTCCTTTTAGTATTGTGTGAAGGGATTTGGAAAACAAAA-3′; M2, 5′-TTTTGTTTTCCAAATCCAGCTGGACAATACTAAAAGGAA-3′; and M2rc, 5′-TTCCTTTTAGTATTGTCCAGCTGGATTTGGAAAACAAAA-3′.
For labeling with γ-32P-ATP, 100 ng of double-stranded probe was mixed with 1 μL of radioactive nucleotide (Perkin-Elmer, BLU502A) and incubated for 1 h at 37°C with T4 polynucleotide kinase (New England Biolabs, M0201S). Excess nucleotide was removed using NucTrap Probe Purification Columns (Stratagene, 400701), according to instructions.
EMSA
Five-hundred nanograms of fusion protein were incubated with binding buffer (10 mM Tris, 1 mM EDTA, 100 mM KCl, 0.1 mM DTT, 5% glycerol), 5 μg/mL salmon sperm, and 50 μg/mL BSA for 10 min at room temperature. A 0.25-ng labeled probe or a 0.25-ng labeled probe with 10×, 25×, 50×, 100×, or 200× cold competitor was added, and the reaction mixture was incubated at room temperature for an additional 15 min. The entire 20-μL binding reaction was then loaded onto a 6% NOVEX DNA Retardation gel (Invitrogen, EC6365BOX) and run for ∼1 h in 0.5× TBE buffer. Experiments were repeated three times.
Acknowledgments
We thank Miri VanHoven for isolating the ky630 mutant; E. Macosko, N. Pokala, C. Maurer, S. Shaham, F. Spagnoli, A. Brivanlou, M. Walhout, and R. McGinty for advice and reagents; and members of the Bargmann laboratory for comments on the manuscript. This work was supported by NIDCD grant DC004089 (C.I.B.) and by NHGRI grant HG003985 (M.L.B.). B.J.L. was supported by MSTP grant GM07739 and by an individual NIMH NRSA, F30MH084482. C.I.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1763509.
Supplemental material is available at http://www.genesdev.org.
References
- Affolter M., Percival-Smith A., Muller M., Leupin W., Gehring W.J. DNA binding properties of the purified Antennapedia homeodomain. Proc. Natl. Acad. Sci. 1990;87:4093–4097. doi: 10.1073/pnas.87.11.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C.I., Hartwieg E., Horvitz H.R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Bauer Huang S.L., Saheki Y., VanHoven M.K., Torayama I., Ishihara T., Katsura I., van der Linden A., Sengupta P., Bargmann C.I. Left–right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural Develop. 2007;2:24. doi: 10.1186/1749-8104-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Bulyk M.L. Universal protein binding microarrays for the comprehensive characterization of the DNA binding specificities of transcription factors. Nat. Protocols. 2009 doi: 10.1038/nprot.2008.195. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Philippakis A.A., Qureshi A.M., He F.S., Estep P.W., III, Bulyk M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Badis G., Gehrke A.R., Talukder S., Philippakis A.A., Pena-Castillo L., Alleyne T.M., Mnaimneh S., Botvinnik O.B., Chan E.T., et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnby D.A., Link E.M., Vowels J.J., Tian H., Colacurcio P.L., Thomas J.H. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics. 2000;155:85–104. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G., Tomlinson A. Transducing the Dpp morphogen gradient in the wing of Drosophila: Regulation of Dpp targets by brinker. Cell. 1999;96:553–562. doi: 10.1016/s0092-8674(00)80659-5. [DOI] [PubMed] [Google Scholar]
- Chaney B.A., Clark-Baldwin K., Dave V., Ma J., Rance M. Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome. Biochemistry. 2005;44:7497–7511. doi: 10.1021/bi0473253. [DOI] [PubMed] [Google Scholar]
- Charney D.S., Nestler E.J. Neurobiology of mental illness. Oxford University Press; Oxford, UK: 2004. [Google Scholar]
- Chen R.Z., Akbarian S., Tudor M., Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chi Y.I. Homeodomain revisited: A lesson from disease-causing mutations. Hum. Genet. 2005;116:433–444. doi: 10.1007/s00439-004-1252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.F., Bargmann C.I. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes & Dev. 2005;19:270–281. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C.F., Vanhoven M.K., Fetter R.D., Verselis V.K., Bargmann C.I. An innexin-dependent cell network establishes left–right neuronal asymmetry in C. elegans. Cell. 2007;129:787–799. doi: 10.1016/j.cell.2007.02.052. [DOI] [PubMed] [Google Scholar]
- Chubb J.E., Bradshaw N.J., Soares D.C., Porteous D.J., Millar J.K. The DISC locus in psychiatric illness. Mol. Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- Costa e Silva J.A. Autism, a brain developmental disorder: Some new pathophysiologic and genetics findings. Metabolism. 2008;57:S40–S43. doi: 10.1016/j.metabol.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Daniels S.A., Ailion M., Thomas J.H., Sengupta P. egl-4 acts through a transforming growth factor-β/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics. 2000;156:123–141. doi: 10.1093/genetics/156.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draganescu A., Tullius T.D. The DNA binding specificity of engrailed homeodomain. J. Mol. Biol. 1998;276:529–536. doi: 10.1006/jmbi.1997.1567. [DOI] [PubMed] [Google Scholar]
- Dragich J., Houwink-Manville I., Schanen C. Rett syndrome: A surprising result of mutation in MECP2. Hum. Mol. Genet. 2000;9:2365–2375. doi: 10.1093/hmg/9.16.2365. [DOI] [PubMed] [Google Scholar]
- Etchberger J.F., Flowers E.B., Poole R.J., Bashliari E., Hobert O. cis-regulatory mechanisms of left/right asymmetric neuron-subtype specification in C. elegans. Development. 2009;136:147–160. doi: 10.1242/dev.030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraenkel E., Rould M.A., Chambers K.A., Pabo C.O. Engrailed homeodomain-DNA complex at 2.2 A resolution: A detailed view of the interface and comparison with other engrailed structures. J. Mol. Biol. 1998;284:351–361. doi: 10.1006/jmbi.1998.2147. [DOI] [PubMed] [Google Scholar]
- Gehring W.J., Affolter M., Burglin T. Homeodomain proteins. Annu. Rev. Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Guy J., Gan J., Selfridge J., Cobb S., Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinska A., Kirov N., Wieschaus E., Roth S., Rushlow C. The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell. 1999;96:563–573. doi: 10.1016/s0092-8674(00)80660-1. [DOI] [PubMed] [Google Scholar]
- Keshavan M., Kennedy J., Murray R. Neurodevelopment and schizophrenia. Cambridge University Press; Cambridge, UK: 2004. [Google Scholar]
- Kishi N., Macklis J.D. MECP2 is progressively expressed in post-migratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell. Neurosci. 2004;27:306–321. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Koga M., Ohshima Y. The C. elegans ceh-36 gene encodes a putative homemodomain transcription factor involved in chemosensory functions of ASE and AWC neurons. J. Mol. Biol. 2004;336:579–587. doi: 10.1016/j.jmb.2003.12.037. [DOI] [PubMed] [Google Scholar]
- L'Etoile N.D., Bargmann C.I. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron. 2000;25:575–586. doi: 10.1016/s0896-6273(00)81061-2. [DOI] [PubMed] [Google Scholar]
- L'Etoile N.D., Coburn C.M., Eastham J., Kistler A., Gallegos G., Bargmann C.I. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans. Neuron. 2002;36:1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- Lanjuin A., VanHoven M.K., Bargmann C.I., Thompson J.K., Sengupta P. Otx/otd homeobox genes specify distinct sensory neuron identities in C. elegans. Dev. Cell. 2003;5:621–633. doi: 10.1016/s1534-5807(03)00293-4. [DOI] [PubMed] [Google Scholar]
- Lans H., Jansen G. Noncell- and cell-autonomous G-protein-signaling converges with Ca2+/mitogen-activated protein kinase signaling to regulate str-2 receptor gene expression in Caenorhabditis elegans. Genetics. 2006;173:1287–1299. doi: 10.1534/genetics.106.058750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H., Rademakers S., Jansen G. A network of stimulatory and inhibitory Gα-subunits regulates olfaction in Caenorhabditis elegans. Genetics. 2004;167:1677–1687. doi: 10.1534/genetics.103.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger D.E., Bulyk M.L. UniPROBE: An online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P.G., Krause M. Transcriptional regulation. 2005 doi: 10.1895/wormbook.1.45.1. WormBook (ed. The C. elegans Research Community), WormBook. doi: 10.1895/wormbook.1.45.1. http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Pokala N., Handel T.M. Energy functions for protein design I: Efficient and accurate continuum electrostatics and solvation. Protein Sci. 2004;13:925–936. doi: 10.1110/ps.03486104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow C., Colosimo P.F., Lin M.C., Xu M., Kirov N. Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes & Dev. 2001;15:340–351. doi: 10.1101/gad.861401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasti A., Hisamoto N., Hyodo J., Tanaka-Hino M., Matsumoto K., Bargmann C.I. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 2001;105:221–232. doi: 10.1016/s0092-8674(01)00313-0. [DOI] [PubMed] [Google Scholar]
- Saller E., Bienz M. Direct competition between Brinker and Drosophila Mad in Dpp target gene transcription. EMBO Rep. 2001;2:298–305. doi: 10.1093/embo-reports/kve068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Simon M.D., Levin A.M., Shokat K.M., Weiss G.A. Dissecting the Engrailed homeodomain–DNA interaction by phage-displayed shotgun scanning. Chem. Biol. 2004;11:1017–1023. doi: 10.1016/j.chembiol.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Schier A.F., Gehring W.J. Functional specificity of the homeodomain protein fushi tarazu: The role of DNA-binding specificity in vivo. Proc. Natl. Acad. Sci. 1993;90:1450–1454. doi: 10.1073/pnas.90.4.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D., Ch'ng Q., Dybbs M., Tavazoie M., Kennedy S., Wang D., Dupuy D., Rual J.F., Hill D.E., Vidal M., et al. Systematic analysis of genes required for synapse structure and function. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Tanaka-Hino M., Sagasti A., Hisamoto N., Kawasaki M., Nakano S., Ninomiya-Tsuji J., Bargmann C.I., Matsumoto K. SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel E.R., Sagasti A., Bargmann C.I. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- Tsunozaki M., Chalasani S.H., Bargmann C.I. A behavioral switch: cGMP and PKC signaling in olfactory neurons reverses odor preference in C. elegans. Neuron. 2008;59:959–971. doi: 10.1016/j.neuron.2008.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden A., Wiener S., You Y.J., Kim K., Avery L., Sengupta P. The EGL-4 PKG acts with the KIN-29 SIK and PKA to regulate chemoreceptor gene expression and sensory behaviors in Caenorhabditis elegans. Genetics. 2008;180:1475–1491. doi: 10.1534/genetics.108.094771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoven M.K., Bauer Huang S.L., Albin S.D., Bargmann C.I. The claudin superfamily protein nsy-4 biases lateral signaling to generate left–right asymmetry in C. elegans olfactory neurons. Neuron. 2006;51:291–302. doi: 10.1016/j.neuron.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Wes P.D., Bargmann C.I. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature. 2001;410:698–701. doi: 10.1038/35070581. [DOI] [PubMed] [Google Scholar]
- Wigle J.T., Eisenstat D.D. Homeobox genes in vertebrate forebrain development and disease. Clin. Genet. 2008;73:212–226. doi: 10.1111/j.1399-0004.2008.00967.x. [DOI] [PubMed] [Google Scholar]
- Yu S., Avery L., Baude E., Garbers D.L. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc. Natl. Acad. Sci. 1997;94:3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]