Abstract
A DNA double-strand break (DSB) is repaired by gene conversion (GC) if both ends of the DSB share homology with an intact DNA sequence. However, if homology is limited to only one of the DSB ends, repair occurs by break-induced replication (BIR). It is not known how the homology status of the DSB ends is first assessed and what other parameters govern the choice between these repair pathways. Our data suggest that a “recombination execution checkpoint” (REC) regulates the choice of the homologous recombination pathway employed to repair a given DSB. This choice is made prior to the initiation of DNA synthesis, and is dependent on the relative position and orientation of the homologous sequences used for repair. The RecQ family helicase Sgs1 plays a key role in regulating the choice of the recombination pathway. Surprisingly, break repair and gap repair are fundamentally different processes, both kinetically and genetically, as Pol32 is required only for gap repair. We propose that the REC may have evolved to preserve genome integrity by promoting conservative repair, especially when a DSB occurs within a repeated sequence.
Keywords: Gene conversion, BIR, SSA, Sgs1, Pol32
Eukaryotic cells have evolved several mechanisms to repair DNA lesions and maintain genome integrity. DNA double-strand breaks (DSBs) can be repaired either by nonhomologous end joining (NHEJ), involving simple religation of the broken DNA ends, or by homologous recombination (HR), in which genetic information from an intact homologous locus is used as a template for repair (Pâques and Haber 1999; Krogh and Symington 2004). Repair by HR involves extensive 5′-to-3′ resection of the broken DNA ends, which leaves 3′-ended single-stranded tails of DNA that subsequently get coated with the Rad51 recombinase protein. This Rad51–DNA filament searches for and base-pairs with (synapses with) a homologous donor, forming a three-strand structure called the displacement loop or D-loop. The 3′ end of the invaded strand primes new DNA synthesis to copy the donor template, leading to repair of the break. When both ends of a DSB share homology with a sister chromatid, a homologous chromosome or an ectopically located donor, the break is usually repaired by gene conversion (GC), in which a relatively short patch of new DNA is synthesized. If the homologies to the two DSB ends are separated by an insertion, the GC event is called gap repair as opposed to simple break repair, which occurs when there is no gap between the homologous donors. In mitotic cells, GC-associated DNA synthesis most often occurs in apparently sequential steps by synthesis-dependent strand annealing (Ira et al. 2006), although repair involving a more stable double-Holliday junction intermediate is also possible (Ira et al. 2003).
When homology with only one of the DSB ends is present, repair occurs by break-induced replication (BIR) (Fig. 1B). The initial steps of Rad51 filament formation, homology searching, and strand invasion are thought to be similar, if not identical, in GC and BIR. However, in the absence of a second end that can either capture the first newly synthesized strand or independently invade the donor sequences, the initial strand invasion intermediate is imagined to be turned into a DNA replication fork, capable of both leading and lagging strand synthesis (Voelkel-Meinman and Roeder 1990; Malkova et al. 1996, 2005; Morrow et al. 1997; Bosco and Haber 1998; Kraus et al. 2001; Davis and Symington 2004; Lydeard et al. 2007; Smith et al. 2007). This recombination-dependent DNA replication may extend to the very end of the template chromosome, or it could be terminated upon meeting a converging replication fork. The presence of only one homologous end can arise at stalled and/or broken replication forks (Haber 1999) or at telomeres that have lost their end protection (Lundblad and Blackburn 1993). BIR is indeed important for the lengthening of telomeres in telomerase-deficient cells (Le et al. 1999; Teng et al. 2000; McEachern and Haber 2006; Lydeard et al. 2007). A single homologous end can also arise when a DSB occurs in generally nonhomologous sequences, but in the vicinity of a sequence such as a transposable element that is present at other chromosome locations (Malkova et al. 2001; VanHulle et al. 2007). In this case, the end closer to the repetitive element can initiate repair by BIR to produce a nonreciprocal translocation, while the other end, which lacks homology, is lost by degradation. When homology with both ends is present, BIR is strongly out-competed by GC (Malkova et al. 2005); however, what actually prevents both ends (participating in GC) from initiating BIR is not clear.
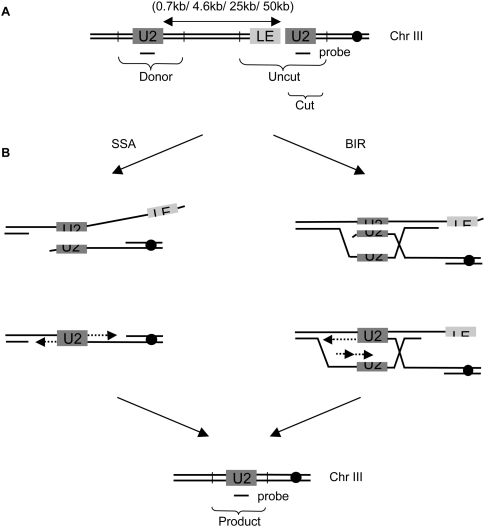
Figure 1.
Intrachromosomal assay system to monitor SSA and BIR. (A) Schematic representation of the strains used. An HO cut site is present within the leu2 gene on chromosome III. Homology with only the right end of the break—the U2 end—is inserted centromere-distal to the cut site in direct orientation at the indicated distances. The endogenous HOcs at MAT, HML, and HMR loci has been deleted. Vertical bars represent the positions of the Asp718 restriction sites when the donor U2 is placed 25 kb away from the HOcs. (B) These strains can repair the break either by SSA, where the two U2 sequences anneal with each other, the noncomplementary tails are clipped off, and the gaps are filled in by new DNA synthesis; or by BIR, where the cut U2 strand-invades the donor U2 and a recombination-dependent replication fork is established to copy all the distal sequences. Repair by either mode gives rise to the same deletion product.
A DSB flanked by direct repeats can be repaired by yet another recombination pathway called single-strand annealing (SSA). Here, the homologous sequences (after they have been rendered single-stranded by resection) simply anneal with each other, the noncomplementary tails are clipped off and the single-stranded gaps are filled in by new DNA synthesis (Fig. 1B). Repair by this mode results in deletion of one of the repeats and all of the intervening DNA sequences. In principle, such a break can also be repaired by Rad51-dependent (intrachromosomal) BIR (Fig. 1B), where the repeat closer to the cut site (which becomes single-stranded first) can strand-invade the repeat that is further away and set up a recombination-dependent replication fork to copy all the distal sequences. Repair by this pathway would also give rise to an SSA-like deletion product. However, the contribution of BIR in repairing such a DSB has never been carefully examined.
GC, BIR, and SSA are kinetically and mechanistically quite different from one another, even though they all require the key HR protein Rad52, which plays a role in both assembling the Rad51 filament as well as in strand annealing (Krogh and Symington 2004). The product of GC, as monitored by intrachromosomal mating-type (MAT) switching in budding yeast, can be seen as early as 1–2 h after formation of an HO endonuclease-induced DSB (Connolly et al. 1988; Sugawara et al. 2003). When GC occurs interchromosomally, product appears after ∼2 h (Ira et al. 2006; Keogh et al. 2006). GC-mediated repair requires the DNA polymerase processivity factor PCNA and apparently either of its associated replicative DNA polymerases—Polε or Polδ (Holmes and Haber 1999; Wang et al. 2004). However, many other components of normal DNA replication—including Cdc45 and the MCM helicase, as well as Polα-primase needed for lagging strand DNA synthesis—are dispensable for GC (Wang et al. 2004). Compared with GC, initiation of BIR is a much slower process, and no repair product is seen until ∼4 to 6 h following HO induction (Malkova et al. 2005; Lydeard et al. 2007). In addition to PCNA, Polα and primase are essential for BIR. Unlike GC, where Polδ and Polε appear to act redundantly, both DNA polymerases are required for BIR, but with distinct roles; while Polδ is required for the initiation of DNA synthesis, Polε plays an important role only during DNA elongation at later stages of the repair process (Lydeard et al. 2007).
Since SSA is dependent on resection for rendering the homologous sequences single-stranded for annealing with each other, the kinetics of product formation by this pathway are governed by the distance between the DSB and the farther repeat (Fishman-Lobell et al. 1992; Sugawara and Haber 1992; Vaze et al. 2002). While Rad52 is essential for the strand-annealing step, and its homolog Rad59 plays an important additional role in annealing shorter regions of homology (Sugawara et al. 2000), other proteins of the Rad52 epistasis group such as Rad51, Rad54, Rad55, and Rad57 are not required for SSA (McDonald and Rothstein 1994; Ivanov et al. 1996).
The presence of these alternative HR pathways necessitates a high level of regulation to ensure correct and efficient DSB repair; but how exactly and at what level this regulation happens, is not clear. For example, how does a cell first assess whether homology with both or only one of the DSB ends is present? If homology with both the ends is present, what exactly channels the break toward repair by GC? When a DSB is flanked by direct repeats, is the break repaired by SSA alone or does BIR also contribute to repair? To address these issues, we built a series of haploid strains in which we could monitor competition between GC and BIR or between SSA and BIR, all initiated by the induction of HO endonuclease. Using a combination of physical monitoring of DSB repair by Southern blots or PCR, and analysis of protein recruitment by chromatin immunoprecipitation (ChIP) assays, we demonstrate that the initial strand pairing events in GC, BIR, and SSA are similar. However, there appears to be a “recombination execution checkpoint” (REC) that regulates the initiation of DNA synthesis during these repair processes. We show that the REC is sensitive to the location, orientation, and distance between the homologous sequences used for repair by the ends of a DSB. The distance-dependent signaling between the ends is modulated by the RecQ family helicase Sgs1. We also conclude that DSB gap repair is distinctly different—kinetically and in its requirement for Pol32 (the nonessential subunit of Polδ)—from DSB break repair.
Results
Cells shift from SSA to BIR ∼6 h after induction of a DSB
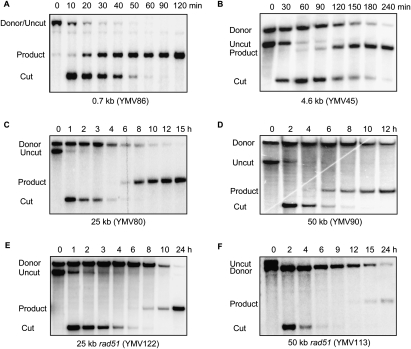
As stated above, a DSB flanked by direct repeats can be repaired, at least in principle, either by Rad51-independent SSA or by Rad51-dependent intrachromosomal BIR (Fig. 1B). However, it is presumed to mostly occur by SSA because repair can be accomplished efficiently even in the absence of Rad51. But whether SSA is indeed the predominant mode of repair, or if BIR also plays a role in repairing such a DSB, has never been carefully examined. Since there is not a good way to eliminate SSA without compromising BIR as well, we built a series of strains to test whether “slowing down” SSA (by progressively increasing the length of the intervening region that needs to be resected before homologous sequences become single-stranded for annealing) would make BIR more visible. These strains contain a 117-base-pair (bp) HO endonuclease recognition sequence (HOcs) at the KpnI site within the leu2 gene on chromosome III, and carry homology with only the right end of the break—i.e., the “U2” end—at different positions centromere-distal to the HO cut site (Fig. 1A; Vaze et al. 2002). HO endonuclease, under the control of a galactose-inducible promoter, was expressed in exponentially growing cultures of these strains, and the kinetics of repair were monitored using Southern blots. We found that repair product appears 20 min, 60 min, and 6 h following DSB induction when the donor U2 is positioned, respectively, 0.7 kb (YMV86), 4.6 kb (YMV45), and 25 kb (YMV80) away from the HOcs (Fig. 2A–C; Vaze et al. 2002; Clerici et al. 2005). This distance-dependent timing of product formation strongly suggests that repair predominantly occurs by SSA, and is consistent with the previously determined 5′-to-3′ resection rate of ∼4 kb/h (Fishman-Lobell et al. 1992; Zhu et al. 2008). Also, as expected for an SSA event, the appearance of the product band is coincident with the disappearance of the donor band as a result of resection.
Figure 2.
Cells shift from SSA to BIR ∼6 h after induction of a DSB. Southern blots showing kinetics of repair in strains carrying the donor U2 sequence 0.7 kb (YMV86) (A), 4.6 kb (YMV45) (B), 25 kb (YMV80) (C), and 50 kb (YMV90) (D) away from the HOcs. (E,F) Southern blots showing kinetics of repair in YMV80 rad51 and YMV90 rad51 cells, respectively. DNA was digested with Asp718 or PstI and probed with a U2-specific probe. Since there is no Asp718 site between the donor U2 and leu2∷HOcs in YMV86, the donor and uncut leu2 sequences run as a single fragment.
However, when the donor U2 is positioned 50 kb (YMV90) away from the HOcs, product still appears ∼6 h after DSB induction (Fig. 2D). This repair must occur by Rad51-dependent BIR, since product is seen long before the donor can become single-stranded by resection to facilitate SSA; moreover, the donor band persists even after the appearance of the repair product. When BIR is eliminated in YMV90 cells by deleting RAD51 (which is required for BIR but not for SSA) (McDonald and Rothstein 1994; Ivanov et al. 1996), no product is seen at 6 h (Fig. 2F). Instead, product appears ∼12 h after HO induction, which is consistent with the time it should take to render the donor U2 sequence single-stranded for repair to occur by SSA. When a similar experiment was done with a rad51Δ derivative of YMV80, the deletion product still appeared between 6 and 8 h (Fig. 2E; Kaye et al. 2004). This could either indicate that BIR does not make a significant contribution to DSB repair when the donor U2 is 25 kb away from the DSB or that the break can be repaired by both SSA and BIR; but since these processes would be kinetically identical in this particular case, deleting either of the repair pathways would not significantly affect the timing of appearance of the repair product. A more detailed analysis of product formation in YMV80 using other techniques has revealed that the latter hypothesis is, in fact, true (N. Sugawara and J.E. Haber, in prep.). Consistent with a SSA mode of repair, deleting RAD51 in YMV45 (where the donor U2 is only 4.6 kb away from the HOcs) did not delay the kinetics of product formation, which still appeared ∼60 min after DSB induction (Supplemental Fig. 1). Together these data suggest that when SSA and BIR are in competition with each other, SSA usually out-competes BIR, which is a kinetically slow process (as also shown before by Malkova et al. 2005; Lydeard et al. 2007) and does not come into effect until ∼6 h after DSB induction.
Initiation of new DNA synthesis is the rate-limiting step in BIR
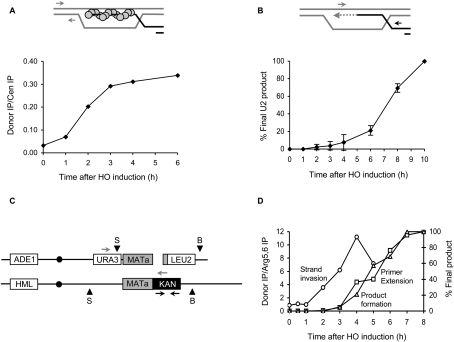
To determine why YMV80 or YMV90 cells wait until ∼6 h to initiate repair by BIR, we took a systematic approach to identify which specific step is slow in the BIR pathway. We first examined kinetics of strand invasion of the donor U2 locus by the cut U2 end. When a Rad51 nucleoprotein filament strand-invades its homologous template, DNA sequences close to the donor locus can be chromatin-immunoprecipitated along with the Rad51 protein, and quantified by PCR using donor-specific primers, to determine the kinetics of synapse formation (Fig. 3A; Sugawara et al. 2003). Using this Rad51 ChIP assay, we found that strand invasion occurs quite rapidly in YMV80 (Fig. 3A), as well as in YMV90 (data not shown) and can be seen ∼2 h after HO induction. Hence, the delay in BIR must occur at some post-synaptic step.
Figure 3.
Initiation of new DNA synthesis is the rate-limiting step in BIR. (A) Rad51 ChIP signal at the donor representing the kinetics of strand invasion in YMV80 (25 kb) cells. The top panel is a schematic showing synapsis between Rad51 nucleoprotein filament and donor U2. Circles represent Rad51 protein, and arrows indicate the position of primers (350 bp and 250 bp upstream of the U2 donor) used for PCR analysis. The bottom panel shows quantification of the PCR product as a function of time following HO induction. IP signal from an independent CEN8 locus was used to normalize for input DNA in the ChIP assay. (B) Kinetics of new DNA synthesis as determined by a primer extension assay. The top panel illustrates the repair step in question. Thick lines indicate synapse formation between cut and donor U2 sequences, and the dotted line represents newly synthesized DNA. Arrows indicate the position of primers (126 bp downstream from leu2∷HOcs and 45 bp upstream of the donor U2) used for PCR analysis. The bottom panel shows quantification of the PCR product as a function of time following HO induction in YMV80 (25 kb). The amount of PCR product obtained from the 10-h time point was set to 100%. Data represent mean ± SD (n = 3). (C) Schematic representation of diploid BIR strain TN001 used for analysis. Black arrows represent the position of primers used for ChIP analysis. Gray arrows represent the position of primers used for primer extension assay. Arrowheads represent the positions of BamHI (B) and SalI (S) restriction sites used for Southern analysis. (D) Graph showing kinetics of strand invasion (circles) as determined by a Rad51 ChIP assay, initiation of new DNA synthesis (squares) as determined by a quantitative PCR-based primer extension assay, and product formation (triangles) as determined by a Southern blot analysis. IP signal from an independent ARG5,6 locus was used to normalize for input DNA in the ChIP assay. The amount of product obtained from the last time point was set to 100% for primer extension and Southern analyses.
Since strand invasion is followed by extension of the synapsed 3′ end by new DNA synthesis, we next determined the kinetics of this step. This was done by a PCR-based primer extension assay (White and Haber 1990) using one primer specific to the cut U2 locus (126 bp downstream from the cut U2) and the other primer specific to the donor U2 locus (45 bp upstream of the donor U2) (Fig. 3B). In this assay, a PCR product can appear only if at least 45 bases of new DNA have been added to the synapsed 3′ end. As seen in Figure 3B, hardly any PCR product is formed until ∼6 h after HO induction. Since strand invasion occurs as early as 2 h but no primer extension can be seen until ∼6 h, initiation of new DNA synthesis is the rate-limiting step in BIR.
We obtained similar results from a diploid BIR system in which a single HO break at MAT locus on one copy of chromosome III can be repaired using its homolog, which shares homology with only the centromere-proximal side of the DSB (Fig. 3C). In this strain, all chromosome III sequences distal to the HOcs have been deleted by creating a LEU2-marked truncation ending in an artificial telomere (Malkova et al. 2005), and the right half of the HOcs on the donor chromosome has been replaced with a KAN-MX cassette. We observed a substantial lag between strand invasion and primer extension in this system as well (Fig. 3D), once again confirming that initiation of new DNA synthesis is indeed the rate-limiting step in BIR.
We note that there is a nearly 2 h difference in the kinetics of BIR product formation between diploids (Fig. 3D; Malkova et al. 2005) and haploids (Figs. 2C,D, 3B; Lydeard et al. 2007). This difference could be attributed either to the relatively high proficiency of HR in diploids (Nickoloff and Haber 2001) or to the presence of virtually unlimited homology with the DSB end in diploids as opposed to only ∼1 kb of homology provided for repair in the haploid systems.
Orientation- and distance-dependent signaling between the synapsed DNA ends determines the kinetics of DSB repair
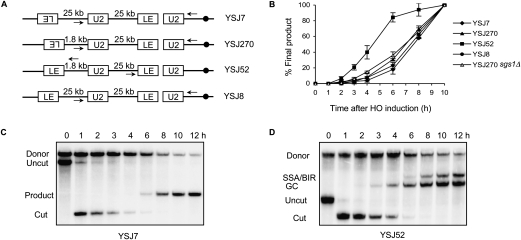
During GC, primer extension can be detected 30 min after synapse formation (White and Haber 1990; Sugawara et al. 2003; W. Hicks and J.E. Haber, unpubl.). We know that GC requires homology with both ends of a DSB, while homology with only one end is present in BIR. We wondered whether the absence of a signal from the unengaged second end is responsible for the delayed initiation of DNA synthesis during BIR. To test this idea, we inserted homology with the left end of the break as well—i.e., the “LE” end—in YMV80. This LE donor was positioned 25 kb centromere-distal to the U2 donor and oriented facing toward the telomere so that even though homology with both ends is present, cells cannot repair the HO break by GC although they can initiate two BIR events (YSJ7) (Fig. 4A). However, engagement of the LE end (as confirmed by a Rad51 ChIP assay) (data not shown) did not accelerate either the initiation (primer extension assay) (Fig. 4B) or the completion (Southern blot analysis) (Fig. 4C) of U2 product formation, which still appeared 6 h after HO induction.
Figure 4.
Orientation- and distance-dependent signaling between the two ends of a break determine the repair kinetics. (A) Schematic representation of the strains used. All these strains were derived from YMV80, which has the donor U2 25 kb away from the HOcs. Homology with left end of the break—the LE end—has been added in different orientations, at indicated distances from the U2 donor. Arrows indicate the position of primers used for the primer extension assay. (B) U2 repair kinetics in these strains as determined by a quantitative PCR-based primer extension assay using a primer 126 bp downstream from leu2∷HOcs and a primer 290 bp upstream of the U2 donor. The amount of PCR product obtained from the last time point was set to 100%. For YSJ52, a primer 126 bp upstream of leu2∷HOcs and a primer 450 bp downstream from the LE donor were used for primer extension analysis (see the text). Data represent mean ± SD (n = 3). (C) Southern blot showing kinetics of repair in YSJ7. DNA was digested with Asp718 and probed with a U2-specific probe. (D) Southern blot showing kinetics of repair in YSJ52. DNA was digested with XbaI and SpeI and probed with a U2-specific probe.
During GC, the two ends of a DSB usually engage rather close to each other, and may therefore be able to cooperatively open/modify the chromatin structure to support DNA synthesis differently from when only one end is engaged or when the two ends are engaged far away from each other. To test this idea, we made a strain where the LE donor is only 1.8 kb away from the U2 donor, although still facing toward the telomere (YSJ270) (Fig. 4A). But putting the donors even in such close proximity did not accelerate the initiation of the repair process (Fig. 4B). However, when we inverted the LE donor at this same locus (YSJ52) (Fig. 4A) so that strand invasions could result in gap repair, the primer extension product accumulated much faster (Fig. 4B). These data clearly demonstrate that a cell can differentiate between the orientations of the engaged ends, and that the ends must be engaged facing toward each other for repair to proceed rapidly. Using a Rad51 ChIP assay we confirmed that even though kinetics of product formation are significantly different in YSJ52 (the 1.8-kb gap repair strain) and YMV80 (the parent strain lacking the LE donor), the kinetics and efficiency of strand invasion are identical in these two strains (Supplemental Fig. 2). We note that in YSJ52, ∼40% of the cells still repair their break by the slower SSA/BIR deletion pathway (Fig. 4D). We attribute this to the presence of a Ty element just upstream of leu2∷HOcs, which may, in a fraction of cells, drive the LE end to other Ty elements elsewhere in the genome (N. Sugawara and J.E. Haber, in prep.). Since the U2 primer extension assay would reflect an average of the kinetics of product formation by the rapid gap repair pathway and the slower SSA/BIR pathways, kinetics of LE product formation are shown for YSJ52 in Figure 4B.
We also found that moving the LE donor 25 kb away from the U2 donor, while maintaining its correct orientation (YSJ8) (Fig. 4A) resulted in a dramatic slowing down of the repair process (Fig. 4B). It should be noted that we assayed initiation of DNA synthesis, and therefore, any delay caused by the time needed to traverse 25 kb DNA to complete the repair process is not seen in this assay. A pulse field gel analysis of DNA isolated from this strain revealed that only ∼25% of the cells repaired the break by GC (Supplemental Fig. 3). However, even these apparent gap repair events may actually have arisen by SSA following BIR from LE and U2 ends, a possibility previously suggested in some other studies (Kang and Symington 2000; Ira and Haber 2002). Nevertheless, these data demonstrate that even when the ends are oriented correctly, they need to be sufficiently close to each other to be able to trigger quick and efficient repair.
Cells shift from GC to BIR as distance between homologies is increased
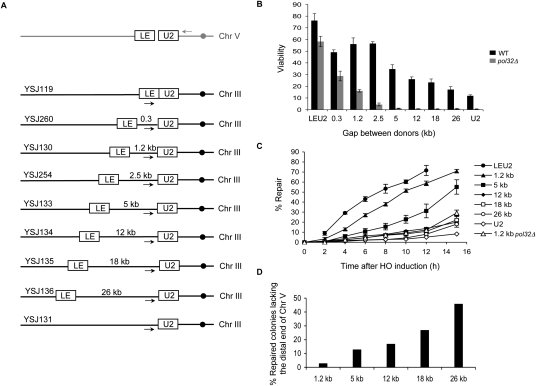
When homologies to DSB ends are positioned very close to each other, most cells repair the break by GC. But separating the donors beyond a certain critical distance may cause the ends to “lose sight” of each other, thereby resulting in BIR-mediated repair (even though homology with both ends is present). In order to determine what this critical distance might be, we built a series of haploid strains (Fig. 5A) carrying a leu2∷HOcs at the can1 locus on chromosome V, and the LE and U2 donor sequences at increasing distances from each other on left arm of chromosome III. (This interchromosomal repair system was constructed to eliminate competition with SSA, and to prevent interference from the Ty element present upstream of the endogenous LEU2 locus.) A strain carrying the entire LEU2 gene for a donor served as the GC control (YSJ119), and a strain carrying only the U2 donor served as the BIR control (YSJ131). In all other strains, repair can occur by either GC or BIR. Even though both the LE and the U2 ends can independently initiate BIR, only the U2-mediated BIR events will give viable colonies because loss of the ∼30 kb nonessential region of chromosome V distal to the DSB is inconsequential. BIR involving the LE end would result in loss of all the essential regions of chromosome V and, hence, would not give viable colonies. Moreover, a BIR event initiated by the LE end may not be able to traverse the centromere on chromosome III (Morrow et al. 1997), leading to premature termination of the repair process. The efficiency of repair, as determined by a viability assay, was found to be ∼75% for GC and ∼12% for BIR (Fig. 5B) as reported previously (Lydeard et al. 2007). Separating the donors by 1.2 kb and 5 kb gave viabilities of ∼57% and ∼35%, respectively. Increasing the gap to 12 kb or more resulted in further reduction in viabilities.
Figure 5.
Cells shift from GC to BIR as distance between homologies is increased. (A) Schematic representation of the strains used. leu2∷HOcs is inserted at the can1 locus on chromosome V. Homology with right end of the break—the U2 end—is present upstream of the SPS22 locus ∼41 kb from the left end of chromosome III. Homology with left end of the break—the LE end—is present at indicated distances from the U2 donor. Arrows indicate the position of primers used for the primer extension assay. (B) Viability of wild-type (black bars) and pol32Δ (gray bars) strains. Data represent mean ± SD (n ≥ 4). (C) U2 repair kinetics as determined by a quantitative PCR-based primer extension assay using a primer 500 bp upstream of the U2 donor and a primer 800 bp downstream from leu2∷HOcs. The amount of PCR product obtained from a repaired colony was set to 100%. For the strain carrying the LE and U2 donors right next to each other, a primer 500 bp upstream of the HOcs and a primer 800 bp downstream from leu2∷HOcs was used. The amount of PCR product obtained from the 0-h time point was set to 100%. Data represent mean ± SD (n = 3). (D) Proportion of repaired colonies lacking the distal fragment of chromosome V, as determined by colony PCR using a primer upstream of leu2∷HOcs and a primer downstream from the LE donor on chromosome III.
The drop in viability with longer gaps could be due to loss of signaling between the ends, resulting in reduced frequency of initiation of repair, or the inability of cells to synthesize longer stretches of DNA required to complete the repair, or a shift from GC to the less efficient and kinetically slower BIR mode of repair. To distinguish among these possibilities, we used a primer extension assay to test the kinetics of initiation of repair synthesis in all these strains. We found that cells shift from the quick GC kinetics to the slower BIR kinetics of initiation as the gap between the LE and U2 donors is increased. This shift is particularly obvious when the donors are separated by a distance of 5 kb or more (Fig. 5C). These data argue that the distance-dependent reduction in viability observed above can mostly be attributed to a defect in the initiation of repair. To test whether this delay in initiating DNA synthesis in strains with larger gaps actually reflects a shift from GC to BIR, we analyzed individual colonies from each strain to determine if the repaired chromosome had lost the distal, nonessential end of chromosome V, which would be indicative of repair by BIR. As shown in Figure 5D, the proportion of cases in which the distal segment of chromosome V is absent increases with the size of the gap until about half the events have this phenotype. These events must have arisen by BIR. We note that at least some of the other events that retained the distal end of chromosome V may also have arisen by BIR-dependent synthesis, perhaps independently from either of the two DSB ends, followed by capture of the other end or a strand-annealing event, resulting in apparent gap repair outcomes (Kang and Symington 2000; Ira and Haber 2002).
SGS1 modulates the efficiency and kinetics of gap repair
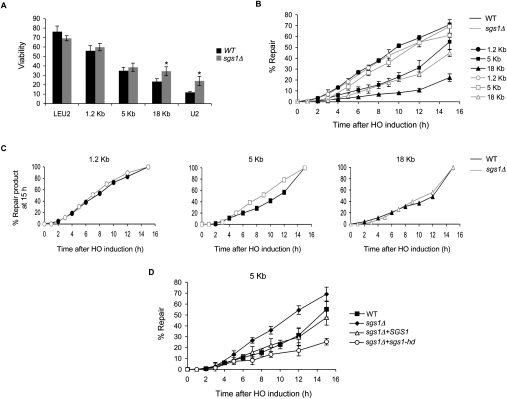
The distance-dependent signaling between the DSB ends could be mediated by the ability of the second end of a DSB to pair up with the D-loop created by strand invasion of the first end (Fig. 7B, below), which should become increasingly difficult as the donors become separated by larger gaps. We reasoned that if this was indeed the case, a helicase that can either disrupt or extend a D-loop might play a role in modulating the balance between GC and BIR as alternate modes of repair when the distance between the donors is increased. We deleted Sgs1, a 3′ → 5′ helicase belonging to the highly conserved RecQ family of helicases, which have been implicated in disruption of HR intermediates (Branzei and Foiani 2007b), resolution of dHJs to yield noncrossovers (Ira et al. 2003, Lo et al. 2006), and regression of stalled replication forks (Branzei and Foiani 2007a; Bachrati and Hickson 2008). Deletion of SGS1 significantly increased the efficiency of BIR and 18-kb gap repair by twofold and 1.5-fold, respectively (Fig. 6A). With a 5-kb gap, there was little effect on the overall efficiency of repair, but sgs1Δ specifically accelerated the kinetics of initiation of repair (Fig. 6B,C). To ensure that the difference in efficiency of repair among strains did not skew our interpretation of the kinetics of repair in these strains, the primer extension data were replotted after normalizing to the total amount of product obtained 15 h after HO induction (Fig. 6C). Together, these data confirm that deleting SGS1 enhances the efficiency but not the kinetics of repair of an 18-kb gap. Conversely, in the 5-kb gap repair strain, SGS1 deletion accelerates the kinetics of repair but has no effect on the efficiency of repair. Finally, in the 1.2-kb gap repair strain, sgs1Δ affects neither the efficiency nor the kinetics of repair.
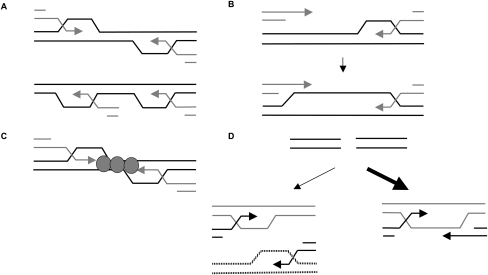
Figure 7.
Proposed models for assessment of relative orientation and position of the homologous sequences used for DSB repair. (A) Topological differences between the D-loops that form when ends engage facing toward each other (top) versus when they engage facing in the same direction (bottom) could provide a clue about the relative orientations of the engaged ends. (B,C) The distance parameter could be communicated either by capture of second end by extension of the D-loop, most likely through the action of a helicase (B) or by bridging of the two ends by some protein factors (C). (D) The requirement for the DSB ends to be engaged close to each other and in the right orientation to signal quick and efficient repair might facilitate conservative repair by preventing gross chromosomal rearrangements that might arise by the uncoordinated initiation of recombination events from the two DSB ends.
Figure 6.
Deletion of SGS1 modulates the kinetics and efficiency of gap repair. (A) Viability of wild-type (WT, black bars) and sgs1Δ (gray bars) strains. Data represent mean ± SD (n ≥ 4). Asteriks denote that viabilities of wild-type and sgs1Δ strains are significantly different (P ≤ 0.001) (B) U2 repair kinetics as determined by a quantitative PCR-based primer extension assay using a primer 500 bp upstream of the U2 donor and a primer 800 bp downstream from leu2∷HOcs. The amount of PCR product obtained from a repaired colony was set to 100%. Data represent mean ± SD (n = 2). (C) U2 repair kinetics obtained in B replotted after normalizing the amount of product obtained at the 15-h time point to 100% for each strain. (D) U2 repair kinetics in YSJ133 (5-kb gap repair strain), YSJ192 (sgs1Δ derivative of YSJ133), YSJ272 (YSJ192 + wild-type SGS1), and YSJ273 (YSJ192 + sgs1-hd). PCRs were performed and quantified as in B.
These data suggest that in wild-type cells, Sgs1 may function to limit the extent of D-loop formation (Bachrati et al. 2006; Lo et al. 2006; Branzei and Foiani 2007a), thus preventing communication between the DSB ends if they are separated by 5 kb or more. However in the absence of Sgs1, a more extensive D-loop may be formed, which can support communication between ends separated by 5 kb but not 18 kb, thereby signaling quicker initiation of new DNA synthesis in the 5-kb gap repair strain but not in the 18-kb strain.
To test whether the helicase domain of Sgs1 is required to disrupt communication between ends separated by a large distance, we complemented the 5-kb gap sgs1Δ strain YSJ192 with a plasmid carrying either wild-type SGS1 or a helicase-dead mutant sgs1-hd (Mullen et al. 2000). Complementation with SGS1 returned the kinetics of initiation of repair to wild-type levels (Fig. 6D). However, addition of the helicase-dead mutant resulted in repair kinetics slower than even in wild-type cells (Fig. 6D) although the viability was not different from wild type.
To examine its role in modulating orientation-dependent signaling between the ends, we deleted SGS1 in YSJ270 (Fig. 4A), in which the LE and U2 donors are oriented in a GC-incompatible configuration. Sgs1 deletion did not alter the kinetics of repair (Fig. 4B); thereby suggesting that it is not responsible for delaying the initiation of DNA synthesis when the ends are not oriented correctly. We did observe a nearly 30% drop in viability in this strain, which we think is attributable to the loss of the SSA component of repair due to the reduced continuing resection far from the DSB ends, in the absence of Sgs1 (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008).
Break repair is fundamentally different from gap repair
We showed recently that Pol32, the nonessential subunit of Polδ (Gerik et al. 1998), is required for BIR but not for GC (Lydeard et al. 2007). As expected, deleting POL32 resulted in a >20-fold reduction in the viability of our BIR strain carrying only the U2 donor (Fig. 5B), whereas it only modestly reduced the efficiency of break repair when there is no gap between the LE and U2 donors. Much to our surprise, deleting POL32 even in the 1.2-kb gap repair strain resulted in a nearly 3.5-fold reduction in viability, which further dropped to <1% in all other longer gap repair strains. This reduction in viability could be due to a defect in initiation of DNA synthesis or a failure to complete repair, especially since Pol32 has been shown to contribute to the processivity of Polδ (Burgers and Gerik 1998). A primer extension assay revealed a severe defect in the initiation of DNA synthesis upon POL32 deletion even in the 1.2-kb gap repair strain (Fig. 5C). However, in this pol32Δ strain, just as in the wild-type parent strain, the amount of primer extension product obtained at 12 h (Fig. 5C) was still comparable with the actual viability of the strain (Fig. 5B), suggesting that the completion of repair is not significantly affected by the deletion of Pol32. Hence, during gap repair—as also during BIR—Pol32 appears to be required for the initiation of new DNA synthesis. This differential requirement of Pol32 for gap repair but not break repair suggests that these are fundamentally different processes.
Discussion
DNA DSBs can be repaired by one of at least three distinct HR pathways: when homology with both the DSB ends is present, repair occurs by GC; when only one DSB end is able to find a homologous donor, repair occurs by BIR; and when a DSB is flanked by direct repeats, repair usually occurs by SSA. But how the initial sensing of engagement of the broken DNA ends with their homologous donors occurs, and how the subsequent decision making to employ one repair pathway or the other happens, is not understood. To address these issues, we studied competition between these different HR pathways in budding yeast Saccharomyces cerevisiae. We show that the initial homology search and strand invasion occur with almost equal efficiency and kinetics in GC, BIR, and SSA (Fig. 3A; Supplemental Fig. 2; Sugawara et al. 2003); however, the next step—i.e., initiation of new DNA synthesis—is differentially regulated during these processes. While the DNA synthesis is initiated ∼30 min after strand invasion during GC (Sugawara et al. 2003; W. Hicks and J.E. Haber, unpubl.), a nearly 4-h lag is observed between strand invasion and DNA synthesis during BIR (Fig. 3), and, during SSA, DNA synthesis is initiated only after both the homologous sequences have been rendered single-stranded by resection (Figs. 2C, 3B; Sugawara and Haber 1992). These data clearly rule out the notion that strand invasion is always immediately followed by DNA synthesis, which could either be terminated upon an encounter with the second end resulting in repair by GC, or—in the absence of a second end—may continue up to the end of the template chromosome resulting in repair by BIR. Instead, these data strongly argue that the choice of the repair pathway is made prior to the actual initiation of DNA synthesis, presumably by a REC that seems to monitor how the DSB ends are engaged in terms of their orientation and distance with respect to each other. When the ends are engaged fairly close to each other and in the right orientation, the break is quickly repaired by GC (Figs. 4B, 5C). In this case, a relatively short patch of new DNA is rapidly synthesized, most likely by the SDSA mechanism (Ira et al. 2006), which does not involve any lagging strand DNA synthesis (Wang et al. 2004). However, if only one of the DSB ends is engaged, or if the two ends are engaged in the wrong orientation or too far apart from each other (Figs. 4B, 5C), a different machinery capable of both leading and lagging strand DNA synthesis (Lydeard et al. 2007) is recruited to carry out repair by BIR. If, however, both the homologous sequences become single-stranded before either of these processes has been initiated (Fig. 2; Ivanov et al. 1996), the SSA machinery is called upon to fill in the single-stranded gaps and seal the break.
Although we did not yet identify a gene that would precisely define the basis of how REC inhibits the initiation of new DNA synthesis from a strand-invaded end, we suggest that there is an active checkpoint and not simply a delay imposed by the more complicated assembly of a DNA replication fork to carry out BIR. Soon after the DSB is made, the resected DSB ends invade their homologous templates; but when the orientation or distance between the ends is incompatible with GC, the next step of primer extension is blocked, even though this should be the same process that happens in GC. We attribute this delay to a surveillance mechanism—the REC—that, by blocking DNA synthesis, may give more time to the DSB ends to search for homology in the vicinity of each other, thereby promoting conservative repair. The REC is distinct from the Mec1-mediated DNA damage checkpoint because we showed previously that deleting MEC1 (and SML1 to maintain viability) does not result in a faster accumulation of repair product in YMV80 (Kaye et al. 2004).
How all this sensing and signaling of the DSB ends actually happens is not entirely clear. We suggest that the REC may be able to distinguish between the orientations of the synapsed ends through some topological features of the D-loops that form when ends engage facing toward each other (YSJ52) versus when they engage facing in the same direction (YSJ14; Fig. 7A). The distance parameter appears to be communicated by the capture of the two ends by a continuous D-loop (Fig. 7B), which seems to be greatly compromised as the distance between the donors is increased to 5 kb or more (Fig. 5). This limitation is imposed, at least in part, by the Sgs1 helicase (Fig. 6B,C), which appears to prevent communication between the DSB ends synapsed with widely separated donors by inhibiting the extension of the D-loop. Sgs1 has previously been implicated in the S-phase checkpoint response induced by DNA damage and/or replication fork arrest (Frei and Gasser 2000; Bjergbaek et al. 2005). Here we provide evidence for its role in another checkpoint pathway, which appears to delay the initiation of DNA synthesis if the DSB ends are not engaged in a GC-compatible configuration.
This function of Sgs1 is independent of its helicase activity—analogous to its previously published role in modulating GC tract lengths (Lo et al. 2006). We also found that helicase-dead Sgs1 impedes repair compared with the wild-type protein (Fig. 6D). It is possible that Sgs1-hd binds to a strand invasion intermediate but prevents the subsequent steps of HR from occurring efficiently. We note that a helicase-independent effect of Mph1 overexpression was recently reported and attributed to the continued association of the helicase to strand invasion intermediates without allowing further steps to occur (Banerjee et al. 2008).
The fact that sgs1Δ causes a quicker initiation of repair in the 5-kb gap repair that is not accompanied by an increased viability may suggest that the enhanced signaling between the ends results in the recruitment of a less processive DNA synthesis machinery that can initiate repair quite rapidly and efficiently but may not be able to go all the way to “complete” it. A similar conclusion was reached by studies in Drosophila, where shorter repair synthesis tracts were observed during gap repair in flies lacking the Sgs1 homolog DmBlm (Adams et al. 2003; McVey et al. 2004).
The increased efficiency of BIR in a sgs1Δ mutant could also be a result of the enhanced stability of the D-loop in the absence of Sgs1. This D-loop stabilization may be more critical during BIR (and thereby also in the longer gap repair events), where DNA synthesis is not initiated for ∼4 h after synapse formation as opposed to GC, where strand invasion is quickly followed by new DNA synthesis.
The distance parameter of REC could also be communicated by a physical bridging of the ends together through one or more protein factors (Fig. 7C) such as the MRX complex or the Rad52 protein (Kaye et al. 2004; Lobachev et al. 2004). Inability to maintain end coordination when the donors are physically separated could then result in loss of signaling between the ends, causing a shift from the quick and efficient GC mode of repair to the slow and inefficient BIR mode of repair (Fig. 5B–D), as the distance between the donors is increased. A somewhat similar correlation between the efficiency of repair and DSB gap length was also seen in Drosophila germ line cells where gaps of ≤11 kb could be repaired with approximately the same efficiency as breaks with no gaps at all; but a gap of 44 kb was repaired only ∼0.1% of the times, while one of 210 kb was not repaired at a measurable frequency (Johnson-Schlitz and Engels 2006). It is possible that the Drosophila case reflects problems in replication across long distances, rather than in the initiation of gap repair events, as we see with yeast.
We also found that Pol32, which is required for BIR but is dispensable for break repair (Lydeard et al. 2007), becomes increasingly important as the gap between the donors is increased (Fig. 5B,C). In fact, the repair of even a small gap of 1.2 kb, which can occur fairly efficiently and quickly in wild-type cells, is significantly compromised in the absence of Pol32. Contrary to the current belief, this differential requirement of Pol32 strongly argues that DSB gap repair is fundamentally distinct from DSB break repair. This idea is also supported by some work done in a previous study where MRE11 deletion was shown to have a differential effect on GC tract lengths during plasmid gap repair compared with plasmid break repair, although this was attributed to the end coordination function of Mre11, and the difference was not seen in a chromosomal context (Krishna et al. 2007).
We suggest that the REC may have evolved to preserve genome integrity by preventing the DSB ends from initiating independent recombination events from two physically unrelated chromosome loci, which could give rise to deleterious translocations and genome rearrangements (Fig. 7D). When a DSB occurs within a repeated sequence, the requirement for the DSB ends to be engaged close to each other and in the right orientation to signal quick and efficient repair should greatly reduce the frequency of illegitimate recombination events involving multiple nonallelic repeats. The REC may also prevent one DSB end from repairing off of a sister chromatid while the other end repairs from a homologous chromosome, resulting in aneuploidy. An analogous problem has been observed in Saccharomyces cerevisiae meiosis in the absence of Sgs1, where the two ends of a DSB engage in promiscuous strand invasions with different templates (Oh et al. 2007). We suggest that in mitotic cells also, Sgs1 may destabilize the D-loop intermediates formed during BIR (or long gap repair events) and, in turn, promote the ends to find homology in the vicinity of each other.
REC may also have some implications for the SDSA model of DSB repair, where often times a single-ended invasion is imagined to prime DNA synthesis, which subsequently dissociates from the donor template and anneals to the other resected, noninvading end. However, our data argue that both the DSB ends must strand-invade their homologous donors, although maybe only transiently, to trigger DNA synthesis from either one or both DSB ends.
Materials and methods
Strains and plasmids
All strains are derived from YFP17 (hmlΔ∷ADE1 mataΔ∷hisG hmrΔ∷ADE1 leu2-cs ade3 ∷GAL ∷HO ade1 lys5 ura3-52). YMV90 was constructed by adding the ∼1.2 kb KpnI–SalI fragment of LEU2 (the “U2” homology) at position 41,430 on left arm of ChrIII. YMV45, YMV80, YMV86, and the rad51Δ derivatives of YMV80 and YMV90 were constructed as described in Vaze et al. (2002). YSJ7, YSJ8, YSJ270, and YSJ52 were derived from YMV80 by adding the ∼700 bp XhoI–KpnI fragment of LEU2 (the “LE” homology) in either orientation (Fig. 4A) at position 41,430 (YSJ7 and YSJ8) or position 67,750 (YSJ270 and YSJ52) on the left arm of ChrIII. Strains used to study competition between GC and BIR were all derived from YSJ131, which itself was derived from YMV90 by moving the leu2-cs to the can1 locus on ChrV. Gap repair strains YSJ133–YSJ136 were constructed by adding the “LE” homology to YSJ131 at positions 37,700, 30,800, 24,500 and 17,050, respectively, on ChrIII upstream of the U2 donor. YSJ119 was independently constructed from YFP17 (Lydeard et al. 2007). YSJ130 was constructed by inserting a 1.2-kb URA3 fragment at the KpnI site within the donor LEU2 in YSJ119. sgs1Δ and pol32Δ strains were made by the standard PCR-based gene disruption method. YSJ192 (YSJ133 sgs1Δ) was transformed with pSJ22 or pSJ23 to obtain strains YSJ272 and YSJ273, respectively. Plasmids pSJ22 and pSJ23 were constructed by subcloning the XhoI–SacI fragments of pSM100 and pSM100-hd (Mullen et al. 2000), respectively, in to pRS414 (Sikorski and Hieter 1989). The diploid BIR strain NT001 was derived from AM729 (Malkova et al. 2005) by replacing homology with the right side of the HOcs on full-length Chr III with a KAN-MX cassette.
HO induction for kinetic analysis of DSB repair
Yeast cells were grown in YEP containing 2% raffinose to a density of ∼1 × 107 cells per milliliter and HO endonuclease was induced by adding galactose to a final concentration of 2%. Samples were collected for DNA analysis just prior to and at different time points following addition of galactose, as described before (Holmes and Haber 1999).
Southern blot analysis
Purified genomic DNA was digested with the appropriate restriction enzyme and probed with a 32P-labeled U2 probe and scanned by Bio-Rad phosphorimager as described before (Vaze et al. 2002). DNA from YMV45 was digested with PstI, from YMV113 with Asp718 and XbaI, from YSJ52 with XbaI and SpeI, and from all other strains with Asp718.
ChIP assay
Rad51 ChIPs were performed as described before (Sugawara et al. 2003). The IP signal from the donor locus was normalized to the IP signal either from the CEN8 locus (immunoprecipitated using anti-Mif2 antibody) (Fig. 3A; Supplemental Fig. 2) or from the ARG5,6 locus (Fig. 3D).
PCR-based primer extension assay
Equal amounts of genomic DNA isolated from samples collected at different time points were amplified within linear range as described before (White and Haber 1990) using the primers specified in the figure legends. The PCR reactions were run on agarose gels and the repair product was quantified using Bio-Rad Quantity One software. PCR signal from an independent locus (ARG5,6 for Figs. 3B, 4B, and LDB16 for Figs. 5C, 6B–D) was used to normalize for input DNA. The ratio of test and reference signals obtained from the 10-h time point (Figs. 3B, 4B), 15-h time point (Fig. 6C), or from a repaired colony (Figs. 5C, 6D) was set to 100%.
Pulse field gel electrophoresis
Pulse field gel electrophoresis was performed by running genomic DNA embedded in plugs of 1% agarose, at 200 V, for 40 h (initial time, 10 sec; final time, 35 sec). DNA plugs were made using a Bio-Rad CHEF yeast genomic DNA plug kit, as per manufacturer's protocol.
Viability measurements
Yeast cells were grown in YEP containing 2% raffinose to a density of ∼1 × 107 cells per milliliter. Equal volumes of appropriate dilutions were plated on YEP containing 2% galacotse (YEPGal; to induce the HO break) and YEP containing 2% dextrose (YEPD; no DSB control). Viability was determined from the ratio of colony-forming units (CFUs) able to survive the break (number of colonies that grew on YEPGal) to the total number of CFUs plated (number of colonies that appeared on YEPD).
Acknowledgments
We are grateful to Akira Shinohara and Douglas Koshland for their generous gift of anti-Rad51 and anti-Mif2 antibodies. We also thank Sue Lovett and members of Haber laboratory for helpful comments and suggestions. This work was supported by NIH grants GM20056 and GM76020. J.R.L. was supported as a Trainee of NIH Training Grant in Genetics T32 GM7122.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1751209.
Supplemental material is available at http://www.genesdev.org.
References
- Adams M.D., McVey M., Sekelsky J.J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- Bachrati C.Z., Hickson I.D. RecQ helicases: Guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- Bachrati C.Z., Borts R.H., Hickson I.D. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Smith S., Oum J.H., Liaw H.J., Hwang J.Y., Sikdar N., Motegi A., Lee S.E., Myung K. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J. Cell Biol. 2008;181:1083–1093. doi: 10.1083/jcb.200711146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjergbaek L., Cobb J.A., Tsai-Pflugfelder M., Gasser S.M. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G., Haber J.E. Chromosome break-induced DNA replication leads to nonreciprocal translocations and telomere capture. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst.) 2007a;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes & Dev. 2007b;21:3019–3026. doi: 10.1101/gad.1624707. [DOI] [PubMed] [Google Scholar]
- Burgers P.M., Gerik K.J. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- Clerici M., Mantiero D., Lucchini G., Longhese M.P. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J. Biol. Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- Connolly B., White C.I., Haber J.E. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A.P., Symington L.S. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J., Rudin N., Haber J.E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei C., Gasser S.M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes & Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Gerik K.J., Li X., Pautz A., Burgers P.M. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998;273:19747–19755. doi: 10.1074/jbc.273.31.19747. [DOI] [PubMed] [Google Scholar]
- Gravel S., Chapman J.R., Magill C., Jackson S.P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes & Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. DNA recombination: The replication connection. Trends Biochem. Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- Holmes A.M., Haber J.E. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Ira G., Haber J.E. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G., Satory D., Haber J.E. Conservative inheritance of newly synthesized DNA in double-strand break-induced gene conversion. Mol. Cell. Biol. 2006;26:9424–9429. doi: 10.1128/MCB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E.L., Sugawara N., Fishman-Lobell J., Haber J.E. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz D.M., Engels W.R. The effect of gap length on double-strand break repair in Drosophila. Genetics. 2006;173:2033–2038. doi: 10.1534/genetics.106.059436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L.E., Symington L.S. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:9162–9172. doi: 10.1128/mcb.20.24.9162-9172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J.A., Melo J.A., Cheung S.K., Vaze M.B., Haber J.E., Toczyski D.P. DNA breaks promote genomic instability by impeding proper chromosome segregation. Curr. Biol. 2004;14:2096–2106. doi: 10.1016/j.cub.2004.10.051. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kim J.A., Downey M., Fillingham J., Chowdhury D., Harrison J.C., Onishi M., Datta N., Galicia S., Emili A., et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- Kraus E., Leung W.Y., Haber J.E. Break-induced replication: A review and an example in budding yeast. Proc. Natl. Acad. Sci. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna S., Wagener B.M., Liu H.P., Lo Y.C., Sterk R., Petrini J.H., Nickoloff J.A. Mre11 and Ku regulation of double-strand break repair by gene conversion and break-induced replication. DNA Repair (Amst.) 2007;6:797–808. doi: 10.1016/j.dnarep.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh B.O., Symington L.S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Le S., Moore J.K., Haber J.E., Greider C.W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y.C., Paffett K.S., Amit O., Clikeman J.A., Sterk R., Brenneman M.A., Nickoloff J.A. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell. Biol. 2006;26:4086–4094. doi: 10.1128/MCB.00136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K., Vitriol E., Stemple J., Resnick M.A., Bloom K. Chromosome fragmentation after induction of a double-strand break is an active process prevented by the RMX repair complex. Curr. Biol. 2004;14:2107–2112. doi: 10.1016/j.cub.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Lundblad V., Blackburn E.H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- Lydeard J.R., Jain S., Yamaguchi M., Haber J.E. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Malkova A., Ivanov E.L., Haber J.E. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Signon L., Schaefer C.B., Naylor M.L., Theis J.F., Newlon C.S., Haber J.E. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes & Dev. 2001;15:1055–1060. doi: 10.1101/gad.875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A., Naylor M.L., Yamaguchi M., Ira G., Haber J.E. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J.P., Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern M.J., Haber J.E. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- McVey M., Larocque J.R., Adams M.D., Sekelsky J.J. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou E.P., Symington L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow D.M., Connelly C., Hieter P. “Break copy” duplication: A model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman V., Brill S.J. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff J.A., Haber J.E. Mating-type control of DNA repair and recombination in Saccharomyces cerevisiae. In: Nickoloff J.A., Hoekstra M.F., editors. DNA damage and repair: Advances from phage to humans. Vol. 3. Humana Press; Totowa, NJ: 2001. pp. 107–124. [Google Scholar]
- Oh S.D., Lao J.P., Hwang P.Y., Taylor A.F., Smith G.R., Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.E., Llorente B., Symington L.S. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Haber J.E. Characterization of double-strand break-induced recombination: Homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Ira G., Haber J.E. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Wang X., Haber J.E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- Teng S.C., Chang J., McCowan B., Zakian V.A. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell. 2000;6:947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- VanHulle K., Lemoine F.J., Narayanan V., Downing B., Hull K., McCullough C., Bellinger M., Lobachev K., Petes T.D., Malkova A. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaze M.B., Pellicioli A., Lee S.E., Ira G., Liberi G., Arbel-Eden A., Foiani M., Haber J.E. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- Voelkel-Meiman K., Roeder G.S. Gene conversion tracts stimulated by HOT1-promoted transcription are long and continuous. Genetics. 1990;126:851–867. doi: 10.1093/genetics/126.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ira G., Tercero J.A., Holmes A.M., Diffley J.F., Haber J.E. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:6891–6899. doi: 10.1128/MCB.24.16.6891-6899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C.I., Haber J.E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]