Abstract
The secreted morphogen, Sonic hedgehog (Shh) is a significant determinant of brain size and craniofacial morphology1–4. In humans, SHH haploinsufficiency results in holoprosencephaly (HPE)5, a defect in anterior midline formation. Despite the importance of maintaining SHH transcript levels above a critical threshold, we know little about the upstream regulators of SHH expression in the forebrain. Here we describe a combination of genetic and biochemical experiments to uncover a critical pair of cis and trans acting determinants of Shh forebrain expression. A rare nucleotide variant located 460kb upstream of SHH was discovered in an individual with HPE that resulted in the loss of Shh brain enhancer-2 (SBE2) activity in the hypothalamus of transgenic mouse embryos. Using a DNA affinity capture assay we screened SBE2 sequence for DNA binding proteins and identified members of the Six3/Six6 homeodomain family as candidate regulators of Shh transcription. Six3 and Six6 showed reduced binding affinity for the mutant compared to wild type SBE2 sequence. Moreover, HPE causing mutations in Six3 failed to bind and activate SBE2, whereas, Shh forebrain expression was unaltered in Six6−/− embryos. These data provide a direct link between Six3 and Shh regulation during normal forebrain development and in the pathogenesis of HPE.
Keywords: Shh, gene expression, forebrain, holoprosencephaly
Shh expression must be regulated in a temporally and spatially restricted manner in order to fulfill its multiple functions during forebrain and craniofacial development (reviewed in refs 6,7). Three tissues, including the prechordal plate, ventral forebrain and facial ectoderm, have been identified as critical sources of Shh that promote distinct aspects of ventral forebrain and craniofacial morphogenesis4,8–10. Interfering with Shh signaling from any of these sites results in HPE, a spectrum of brain and craniofacial malformations, the severity of which correlates with the timing of Shh perturbation1,10,11. In humans, SHH haploinsufficiency is the predominant cause of HPE, indicating that the level of SHH expression is important for proper forebrain and craniofacial development5. Several downstream effectors of SHH and NODAL signaling pathways have also been identified as targets of mutation in HPE, whereas mutations in other genes such as SIX3 and ZIC2 cause HPE through poorly defined mechanisms12. While much is known about the signal transduction pathway functioning downstream of Shh, we know relatively little of the genes operating upstream in the pathway that regulate Shh transcription in key signaling centers mediating forebrain and craniofacial development.
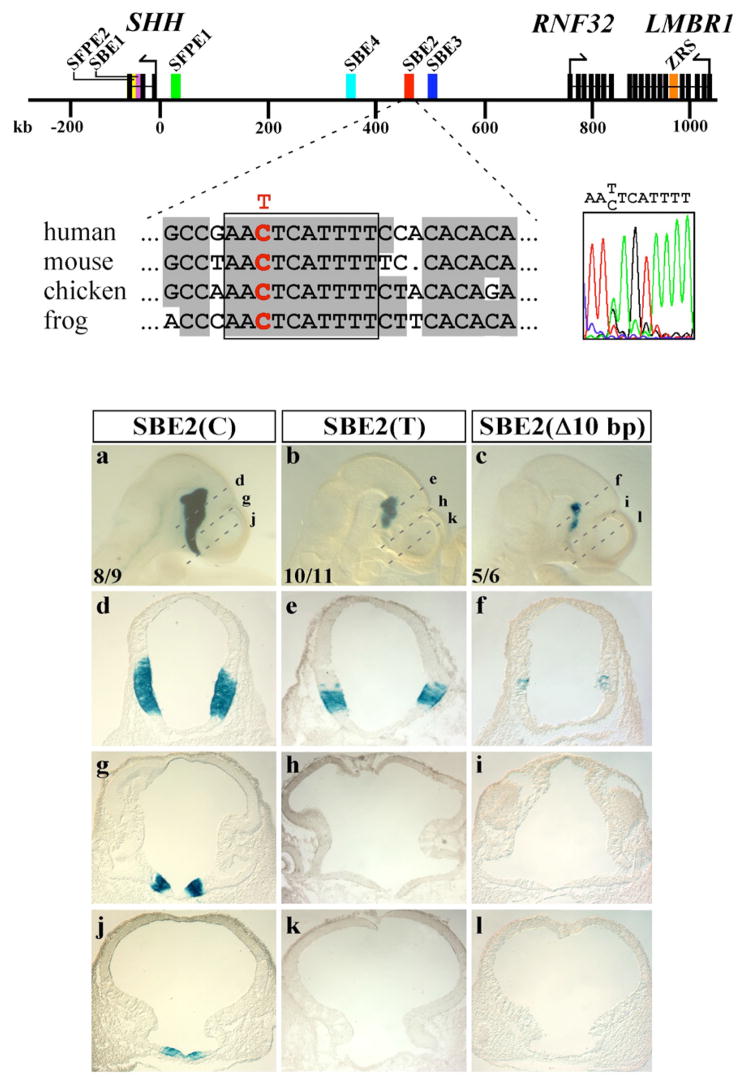
Previous efforts to address this issue focused on determining the genomic location of functional Shh regulatory elements13. These experiments identified six enhancers distributed over a 500 kb interval surrounding the Shh gene that directed reporter activity to most areas of Shh expression in the mouse CNS, including the ventral forebrain (Fig. 1). In particular, the highly conserved Shh brain enhancer-2 (SBE2), located 460 kb upstream of the SHH coding sequence, was identified as unique in its ability to regulate Shh-like expression throughout the hypothalamus.
Figure 1. SBE2 activity in the rostral hypothalamus is compromised by a sequence variant found in an individual with HPE.
(Top) Physical map displaying the distribution of genes (black rectangles) and regulatory sequences (colored rectangles) spanning 1 Mb upstream of SHH on human chromosome 7 (from ref. 13). The sequence tracing to the right is from an individual with lobar HPE who is heterozygous for a C/T transition mutation in SBE2. The mutation resides within a 10 bp-block of SBE2 sequence that was 100% conserved in human, mouse, chicken and frog (red base in boxed sequence alignment). (a-l) X-gal staining of representative embryos carrying wild type SBE2(C) (a, d, g, j), mutant SBE2(T) (b, e, h, k) or a 10 bp deletion SBE2(Δ10 bp) (c, f, i, l) at E10.5. Dashed lines in (a–c) indicate the planes of section shown in panels (d-l). The number of embryos showing representative reporter activity over the total number of transgenic embryos is indicated for each construct (a–c). Abbreviations: SBE, Shh brain enhancer; SFPE, Shh floor plate enhancer; ZRS, zone of polarizing activity regulatory sequence.
To identify functionally relevant nucleotides in SBE2, we screened the 1.1 kb sequence mediating its activity for mutations in humans with HPE. We reasoned that HPE causing variants in SBE2 could aid in identifying critical cis and trans determinants of SHH expression in the forebrain. Similar resequencing approaches have been successful in identifying common and rare coding sequence variants in genes associated with common diseases, but have not been routinely applied to the study of remote noncoding regions in rare diseases such as HPE (1:16,000 livebirths)12,14.
From 474 HPE patients, we identified one individual who was heterozygous for a C to T base change at nucleotide position 444 of the enhancer sequence. The C/T variant is situated within a block of 10 nucleotides that have been maintained in human, mouse, chicken and frog for over 350 million years (Fig. 1). This C/T nucleotide variant was not observed in DNA samples from 450 unrelated control individuals. The affected female exhibited features of semilobar HPE including microcephaly, midfacial hypoplasia, cleft-lip and palate, diabetes insipidus, and moderate fusion of the hypothalamus and basal ganglia. The parents’ genotype revealed that the father is an unaffected carrier, while the mother is homozygous for the wild type SBE2(C) allele. It is known that approximately 30% of individuals heterozygous for loss-of-function mutations in Shh show no evidence of HPE12. That is, these mutations are often non-penetrant. So the finding that the carrier father is unaffected does not discount the possibility that SBE2(T) confers an increased risk of HPE. As mutations in known HPE genes were not detected in the affected female, we sought to determine whether the single nucleotide change could alter SBE2 activity and thus, provide a molecular basis for her phenotype.
Human SBE2 sequences containing either the wild type SBE2(C) or variant SBE2(T) residue were tested for their ability to drive lacZ expression in transgenic embryos. Embryos carrying the wild type SBE2(C) reporter construct showed little variability in the spatial distribution of X-gal staining, recapitulating Shh expression in the hypothalamus from the mammillary region caudally to the preoptic area rostrally (n= 8/9, Fig. 1a, d, g, j). In contrast, embryos carrying SBE2(T) consistently showed a loss of reporter activity from the level of the optic vesicles to the rostral extent of the diencephalon (n= 10/11, Fig. 1b, e, h, k). The reduction in SBE2 reporter activity was similar in embryos carrying SBE2(Δ 10bp), a construct in which the highly conserved 10 bp sequence overlapping the C/T substitution was deleted (n=5/6, Fig. 1c, f, i, l). Notably, the area of the ventral diencephalon that showed decreased X-gal staining in embryos carrying SBE2(T) correlated with the sites of malformation exhibited by the individual with HPE. The anterior region of the ventral diencephalon is an important source of Shh for the development of the face and pituitary gland11,15,16.
Based on these findings, we hypothesized that the conserved 10 bp SBE2 sequence serves as a binding site for a transcriptional regulator whose function is required for the activation of Shh expression in the anteroventral portion of the hypothalamus. In the presence of the SBE2(T) variant, assembly of this transcriptional activation complex is compromised, likely resulting in the reduction of Shh expression below a critical threshold. The analysis of the 10 bp sequence in question did not reveal informative transcription factor binding sites in the TRANSFAC database, therefore, we sought to identify the putative SBE2-binding protein using a DNA affinity capture assay17.
A biotinylated 18 bp double-stranded SBE2 probe was incubated with nuclear extracts prepared from adult mouse brain. DNA/protein complexes were pulled-down with streptavidin-coated agarose beads and their protein content analyzed by mass spectrometry (see methods). To control for nonspecific DNA-binding proteins, SBE2 extracted proteins were compared to those pulled down with an SBE2 probe carrying multiple nucleotide mismatches in highly conserved residues. Only DNA-binding proteins specific for SBE2 were considered further. Of the six transcription factors identified (Table S1), the one of greatest interest was Six6, a homeodomain-containing protein belonging to the optix family of transcriptional regulators that includes Six3, a gene associated with HPE18,19.
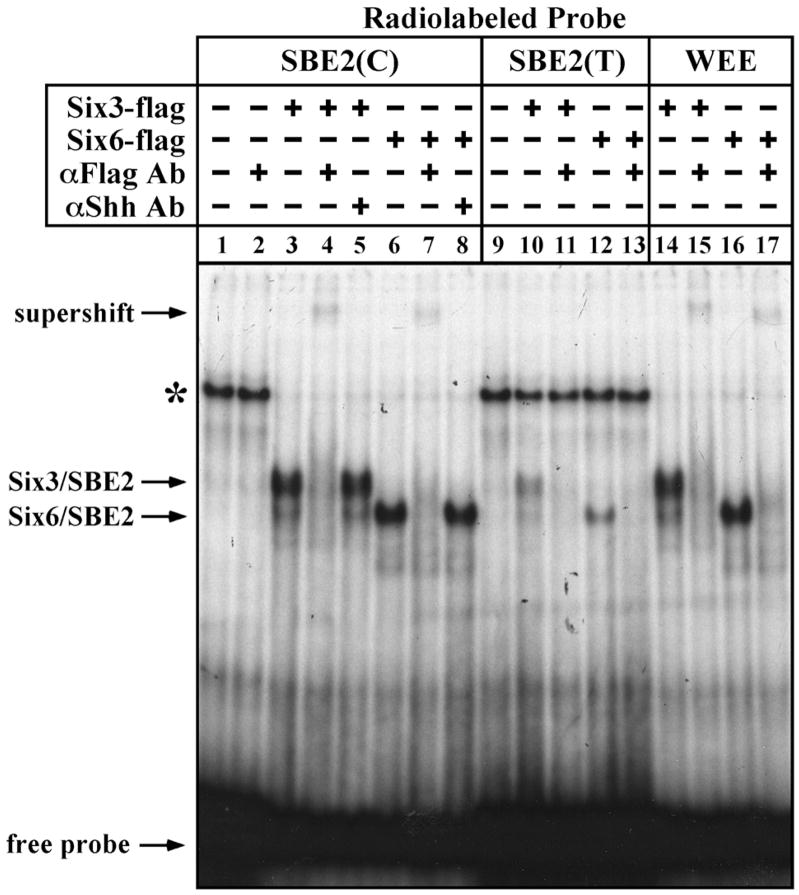
In order to validate the binding of Six3/6 proteins to SBE2 we performed electrophoretic mobility shift assays (EMSAs). Cos-1 cell lysates transfected with Flag-tagged versions of full length Six3 and Six6 formed specific complexes when incubated with a radiolabeled SBE2(C) probe (Fig. 2 lanes 1,3,6). These protein/DNA complexes were supershifted when exposed to an αFlag antibody (Fig. 2 lanes 2,4,7), but not a nonspecific antibody (Fig. 2 lanes 5,8), indicating that the binding of Six3 and Six6 to SBE2 was direct. Specific complexes formed with similar mobility when an SBE2(T) probe was used, however, the intensity of the bands was noticeably weaker (Fig. 2 lanes 9–13). We also observed similar protein/DNA complexes using a probe overlapping a Wnt1 enhancer element (WEE) that was previously shown to contain a consensus Six3 binding motif20,21 (Fig. 2 lanes 14–17). Interestingly, other than the enrichment of AT sequence, the Six3 binding site in the WEE (ATTA) showed little resemblance to the one identified in SBE2 (AACTCATTTT). These results confirm the existence of a unique Six3/6 binding site in SBE2.
Figure 2. Six3/6 proteins bind directly to SBE2.
EMSAs performed with Cos-1 cell extracts transfected with flag-tagged Six3 (lanes 3–5,10,11) or Six6 (lanes 6–8,12,13) expression vectors and incubated with SBE2(C) (lanes 1–8), SBE2(T) (9–13) or WEE (14–17) radiolabeled probes. Specific protein/DNA complexes were supershifted in the presence of an αFlag antibody (lanes 4,7,15,17), but not a nonspecific antibody (lanes 5,8). Note that in addition to the supershift, incubation with the αFlag antibody also disrupted Six3/6-SBE2 and Six3/6-WEE complex formation (lanes 4,7,11,13,15,17). The asterisk indicates the formation of a nonspecific complex that is more effectively competed away in the presence of Six3/6 and high affinity probes.
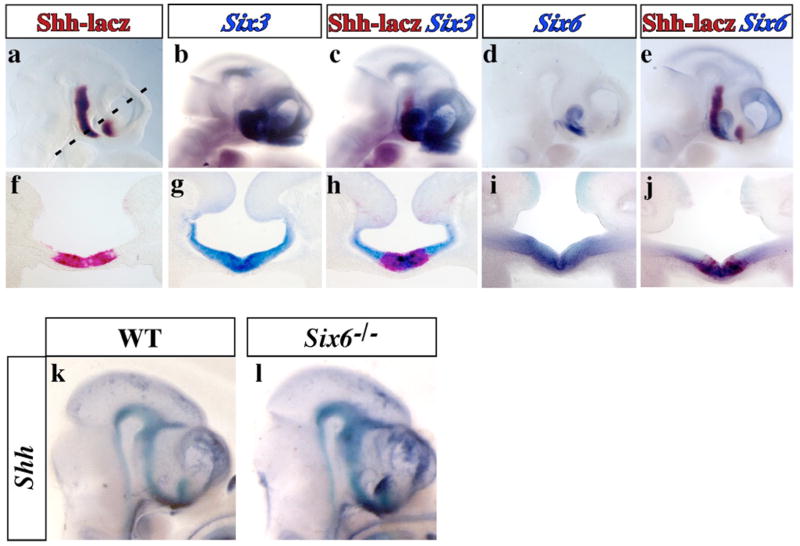
Six3 and Six6 show dynamic patterns of expression in the developing forebrain18,22. For Six3 and/or Six6 to be considered direct regulators of Shh, their spatial and temporal expression profiles should overlap. In comparing Six3 and Six6 expression with SBE2 dependent Shh-like reporter activity in mouse embryos at E10.5, we noted that in the ventral forebrain, from the level of the optic vesicles to the rostral extent of the diencephalon, Shh-lacZ was embedded within the Six3 and Six6 expression domains (Fig. 3). Both Six3 and Six6 showed a broad distribution throughout the ventral portion of the anterior hypothalamus, whereas Shh-lacZ expression was restricted medially within the Six3/6 domains (Fig. 3f-j). Similar observations were made at E9.5 (data not shown). Interestingly, the region of overlap between Shh-lacZ and Six3/6 is precisely where mutant SBE2(T) reporter activity was diminished (Fig. 1b).
Figure 3. Overlap of Shh and Six3/Six6 expression in the ventral diencephalon.
Whole mount (a–e) and transverse sections (f–j) showing the colocalization of Shh (salmon-gal), Six3 and Six6 (alkaline phosphatase) expression in the mouse embryonic forebrain at E10.5. The expression of Shh was monitored using a lacZ reporter line (447L17βlacZ) that recapitulates endogenous Shh expression in the hypothalamus in an SBE2 dependent manner13. (k-l) Shh expression (alkaline phosphatase) in wild type and Six6−/− embryos at E10.5.
We next determined whether Six6 is required to regulate Shh expression. Six6−/− mouse embryos show reduced proliferation of retinal and pituitary progenitors but do not show overt signs of HPE23. Consistent with this milder phenotype, Shh expression was unaffected in the ventral forebrain of Six6−/− embryos (Fig. 3k-l). In contrast, Six3−/− mouse embryos display severe forebrain truncations including rostral regions of the diencephalon20. Moreover, the combination of findings that mutations in SIX3 cause HPE in humans19, and mouse embryos carrying a knock-in allele of an HPE causing point mutation in Six3 show reduced Shh expression in the forebrain (X. Geng and G. Oliver, unpublished observations), are consistent with an essential role for Six3 in the direct regulation of Shh transcription.
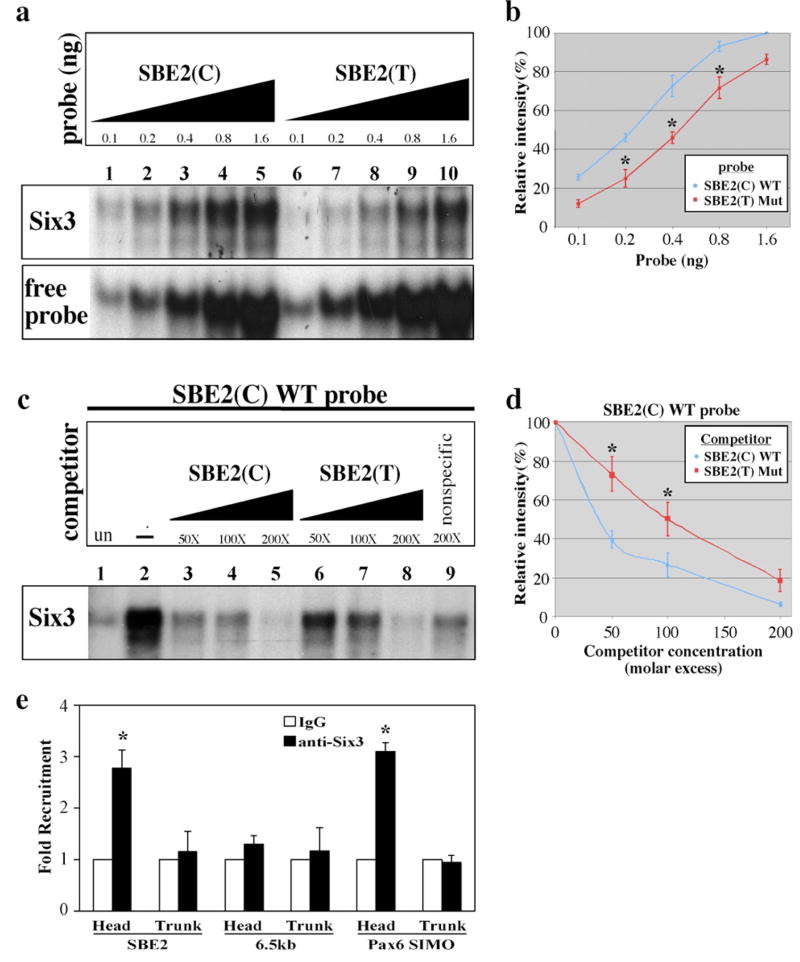
Our finding that the DNA sequence containing SBE2(C) functions as a Six3/6 binding site raised the possibility that the SBE2(T) variant interferes with the recruitment of Six3 to this site. To test this hypothesis, we evaluated the affinity of Six3 for SBE2(C) compared to SBE2(T). A dose response curve was generated by varying the amount of radiolabeled probe exposed to a constant amount of Six3 protein and quantifying band intensity as a measure of Six3/SBE2 complex formation. SBE2(C) consistently showed a stronger association with Six3, compared to SBE2(T), at all doses of probe tested (Fig. 4a, b). Similar results were observed with Six6 (data not shown).
Figure 4. Six3/Six6 proteins bind SBE2(C) with higher affinity than SBE2(T).
(a) EMSAs performed with increasing amounts of radiolabeled SBE2(C) (lanes 1–5) and SBE2(T) (lanes 6–10) probes incubated with Cos-1 cell lysates transfected with pCDNA3-Six3-Flag. (b) Dose responsive curves for data shown in (a). Each point along the curve is the average band intensity from three independent experiments (*p<0.05, Student’s t-test). (c) Competitive EMSAs showing the binding of Six3 to a radiolabeled SBE2(C) probe. Cos-1 cell lysates transfected with pCDNA3-Flag (lane 1) or pCDNA3-Six3-Flag (lanes 2–8) were analyzed for binding to a 35 bp probe overlapping wild type SBE2(C). Increasing concentrations of cold wild type SBE2(C) competitor (lanes 3–5), was more efficient at displacing radiolabeled SBE2(C) probe from Six3, compared to increasing concentrations of cold SBE2(T) competitor (lanes 6–8). A nonspecific probe (lane 9) did not significantly alter the shifted complex. (d) Graphical representation of the data in (a). The relative intensities of the retarded bands were quantified and plotted against competitor concentration. Each data point on the curve is an average of five independent experiments. At sub-saturating concentrations of competitor (50x, 100x), SBE2(C) (blue line) was significantly more effective at interfering with complex formation than SBE2(T) (red line) (*p<0.05, Student’s t-test). (e) ChIP from embryos using anti-Six3 or anti-IgG antibodies. QPCR results from three independent experiments reveal a significant enrichment of SBE2 DNA in Six3 versus IgG bound chromatin isolated from forebrain but not posterior trunk regions of E8.75 mouse embryos(*p<0.01, Student’s t-test). A negative control sequence, 6.5 kb downstream of SBE2, was not enriched in Six3 bound chromatin, whereas a positive control sequence, Pax6 SIMO, was enriched to a similar degree as SBE2.
We also quantified the amount of radiolabeled probe displaced from Six3 in the presence of increasing amounts of unlabeled (cold) competitor. In the presence of 50 and 100 fold excess wild type SBE2(C) unlabeled competitor, the majority, 60% and 75% respectively, of the radiolabeled SBE2(C) probe was displaced from Six3 (Fig. 4c lanes 2–4 and Fig. 4d). In comparison, when 50 and 100 fold excess SBE2(T) unlabeled competitor was introduced, significantly less, 27% and 50% respectively, of the radiolabled SBE2(C) probe was displaced from Six3 (Fig. 4c lanes 2,6,7 and Fig. 4d). Similar results were obtained when SBE2(T) was used as the radiolabeled probe (data not shown). These results indicate that the SBE2(T) variant weakens the affinity of Six3 binding by approximately two-fold in relation to SBE2(C) (Fig. 4b, d). In agreement with this conclusion is the additional observation that Six3 dependent activation of SBE2(T)-lacZ expression in Cos-1 cells was significantly reduced compared to SBE2(C)-lacZ (Fig. 5b).
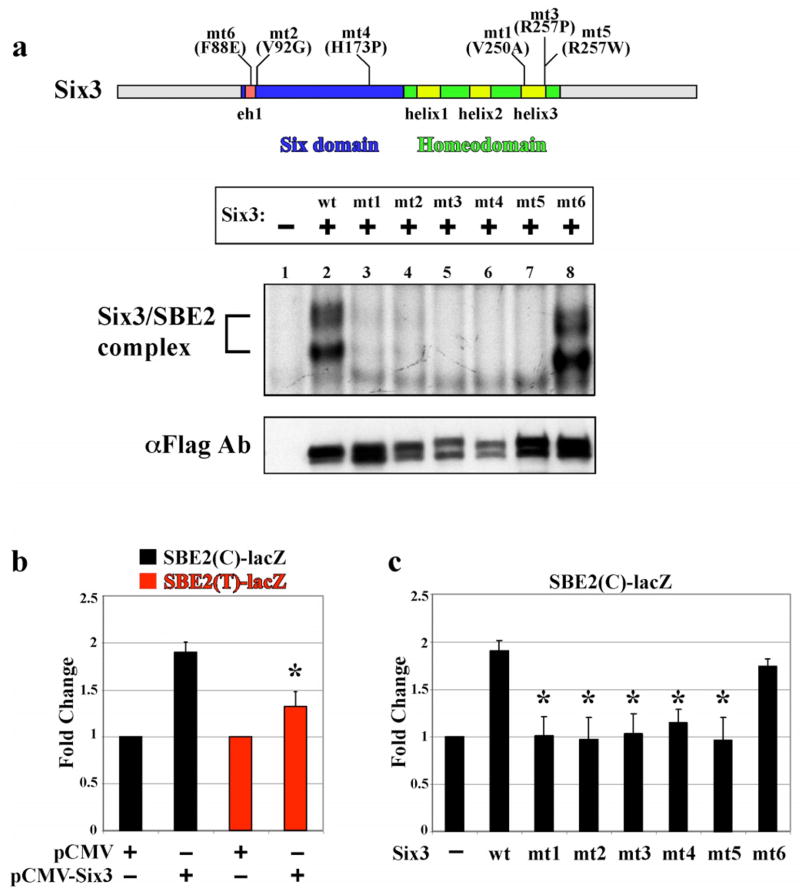
Figure 5. HPE causing mutations in Six3 alter binding and activation of SBE2.
(a) Top: Schematic of Six3 protein showing the location of amino acid substitutions resulting from five different HPE causing point mutations (mt1-5) in either the six domain or homeodomain. An additional mutation (mt6) in the groucho interaction domain interferes with Six3 repressor activity21 but was not identified in HPE patients. Bottom: Cell lysates transfected with pCDNA3-Flag (Lane1), pCDNA3 (wt)Six3-Flag (Lane 2), or pCDNA3 (mt1-6)Six3-Flag (Lanes 3–8) were analyzed for binding to a 35 bp probe overlapping wild type SBE2(C). The Six3/SBE2 complex is indicated with a bracket. Weak or no complex formation was observed for mt1,2 (lanes 3–4) and mt3–5 (lanes 5–7), respectively. Whereas, DNA binding activity was retained by mt6 (lanes 8). αFlag immunoblot demonstrates that wild type and mutant Six3 proteins were expressed at equivalent levels. (b) Six3 regulates SBE2 activity in Cos-1 cells. pCMV-Six3 stimulated wild type SBE2(C)-lacZ expression (black bars), compared to the empty expression plasmid. This transcriptional activation by Six3 was attenuated in cells expressing SBE2(T)-lacZ (red bars). Each bar represents an average of six replicates. (c) SBE2(C)-lacZ activation is compromised by mutations in Six3. Wild type and F88E (mt6) forms of Six3 activated reporter expression while the other Six3 mutants (mt1-5) showed reduced capacity to stimulate SBE2 (black bars). Each bar represents an average of three replicates. Asterisk indicates significant difference from wild type (p<0.001).
Six3 binding to SBE2(C) was also confirmed in vivo, by chromatin immunoprecipitation (ChIP) experiments. SBE2 was significantly enriched in Six3 bound chromatin isolated from forebrain compared to posterior trunk regions of E8.75 mouse embryos (Fig. 4e). A control sequence, 6.5 kb downstream of SBE2, was not enriched in Six3 bound chromatin. The extent of Six3 binding to SBE2 is comparable to that of another Six3 target sequence identified in the Pax6 SIMO lens enhancer26 (Fig. 4e). Taken together, these data are consistent with our hypothesis that Six3 is a direct regulator of SBE2 activity, and that the SBE2(T) variant compromises the recruitment of Six3 and subsequent activation of Shh transcription. While our data suggest that the SBE2(T) variant may cause HPE, formal proof of this claim must await the evaluation of mice carrying a targeted knock-in of SBE2(T) into the mouse genome.
Six3 functions as a context dependent activator or repressor of target gene expression in the developing eye and forebrain20,21,24–26. In addition, Six3 promotes the proliferation of forebrain progenitors by antagonizing Geminin, a DNA replication inhibitor27. This aspect of Six3 function is independent of its DNA binding properties. Hence, the mechanism by which individuals carrying mutations in SIX3 develop HPE may be due to heightened Geminin function, improper regulation of target gene expression, or both. To determine whether HPE causing mutations in Six3 impairs their ability to bind SBE2 we performed EMSAs. Cos-1 cell extracts expressing equivalent amounts of wild type or mutant forms of mouse Six3 were incubated with radiolabeled SBE2(C) probe (Fig. 5). Three independent mutations in the homeodomain either reduced (V250A), or prevented (R257P, R257W), Six3/SBE2 complex formation (Fig. 5 lanes 2,3,5,7). Unexpectedly, two of three mutations in the six domain also showed greatly reduced (V92G), or absent (H173P), Six3/SBE2 complex formation (Fig. 5 lanes 2,4,6). Only the F88E mutation in the Groucho interaction domain retained the ability to bind SBE2 (Fig. 5 lanes 2,8). The Six3 mutants that failed to bind SBE2(C) were also impaired in their ability to stimulate SBE2(C)-lacZ expression in Cos-1 cells (Fig. 5c). How mutations in the six domain interfere with the DNA binding properties of Six3 is unclear, but may indicate a previously unappreciated interaction between the six domain and the homeodomain. Based on these results, we deem it likely that the mechanism by which HPE manifests in individuals carrying point mutations in SIX3 is due, in part, to a failure in the binding of SHH regulatory sequences and subsequent activation of SHH transcription in the ventral forebrain.
Results from this study provide a better understanding of the transcriptional control mechanisms regulating SHH expression during normal forebrain development and in the pathogenesis of HPE. Our data suggest that Six3 is a direct regulator of Shh expression in the anterior diencephalon. Moreover, the approaches taken in assigning function to a putative HPE causing variant in a remote SHH regulatory element should be generally applicable for studying the growing number of rare, as well as common regulatory SNPs that modulate gene expression levels in normal and disease states28–30.
Materials and methods
Sequence analysis of SBE2
The genomic DNA from 474 sporadic and familial HPE patients registered at the National Institutes of Health (NIH) and 450 normal controls were screened for mutations in SBE2. The genomic DNA was extracted from either lymphocytes or established lymphoblastoid cell lines by routine methods. All samples were obtained by informed consent according to the guidelines of the NHGRI Institutional Review Board.
PCR methods and direct DNA sequence
One pair of primers was designed to amplify the 1.1 kb SBE2 region: FBE-F1 5′ GCC TAG CGT TTC CAA CAT GCA GCC 3′ and FBE-R1 5′ TAC GGC TCT AAC AGT AAA GCA CTC 3′. Sequencing was performed using five internal primers (available upon request) allowing sequence reads of both strands. Amplification of patient genomic DNA was performed in a 35μl reaction volume, using 60–100ng DNA template, 3.5μl of 10× PCR Amplification Buffer (Invitrogen, CA), 1.75μl of PCR Enhancer solution (Invitrogen, CA), 1μl of 50mM MgSO4 (Invitrogen, CA), 0.3μl of 25mM dNTP stock mixture (Amersham Biosciences, NJ), 1μl of each 20 pmol primer (Invitrogen, CA) and 0.5μl of AmpliTaq 5U/μl (Applied Biosystems, CA). The PCR cycling parameters used for amplification were 95°C for 4 min followed for 30 cycles at 95°C for 30 sec, annealing at 60°C for 30 sec, 72°C for 1 min and a final extension of 72°C for 7 min. Direct DNA sequencing was performed using the Big Dye™ terminator cycle sequencing kit 3.1 (Applied Biosystems, CA) and the reactions were analyzed on an ABI 3100 Genetic Analyzer.
DNA affinity capture assay
500 ul of brain nuclear extracts (2 ug/ul) prepared from adult mice (Sigma NuCLEAR Extraction kit) were preincubated for 20 min at 4°C in 300 ul of binding buffer (10mM Tris-Cl pH 7.4, 50mM NaCl, 1mM EDTA, 1mM DTT and 5% glycerol), 25 ul of dIdC (200 ng/ul), 25 ml of BSA (1ug/ul), and 550 ul of H20. The extracts were precleaned with 100 ul of a 50% slurry of streptavidin agarose beads (Invitrogen) at 4°C for 30 min with rotation, centrifuged at high speed for 30 sec and transferred (supernatant) into a new tube. A double-stranded DNA oligonucleotide (18mer) overlapping a conserved 10 bp segment of SBE2 (5′-biotin GCCTAATTCATTTTTCCA-3′) was synthesized with a terminal 5′ biotin modification (Invitrogen) and incubated with the brain nuclear extracts for 3 hrs at 4°C with rotation. An SBE2 mismatch oligo (5′-biotin GCCAATCTCTTATTTCCA-3′ was also used in a parallel experiment as a negative control for nonspecific binding proteins. 50 ul of a 50% slurry of streptavidin agarose beads was added to the mixture for 30 min at 4°C with rotation. The beads were centrifuged, washed twice in binding buffer, twice in wash buffer (10mM Tris-Cl pH 7.4, 100mM NaCl, 1mM EDTA, 1mM DTT, 0.1% NP40) and twice in PBS. The beads were stored at −20°C until trypsin digestion and subjected to reversed-phase liquid chromatography/tandem mass spectrometry analysis at the University of Pennsylvania proteomics core facility. The raw mass spectrometry data were submitted to Bioworks Browser (Thermo Electron, San Jose, CA) and batch searched through TurboSEQUEST™ against an indexed mouse RefSeq database (version updated 12/04). (details available upon request).
Electromobiliy Shift Assays (EMSA)
pCDNA3-Flag, pCDNA3-Six3-Flag, pCDNA3-(mt1-6)-Six3-Flag and pCDNA3-Six6-Flag plasmids were transfected into Cos-1 cells using FuGENE 6 transfection reagent (Roche). After 48 hours, whole cell lysates were prepared in a buffer containing 50 mM Tris-Hcl, pH 7.4, 1 mM EDTA, 1 mM DTT, protease inhibitor cocktail and 25% glycerol. For EMSA, 10 μg of protein from the cell lysates was incubated for 10 min at room temperature in a DNA binding buffer containing 10 mM Tris-HCl, pH7.4, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 5% glycerol, 200 ng poly(dI-dC), 1 μg BSA in the presence or absence of competitor double stranded oligonucleotides. After 0.1 ng (5×104 to 10×104 cpm) of probe was added to the mixture, incubation was continued for an additional 20 minutes. Supershifts were performed by incubating protein/DNA mixtures with 0.5ul of mouse monoclonal αFlag M2 antibody (F3165; Sigma-Aldrich, St. Louis, MO), or rabbit polyclonal αSix3 antibody (G. Oliver, St. Jude Children’s Hospital) for 5 min prior to gel loading. Protein-bound DNA complex was separated from free probe on a 6.5% acrylamide gel run in 1× TBE (Tris-borate-EDTA) buffer. The nucleotide sequence of the sense strand of probes and competitors is as follows: SBE2(C): 5′-AAGTAGGCCGAACTCATTTTCCACACACAG-3′, SBE2(T): 5′-AAGTAGGCCGAATTCATTTTCCACACACAG-3′, WEE: 5′-GACTAGCACATCTAATGATAAGCACAGGTTGA-3′. Competitive EMSAs were performed by incubating protein/DNA complexes with 50, 100 or 200 molar excess of cold probe. After overnight exposure, autoradiographs were scanned and the intensity of individual bands corresponding to the protein/DNA complex of interest was quantified using NIH ImageJ. Values were plotted as the ratio (percentage) of band intensities in the presence and absence of specific competitor and compared using the Student’s t-test.
Transient transfection and dual reporter assay
COS-1 cells were seeded at 50–70% confluence, and transfection was performed using Fugene 6 (Roche Applied Science) according to the manufacture’s instructions. 20 ng of pRL-TK vector (Promega), which contains the Renilla luciferase gene as a transfection efficiency control, and 500 ng of SBE2(C) or SBE2(T) LacZ reporter plasmids, were mixed with 125 ng of empty pCMVor pCMV-Six3, or pCMV-mutant Six3 (mt1-mt6). Lysates were prepared 36 h after transfection by adding 100 μL of lysis solution (Dual-Light System; Applied Biosystems). β-galactosidase activity was determined by the accumulated product of Galacton-Plus substrate reaction (Applied Biosystems), and was normalized to that produced by Renilla luciferase. Enhancer activity was expressed as fold induction relative to that of cells transfected with the empty pCMV vector. At least three independent experiments were performed for each construct.
Chromatin Immunoprecipitation
The chromatin immunoprecipitation (ChIP) assay followed a modified version of a previously described protocol31. Briefly, pooled embryos at the 13–17 somite stage were fixed with 1% formaldehyde for 15 minutes with shaking. After a 5 min incubation with 100 mM glycine, heads and trunks were separated and disrupted in lysis buffer (20 mM Tris-HCl, pH7.5, 1% Triton X-100, 1mM EDTA) and protease inhibitor cocktail (Sigma) by passing through 30G needles. Chromatin was sonicated and diluted with 20mM Tris-HCl, pH7.5, 140mM NaCl, 0.5% Triton X-100, and protease inhibitor cocktail. After preclearing with protein A agarose beads (Upstate), the chromatin was incubated overnight with 3 ul of anti-Six3 (Rockland) or anti-IgG (Santacruz) antibodies, followed by incubation with protein A agarose beads, and washed with 20mM Tris-HCl, pH7.5, 140 mM NaCl and 0.5 % Triton X-100. After elution with 20 mM Tris-HCl, pH7.5, 10 mM EDTA and 1% SDS and decrosslinking, DNA was purified with QIAquick kit (Qiagen) and subject to quantitative PCR using the following primer pairs: SBE2:
(F)5′CAGCTCTCCAAAATTACTGCC3′,
(R)5′CTAAAAGCAGGGGATCAGATG 3′.
6.5 kb 3′ of SBE2: (F) 5′ CACATCAGCATCCTAGCCTAC 3′,
(R)5′ GGTACATTTCTTGTAGCTTCG 3′.
Pax6 SIMO: (F)5′ TCTCTGTGTCATTCCTAATGCACTT 3′,
(R)5′ TCCCAAGATAAACTTTCCCATTG-3′.
QPCR was performed using Brilliant SYBR Green QPCR Master Mix (Stratagene, Cat no.600548) on an Mx4000 instrument (Stratagene). Each reaction contained 0.6 ul of a 1:1600 diluted reference dye, 2 ul of chromatin DNA, 1 ul of 2uM primer A, 1 ul of 2uM primer B and 10 ul of 2x Master Mix in a final reaction volume of 20 ul. PCRs were amplified for 1 cycle at 95°C for 10min and 40 cycles at 95°C for 30sec, 57°C for 1min and 72°C for 30 sec. PCRs of three independent replicates were each performed in triplicate. Differences in threshold cycle (Ct) number were used to quantify relative amounts of target DNA template. Ct number for each chromatin sample was normalized to Ct number for input PCR. Relative enrichment of target chromatin DNA was determined by 2−(Ct1-Ct2), assuming that one Ct number difference represents a two-fold difference in the amount of starting template.
Plasmid Construction
SBE2 reporter constructs were cloned into a vector containing the Shh minimal promoter, lacZ gene and SV40 poly(A) signal. A construct harboring a deletion of the 10 bp sequence (AACTCATTTT) from human SBE2 was generated by ligating two PCR products flanking the region of interest that were amplified with the following primer pairs: E311 (5′-ATTAGCGGCCGCCGAGCAGGCTAACCTGGAGGCCAC-3′) and E315 (5′-ATTATACGTACGGCCTACTTGAGTTTTTCCTTC-3′); E313 (5′-ATTAGGATCCCTCCCATTCATTCCTTTCTCCCTC-3′) and E316 (5′-ATTATACGTACCACACACAGAGAGATAATTG-3′). The cloning of full length human Six3 and Six6 cDNAs into the pCDNA3-Flag expression vector was described previously22.
Production and genotyping of transgenic mice
Transgenes were prepared for microinjection as described8. Transient transgenic embryos were generated by pronuclear injection into fertilized eggs derived from the (BL6×SJL) F1 mouse strain (Jackson Labs). The generation of Six6−/− embryos was described previously20.
Whole-mount β-galactosidase staining and in situ hybridization
The assessment of β-galactosidase activity was detected in whole-mount embryos by using X-gal (Sigma) or Salmon-gal (Biosynth) as substrates. Whole-mount RNA in situ hybridization was performed using digoxygenin-UTP-labeled riboprobes against Shh, Six3, and Six6 according to a previously described protocol13. For double labeling experiments, embryos were initially fixed for 60 min in 4% paraformaldehyde, stained in salmon-gal substrate for 2 hours then post-fixed overnight in 4% paraformaldehyde. Whole-mount in situ hybridization was then performed essentially as described13. Stained embryos were photographed after dehydration in methanol and clearing in benzyl alcohol:benzyl benzoate (1:1). Representative embryos were rehydrated, sunk in 30% sucrose overnight, embedded and frozen in OCT and sectioned at 20 μm on a cryostat.
Acknowledgments
We thank the families for their participation in these studies. We also thank Dr. Jean Richa and his staff at the University of Pennsylvania Transgenic and Mouse Chimeric Facility for their assistance in transgenic mouse production and Drs. Vivian Cheung, Dan Kessler and Tom Kadesch for helpful comments on the manuscript. We are grateful to Kathy Ewens and Wendy Ankener (R. Spielman lab) for the control human genotyping data and Dr. P. Bovolenta (Instituto Cajal, CSIC, Madrid, Spain) for kindly providing the human Six3 and Six6 expression constructs. This work was supported by NIH grants R01 NS39421 from NINDS (DJE), R01 NS052386 (GO), March of Dimes grant #1-FY05-112 (DJE), a Pew Scholar Award in the Biomedical Sciences (DJE), Cancer Center Support CA-21765 (GO), the American Lebanese Syrian Associated Charities (ALSAC) (GO) and the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health (MM).
References
- 1.Chiang C, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 2.Britto J, Tannahill D, Keynes R. A critical role for sonic hedgehog signaling in the early expansion of the developing brain. Nat Neurosci. 2002;5:103–10. doi: 10.1038/nn797. [DOI] [PubMed] [Google Scholar]
- 3.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–51. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcucio RS, Cordero DR, Hu D, Helms JA. Molecular interactions coordinating the development of the forebrain and face. Dev Biol. 2005;284:48–61. doi: 10.1016/j.ydbio.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Roessler E, et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- 6.Helms JA, Cordero D, Tapadia MD. New insights into craniofacial morphogenesis. Development. 2005;132:851–61. doi: 10.1242/dev.01705. [DOI] [PubMed] [Google Scholar]
- 7.Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–83. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- 8.Shimamura K, Rubenstein JL. Inductive interactions direct early regionalization of the mouse forebrain. Development. 1997;124:2709–18. doi: 10.1242/dev.124.14.2709. [DOI] [PubMed] [Google Scholar]
- 9.Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130:1749–58. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- 10.Fuccillo M, Rallu M, McMahon AP, Fishell G. Temporal requirement for hedgehog signaling in ventral telencephalic patterning. Development. 2004;131:5031–40. doi: 10.1242/dev.01349. [DOI] [PubMed] [Google Scholar]
- 11.Cordero D, et al. Temporal perturbations in sonic hedgehog signaling elicit the spectrum of holoprosencephaly phenotypes. J Clin Invest. 2004;114:485–94. doi: 10.1172/JCI19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubourg C, et al. Holoprosencephaly. Orphanet J Rare Dis. 2007;2:8. doi: 10.1186/1750-1172-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong Y, El-Jaick K, Roessler E, Muenke M, Epstein DJ. A functional screen for Sonic hedgehog regulatory elements across a 1 Mb interval identifies long range ventral forebrain enhancers. Development. 2006;133:761–772. doi: 10.1242/dev.02239. [DOI] [PubMed] [Google Scholar]
- 14.Topol EJ, Frazer KA. The resequencing imperative. Nat Genet. 2007;39:439–40. doi: 10.1038/ng0407-439. [DOI] [PubMed] [Google Scholar]
- 15.Treier M, et al. Hedgehog signaling is required for pituitary gland development. Development. 2001;128:377–86. doi: 10.1242/dev.128.3.377. [DOI] [PubMed] [Google Scholar]
- 16.Roessler E, et al. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc Natl Acad Sci U S A. 2003;100:13424–9. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SS, Ko BJ, Kim BG. Mass spectrometric screening of transcriptional regulators using DNA affinity capture assay. Anal Biochem. 2005;344:152–4. doi: 10.1016/j.ab.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 18.Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev. 1999;84:31–40. doi: 10.1016/s0925-4773(99)00068-4. [DOI] [PubMed] [Google Scholar]
- 19.Wallis DE, et al. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–8. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- 20.Lagutin OV, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–79. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu C, et al. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development. 2002;129:2835–2849. doi: 10.1242/dev.129.12.2835. [DOI] [PubMed] [Google Scholar]
- 22.Conte I, Morcillo J, Bovolenta P. Comparative analysis of Six 3 and Six 6 distribution in the developing and adult mouse brain. Dev Dyn. 2005;234:718–25. doi: 10.1002/dvdy.20463. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–3. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- 24.Carl M, Loosli F, Wittbrodt J. Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye. Development. 2002;129:4057–63. doi: 10.1242/dev.129.17.4057. [DOI] [PubMed] [Google Scholar]
- 25.Lopez-Rios J, Tessmar K, Loosli F, Wittbrodt J, Bovolenta P. Six3 and Six6 activity is modulated by members of the groucho family. Development. 2003;130:185–95. doi: 10.1242/dev.00185. [DOI] [PubMed] [Google Scholar]
- 26.Liu W, Lagutin OV, Mende M, Streit A, Oliver G. Six3 activation of Pax6 expression is essential for mammalian lens induction and specification. EMBO J. 2006;25:5383–95. doi: 10.1038/sj.emboj.7601398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–9. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- 28.Cheung VG, et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–9. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emison ES, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–63. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 30.Haiman CA, et al. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cawley S, et al. Unbiased Mapping of Transcription Factor Binding Sites along Human Chromosomes 21 and 22 Points to Widespread Regulation of Noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]