Abstract
We generated mutant alleles of Drosophila melanogaster in which expression of the linker histone H1 can be down-regulated over a wide range by RNAi. When the H1 protein level is reduced to ∼20% of the level in wild-type larvae, lethality occurs in the late larval – pupal stages of development. Here we show that H1 has an important function in gene regulation within or near heterochromatin. It is a strong dominant suppressor of position effect variegation (PEV). Similar to other suppressors of PEV, H1 is simultaneously involved in both the repression of euchromatic genes brought to the vicinity of pericentric heterochromatin and the activation of heterochromatic genes that depend on their pericentric localization for maximal transcriptional activity. Studies of H1-depleted salivary gland polytene chromosomes show that H1 participates in several fundamental aspects of chromosome structure and function. First, H1 is required for heterochromatin structural integrity and the deposition or maintenance of major pericentric heterochromatin-associated histone marks, including H3K9Me2 and H4K20Me2. Second, H1 also plays an unexpected role in the alignment of endoreplicated sister chromatids. Finally, H1 is essential for organization of pericentric regions of all polytene chromosomes into a single chromocenter. Thus, linker histone H1 is essential in Drosophila and plays a fundamental role in the architecture and activity of chromosomes in vivo.
Keywords: Linker histone H1, heterochromatin, histone methylation, polytene chromosomes, chromocenter, position effect variegation
The genomes of eukaryotes are packaged into a highly compact nucleoprotein complex called chromatin. The histones constitute a family of proteins that are intimately involved in organizing chromatin structure. There are five major classes of histones: the core histones H2A, H2B, H3, and H4, and the linker histones usually referred to as H1. The nucleosome core particle is the highly conserved repetitive unit of chromatin organization. It consists of an octamer of the four core histones around which ∼145 base pairs (bp) of DNA are wrapped and protected from nuclease digestion (Van Holde 1988; Wolffe 1998). The linker histone H1 binds to core particles and protects an additional ∼20 bp of DNA (linker DNA). In metazoans, the abundance of linker histones, although variable during development, approaches that of core histones (Woodcock et al. 2006), suggesting that they play an important role in establishing and maintaining the structure of the chromatin fiber.
Much of our knowledge about the roles of linker histones comes from in vitro studies. These studies indicate that two principal functions of linker histones are to stabilize the DNA entering and exiting the core particle and to facilitate the folding of nucleosome arrays into more compact structures (Ramakrishnan 1997; Wolffe 1997). H1 also affects nucleosome core particle spacing and mobility. In vitro studies also suggest that H1 acts primarily as a transcriptional repressor (Laybourn and Kadonaga 1991; Brown et al. 1996). Recent attempts to study the functions of linker histones in vivo have used gene inactivation approaches. Elimination of the linker histone-like protein Hho1p in Saccharomyces cerevisiae did not cause any major phenotypic effects, nor were any perturbations in chromosome structure apparent (Ushinsky et al. 1997; Patterton et al. 1998; Hellauer et al. 2001; Downs et al. 2003). This finding may be explained by the much lower abundance of Hho1p in yeast (1:37 core particles). The absence of any phenotypic effects was also reported upon elimination of H1 from Aspergillus nidulans, which has a more typical linker histone (Ramon et al. 2000). H1 also was found to be dispensable for growth and viability of Tetrahymena (Shen et al. 1995). However, its elimination by gene inactivation led to partially decondensed chromatin, supporting an in vivo role for H1 in chromatin folding.
Mammals express at least eight nonallelic H1 subtypes that differ in their expression during development. Although none of the eight individual subtypes appear to be essential, mouse embryos in which the stoichiometry of H1 to the core particles has been reduced ∼50% by inactivation of three of the five somatic linker histone subtype genes die at midgestation. This observation indicates that the total amount of linker histones is important for normal mammalian development (Fan et al. 2003). In the knockout embryos and embryonic stem (ES) cells derived from them, H1 is an important determinant of nucleosome spacing and of local chromatin folding in vivo (Fan et al. 2005). However, despite these advances, the role of H1 in higher-order chromatin folding and long-range chromosome structure remains enigmatic.
Further advances in our understanding of linker histone functions would be greatly facilitated by studies in a genetically tractable organism where H1 may prove to play an essential role. As mentioned, deletion of the yeast HHO1 gene does not lead to obvious phenotypic effects. Although linker histones are essential for embryonic development in mice, the existence of multiple, nonallelic mouse H1 variant genes, as well as apparent compensatory gene expression mechanisms within this gene family, has hindered attempts to study the effects of decreasing H1 expression in this species (Fan et al. 2005). On the other hand, Drosophila offers an attractive alternative for such studies because it contains a single linker histone protein. However, it is encoded by ∼100 copies of the H1 gene positioned within a tandemly repeated array of five sequentially arranged histone genes (Lifton et al. 1978). Thus, specific elimination of Drosophila H1 histone by classical genetic approaches is not feasible. Here we describe the use of specific RNAi to nearly completely deplete H1 in Drosophila in vivo. This approach has allowed us to demonstrate that the H1 linker histone is (1) an essential protein in Drosophila, (2) a major determinant of heterochromatin formation and function, and (3) an important biochemical component of the machinery that maintains sister chromatid alignment in Drosophila polytene chromosomes.
Results
Linker histone H1 is essential for Drosophila development
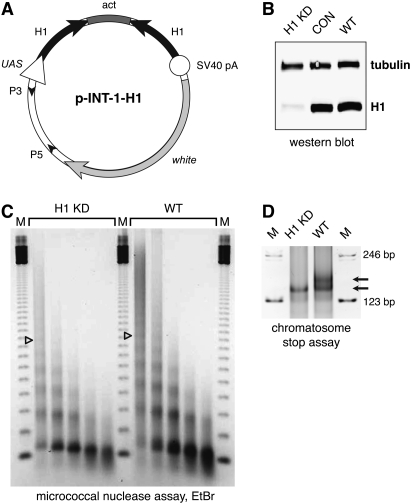
To deplete H1 histone in vivo in Drosophila, we used RNAi. The H1 RNAi expression vector is depicted in Figure 1A. It was constructed by inserting PCR fragments encompassing the first 600 bp of the Drosophila melanogaster H1 coding sequence (encoding the first 200 amino acids of the 256-amino-acid full-length H1 protein) in opposite orientations on both sides of the first intron of the actin 5C gene in pINT-1 (Wei et al. 2007). pINT-1 is a modified pUAST vector in which transcription is driven by the GAL4-responsive UAS promoter. We prepared six individual pINT-1-H1 transgenic lines, and homozygous transgenic flies were mated at various temperatures to counterparts of the opposite sex that carry the Tubulin-GAL4 driver (Tub∷GAL4/TM3, Sb) (see the Materials and Methods). Another pINT-1 transgenic line (pINT-1-Nau) (Wei et al. 2007) encoding a dsRNA for D. melanogaster Nautilus protein was used as a control. In the adult offspring of the experimental cross at 29°C, we did not observe flies that carry both the pINT-1-H1 and Tubulin-GAL4 transgenes, and we observed few offspring at 26°C. At the same time, expected numbers of flies with pINT-1-Nau and Tubulin-GAL4 transgenes were obtained at both temperatures (Table 1). Thus, the expression of H1-specific dsRNA under control of the ubiquitous tubulin promoter causes lethality before eclosion.
Figure 1.
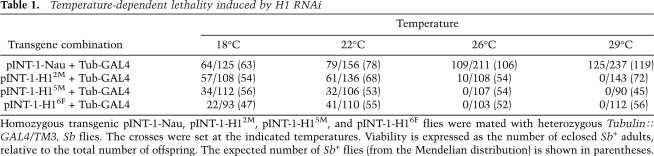
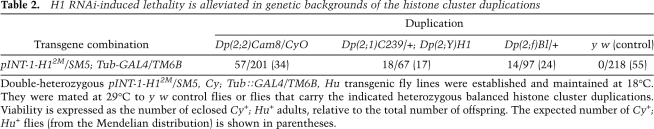
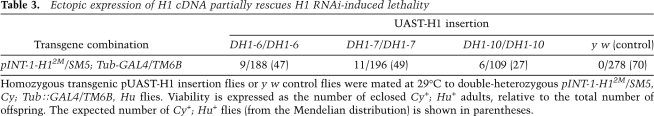
Depletion of histone H1 in Drosophila. (A) Transgenic RNAi construct for sequence-specific post-transcriptional silencing of the Drosophila linker histone H1 gene. Two identical fragments of the H1 coding sequence (the first 600 bp) were inserted in opposite orientations on both sides of the first intron of the actin 5C gene in the pINT-1 vector. The expression of H1-specific dsRNA is driven by the GAL4-responsive UAS promoter. (Open triangle) UAS promoter; (black arrows) H1 cDNA fragments; (dark-gray box) act5C intron; (open circle) SV40 polyadenylation site; (light arrow) white gene; (small black arrowheads) P-element sequences. (B) The expression of H1 protein in the third instar (L3) larval salivary glands is abrogated by RNAi. H1 synthesis was inhibited by H1-specific dsRNA transcribed from p-INT-1-H15M transgene driven by the GAL4 transactivator encoded by the Tubulin-GAL4 transgene at 29°C (from the beginning of embryonic development to L3). H1 protein levels in salivary gland lysates of H1-depleted animals (H1 KD) were compared with those in salivary glands of the wild-type (WT) and pINT-1-Nautilus (CON) animals by immunoblotting using an antiserum against Drosophila histone H1. Protein loading was controlled by immunoblotting for tubulin. (C) Nucleosome repeat length (NRL) is reduced in L3 larval chromatin upon depletion of H1 by RNAi. H1 synthesis was inhibited by H1-specific dsRNA as described in B. Nuclei from L3 larvae were subjected to partial micrococcal nuclease digestion, and the DNA was analyzed by agarose gel electrophoresis and EtBr staining. The NRL in knockdown animals (H1 KD) was calculated to be ∼176 bp compared with ∼188 bp in the wild-type control (WT). Open triangles indicate the positions of the hexanucleosome bands in each sample. (M) 123-bp DNA ladder. (D) Chromatosome particles are depleted in the chromatin of H1 knockdown larvae. Chromatin from H1 knockdown (H1 KD) and wild-type (WT) larvae was prepared and digested extensively with MNase as in C. The DNA was analyzed by native PAGE and EtBr staining. In H1 knockdown larvae, the chromatosome band (top arrow) is substantially depleted in comparison with the wild-type control larvae. (Bottom arrow) Core particle; (M) 123-bp DNA ladder.
Table 1.
Temperature-dependent lethality induced by H1 RNAi
Homozygous transgenic pINT-1-Nau, pINT-1-H12M, pINT-1-H15M, and pINT-1-H16F flies were mated with heterozygous Tubulin∷GAL4/TM3, Sb flies. The crosses were set at the indicated temperatures. Viability is expressed as the number of eclosed Sb+ adults, relative to the total number of offspring. The expected number of Sb+ flies (from the Mendelian distribution) is shown in parentheses.
To confirm that the combination of the pINT-1-H1 and Tubulin-GAL4 transgenes causes depletion of H1 histone protein, larvae of this genotype were collected at 29°C, salivary glands were dissected, and tissue lysates were examined by SDS-PAGE and immunoblotting with histone H1 antiserum. We observed that lysates from pINT-1-H15M + Tubulin-GAL4 salivary glands contained <5% of the H1 protein present in control lysates (Fig. 1B). We also determined the extent of H1 depletion in animals with other H1 RNAi transgene insertions at various temperatures (Supplemental Fig. 1; Supplemental Table 1). Depending on the temperature and the insertion allele, the expression of H1 protein in salivary glands of transgenic larvae was decreased by 14%–95% in salivary glands and by 13%–99% in whole larvae. GAL4 is known to exert higher transcriptional activity at elevated temperatures (Brand et al. 1994). Consistent with this observation, the lethal effect of combining the pINT-1-H1 and Tubulin-GAL4 transgenes was alleviated at lower temperatures (18°C and 22°C), due to reduced dosage of the H1-specific siRNA (Table 1; Supplemental Fig. 1; Supplemental Table 1). It appears from our analyses that lethality is caused by the knockdown when H1 protein levels are decreased below a certain threshold, between ∼20% and 60% of the wild-type level (Supplemental Table 1). This result is similar to the effect of H1 dosage reduction in mice (Fan et al. 2003), in which reducing the total level of H1 protein below a certain threshold value causes lethality at early stages of development.
To demonstrate that histone H1 is depleted from the chromatin of RNAi-treated animals, we treated nuclei prepared from whole L3 larvae with micrococcal nuclease (MNase) (Fig. 1C). The nucleosome repeat length in the chromatin of the knockdown animals was reduced from ∼188 bp to ∼176 bp. Furthermore, we observed depletion of the chromatosome band in the H1-depleted chromatin as compared with the control chromatin (Fig. 1D). Thus, bulk native chromatin contains substantially less H1 in pINT-1-H1 + Tubulin-GAL4 animals.
To confirm that the lethality of pINT-1-H1 + Tubulin-GAL4 flies is due to reduced expression of histone H1, rather than an off-target effect of RNAi, we tested whether duplication of the histone gene cluster (which encompasses ∼100 copies of the H1 gene) or expression of wild-type Drosophila histone H1 cDNA from an ectopic pUAST-H1 transgene could rescue the lethal phenotype. We observed that H1 RNAi in the genetic background of several different duplications encompassing the histone gene cluster caused a significantly less detrimental effect (Table 2). Consistent with this result, larvae that contained the duplications expressed H1 protein at levels above the lethality threshold (Supplemental Fig. 2). The pUAST-H1 transgenes also rescued the lethality, albeit with a substantially reduced effectiveness compared with the histone gene cluster duplications (Table 3). We conclude that elevated expression of H1 mRNA can counteract the defect that is caused by the expression of double-stranded H1-specific RNA.
Table 2.
H1 RNAi-induced lethality is alleviated in genetic backgrounds of the histone cluster duplications
Double-heterozygous pINT-1-H12M/SM5, Cy; Tub∷GAL4/TM6B, Hu transgenic fly lines were established and maintained at 18°C. They were mated at 29°C to y w control flies or flies that carry the indicated heterozygous balanced histone cluster duplications. Viability is expressed as the number of eclosed Cy+; Hu+ adults, relative to the total number of offspring. The expected number of Cy+; Hu+ flies (from the Mendelian distribution) is shown in parentheses.
Table 3.
Ectopic expression of H1 cDNA partially rescues H1 RNAi-induced lethality
Homozygous transgenic pUAST-H1 insertion flies or y w control flies were mated at 29°C to double-heterozygous pINT-1-H12M/SM5, Cy; Tub∷GAL4/TM6B, Hu flies. Viability is expressed as the number of eclosed Cy+; Hu+ adults, relative to the total number of offspring. The expected number of Cy+; Hu+ flies (from the Mendelian distribution) is shown in parentheses.
Our results indicate that linker histone H1 has an essential function in vivo. Substantially reduced H1 expression results in lethality. To investigate the developmental stage at which lethality occurs, we performed crosses, similar to those described in Table 1, at 18°C. At various developmental stages, the offspring of the cross were transferred to a higher, nonpermissive temperature (29°C), and the offspring were later examined for the presence of pINT-1-H1 + Tubulin-GAL4 adults (data not shown). The lethal effect of H1 RNAi was observed only when the offspring of the cross were exposed to the elevated temperature at or prior to pupariation. When pINT-1-H1 + Tubulin-GAL4 pupae were transferred to 29°C, expected numbers of flies survived to adulthood. Also, elevated temperature did not affect viability or longevity of eclosed adults. Thus, expression of the wild-type levels of H1 is essential in vivo before or during metamorphosis.
The histone H1 gene is a suppressor of position effect variegation (PEV)
In vitro analyses and overexpression studies in cultured mammalian cells indicate that H1 may function primarily as a transcriptional repressor (Laybourn and Kadonaga 1991; Brown et al. 1996). However, other studies suggest that H1 can have both positive and negative effects on gene expression (Shen and Gorovsky 1996; Lin et al. 2004; Fan et al. 2005). To test the role of H1 on transcriptional silencing in vivo in Drosophila, we analyzed the effect of H1 depletion on PEV. PEV is a phenomenon in which expression of a gene is altered in a stochastic manner when the native genomic location of the gene is altered, usually by positioning it within or near large blocks of heterochromatin. Many mutations that are known to suppress PEV affect factors that are involved in transcriptional repression as well as the establishment and/or maintenance of heterochromatin (Wallrath 1998; Richards and Elgin 2002; Ebert et al. 2006). In Drosophila, PEV can be assayed readily by studying mosaic silencing of genes that affect visible phenotypes in adult flies.
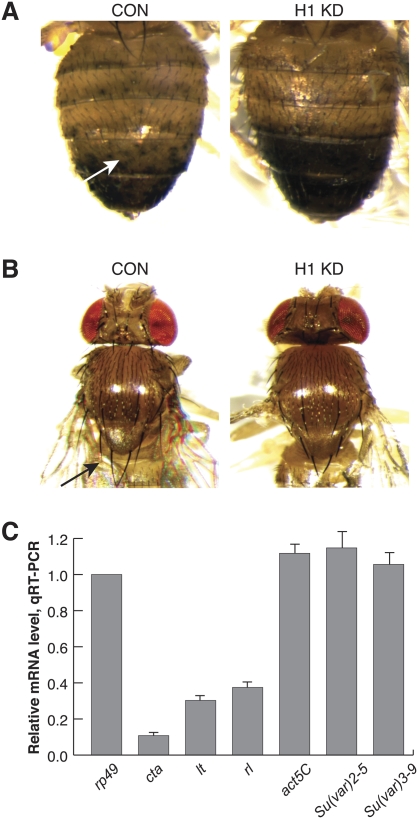
We examined the effect of H1 histone depletion on PEV using the variegated yellow gene in the KV111 P-element insertion line (Konev et al. 2003) and the variegated T(2;3) Sb[V] allele (Csink et al. 1994). We performed these analyses with H1 knockdown animals incubated at 22°C, which is permissive for fly viability. When a recessive yellow transgene is positioned near pericentric heterochromatin, its partial silencing results in the appearance of patches of cells that do or do not contain dark pigment in the dorsal abdominal cuticle (Fig. 2A, left panel). When combined with pINT-1-H1 + Tubulin-GAL4 transgenes, variegation of yellow was suppressed, which was manifested as an almost complete restoration of the yellow + phenotype in the dorsal abdomen (Fig. 2A, right panel). In another experiment, we used Sb[V], a dominant, gain-of-function allele. When Sb is positioned in euchromatin, adult flies have short thoracic sensory bristles. Silencing of heterochromatin-proximal Sb[V] causes a variegated increase of the bristle length toward the wild-type phenotype (Fig. 2B, left panel). The combination of this PEV reporter gene with the pINT-1-H1 + Tubulin-GAL4 RNAi transgenes also caused a suppression of PEV, as indicated by an increased number of short bristles due to derepression of the dominant Sb[V] allele (Fig. 2B, right panel). We conclude that, similar to other suppressors of PEV, the H1 linker histone stimulates silencing in pericentric heterochromatin, either by its general activity in transcriptional repression or through a specific biochemical and/or structural function in heterochromatic silencing. For instance, H1 may serve as an essential protein component of the pathway that is necessary for the assembly and/or maintenance of the heterochromatin structure.
Figure 2.
H1 is a suppressor of PEV and an activator of heterochromatic gene expression. (A) Variegation of a yellow transgene transposed into pericentric heterochromatin in 2R is suppressed by partial depletion of H1. The recessive yellow gene within the SUPorP P-element is inserted into pericentric heterochromatin (KV111, 2h46) (Konev et al. 2003). (Left panel) Variegated expression of yellow manifests in the dorsal abdominal cuticle as dispersed patches of cells with or without the dark pigment (arrow). (Right panel) In H1-depleted adult flies (pINT-1-H12M + Tubulin-GAL4), variegation of yellow is suppressed, which is indicated by intensification of the dark pigment in the dorsal abdomen. The animals were incubated at 22°C throughout their life cycle. (H1 KD) H1-depleted animals; (CON) Nautilus RNAi control. (B) Variegation of Sb[V] allele in a rearranged Chromosome 3, T(2;3)Sb[V], is suppressed by partial depletion of H1. (Left panel) Silencing of heterochromatin-proximal Sb[V] causes a variegated increase of bristle length toward the wild-type phenotype (arrow). (Right panel) H1 depletion (pINT-1-H12M + Tubulin-GAL4) causes a suppression of PEV, which is indicated by an increased number of short bristles. The animals were incubated at 22°C throughout their life cycle. (H1 KD) H1-depleted animals; (CON) Nautilus RNAi control. (C) H1 depletion causes repression of pericentric genes concertina, light, and rolled. Total RNA was prepared from dissected salivary glands of H1-depleted (pINT-1-H15M + Tubulin-GAL4) and control (pINT-1-Nau + Tubulin-GAL4) L3 larvae, and mRNA levels were analyzed in triplicate by real-time qRT–PCR. The measurements were normalized to rp49. Gene expression levels in H1-depleted larvae are expressed relative to the Nautilus RNAi control. The animals were incubated at 29°C throughout their life cycle. act5C, Su(var)2-5, and Su(var)3-9 gene expression levels were measured as controls.
To determine whether the effects of H1 on PEV are transcriptional, we analyzed the effect of H1 depletion on expression of genes that depend on their position in heterochromatin for their transcriptional activity. Unlike typical euchromatic genes, concertina, light, and rolled, which are embedded into proximal heterochromatin of the second chromosome (Hilliker and Holm 1975), are silenced when taken out of their normal heterochromatic context (Wakimoto and Hearn 1990; Eberl et al. 1993). Notably, these genes have distinct regulatory requirements and are transcriptionally activated by dominant suppressors of variegation (Lu et al. 2000). We used quantitative real-time RT–PCR (qRT–PCR) to analyze the expression levels of these heterochromatic genes in H1-depleted and control salivary glands. We found that concertina, light, and rolled are strongly repressed (threefold to 10-fold) upon abrogation of H1 (Fig. 2C). We performed quantitative chromatin immunoprecipitation (qChIP) and discovered that whereas H1 is highly abundant in chromatin in the promoters and transcription units of concertina and light in the control larvae, it is almost entirely eliminated in the knockdown larvae (Supplemental Fig. 3). As a control, we measured mRNA levels of several highly and ubiquitously expressed genes. In contrast to heterochromatic genes, the expression of rp49, act5C, Su(var)2-5, and Su(var)3-9 was not substantially affected by H1 depletion (Fig. 2C). Thus, similar to suppressors of variegation, H1 functions in transcriptional activation of heterochromatic loci. In contrast, H1 does not appear to play a significant role in expression of at least some euchromatic genes, which may be positioned in chromatin loci that normally contain low levels of H1 (Fig. 2C; Supplemental Fig. 3).
Depletion of H1 histone disrupts polytene chromosome structure
pINT-1-H1 + Tubulin-GAL4 animals survive through the third instar larval stage of development at 29°C. H1 depletion causes a range of morphological defects in larvae, including smaller salivary glands that contain fewer cells with enlarged nuclei (Fig. 3A, left panel; Supplemental Fig. 4). Furthermore, H1-depleted larvae have smaller imaginal discs (Fig. 3A, right panel) that contain reduced numbers of cells. These changes could be due to decreased cell proliferation or increased cell death under the conditions of very low H1 protein levels. Additionally, they may result from altered gene expression programs and/or chromosome structure in H1-depleted animals.
Figure 3.
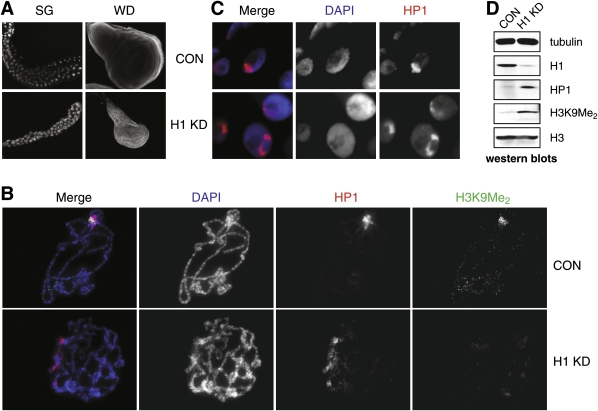
H1 depletion affects HP1 localization and the status of H3K9 dimethylation in Drosophila polytene chromosomes. (A) H1 depletion by RNAi leads to a reduction in the number of cells and the size of salivary glands and imaginal discs in L3 larvae. Whole salivary glands and wing imaginal discs were dissected from H1-depleted (pINT-1-H15M + Tubulin-GAL4) and control (pINT-1-Nau + Tubulin-GAL4) L3 larvae. The animals were incubated at 29°C throughout their life cycle. Tissues were fixed and stained with DAPI to visualize the nuclei. (SG) Salivary gland; (WD) wing disc; (CON) pINT-1-Nau + Tubulin-GAL4 control larvae; (H1 KD) pINT-1-H15M + Tubulin-GAL4 H1 RNAi larvae. (B) H1 depletion causes abnormal polytene chromosome morphology and the loss of the H3K9Me2 marker in pericentric heterochromatin. Salivary glands from control (pINT-1-Nau + Tubulin-GAL4) and H1-depleted (pINT-1-H15M + Tubulin-GAL4) L3 larvae were squashed, and polytene spreads were stained with DAPI and antibodies against HP1 and H3K9Me2. Control chromosomes (CON) have a uniform regular structure of bands and interbands as well as a prominent chromocenter brightly stained with DAPI and both antibodies, whereas no such uniform banding pattern or the chromocenter are apparent in the H1-depleted polytene chromosomes (H1 KD). In contrast to the control chromosomes, the H1-depleted polytene chromosome structure is severely disturbed, HP1 is dispersed broadly, and the H3K9Me2 cannot be readily detected, including at the loci where HP1 is present. The animals were incubated at 29°C throughout their life cycle. (Blue) DAPI; (red) HP1; (green) H3K9Me2. (C) HP1 is distributed in more than one chromosomal locus in salivary gland cells with depleted H1. Whole-mount salivary glands from control (pINT-1-Nau + Tubulin-GAL4, CON) and H1-depleted (pINT-1-H15M + Tubulin-GAL4, H1 KD) larvae were fixed and stained with anti-HP1 antibody. Whereas in control cells HP1 is mostly concentrated in a single region (chromocenter), in H1-depleted cells HP1 is dispersed broadly. The animals were incubated at 29°C throughout their life cycle. (Blue) DAPI; (red) HP1. (D) Total cellular HP1 and H3K9Me2 are increased upon abrogation of H1 expression in larvae. HP1 and H3K9Me2 were detected by Western blotting of crude lysates of salivary glands from control (pINT-1-Nau + Tubulin-GAL4, CON) and H1-depleted (pINT-1-H15M + Tubulin-GAL4, H1 KD) L3 larvae. Western blotting for tubulin and total histone H3 were used as loading controls. The animals were incubated at 29°C throughout their life cycle.
To analyze possible altered chromatin structure in H1 knockdown larvae, we examined the effects of H1 histone depletion on polytene chromosome structure. Figure 3B shows a comparison of polytene chromosomes prepared from control and H1-depleted salivary glands. Unlike wild-type polytene chromosomes, the polytene chromosomes from H1 RNAi animals have a severely perturbed pattern: They do not exhibit the normal regular structure of intensely stained bands and dark interband regions. In addition, the chromosome arms are often tangled into unstructured clumps of chromatin and occasionally appear substantially thinner than normal. The loss of discernable polytene chromosome banding is reminiscent of that observed in mutants of the general transcription factor TRF2, a positive regulator of H1 transcription in flies (Isogai et al. 2007), and in strains carrying dominant-negative alleles of Iswi, in which H1 deposition into chromosomes is compromised (Corona et al. 2007).
A prominent feature of salivary gland nuclei is the chromocenter, a single coalesced region of underreplicated pericentric heterochromatin contributed by all chromosomes. Although certain components, such as Heterochromatin Protein 1 (HP1), enzymes that mediate histone H3 Lys 9 dimethylation (H3K9Me2) and the RNAi machinery factors, are known to be associated with the chromocenter, the complete molecular details of its establishment are not known. DAPI staining of squashed salivary gland polytene chromosomes showed that H1 depletion caused the loss of a well-defined chromocenter (Fig. 3B). In fact, when salivary gland squashes from H1 knockdown animals are stained with DAPI, visual inspection does not allow unambiguous identification of pericentric regions in these polytene chromosomes.
HP1 is a major marker of the pericentric heterochromatin and is normally highly concentrated in the chromocenter and also associated with telomeres, the fourth chromosome, and several euchromatic loci (Fanti and Pimpinelli 2008). Indirect immunofluorescence staining of polytene chromosomes from H1-depleted salivary glands showed that pericentric HP1 is dispersed much more broadly through the plane of the polytene spread (Fig. 3B). These HP1-enriched foci resemble two or more “partial chromocenters.” These changes in HP1 localization were also apparent in stained whole-mount preparations of salivary gland cells (Fig. 3C). Instead of a typical single locus of strong pericentric HP1 staining, H1-depleted salivary gland cells invariably contain two or more separate HP1 foci. In addition, the overall size of HP1-enriched foci in salivary gland cells from H1 knockdown larvae appeared somewhat smaller than those in the wild-type controls.
H3K9Me2 is another major marker of the chromocenter in salivary gland polytene chromosomes. Pericentric heterochromatin in Drosophila contains core histone H3 that is dimethylated at Lys 9 by the histone methyltransferase enzyme Su(var)3-9 (Schotta et al. 2002). When H1-depleted chromosomes were stained with H3K9Me2-specific antibodies, virtually no signal above the background could be observed anywhere in the polytene chromosomes (Fig. 3B). This is in stark contrast to the wild-type polytene chromosomes that exhibit a very strong focus of H3K9Me2 staining overlapping with the HP1 signal in the chromocenter. Since very little H3K9Me2 could be detected in polytene chromosomes of H1-depleted cells, including in the region of the foci where HP1 was still present, we conclude that in these cells HP1 is deposited in the pericentric region in a manner that is independent of histone H3 dimethylation.
The observed changes in chromocenter morphology as well as abundance and localization of HP1 and H3K9Me2 were not due to changes in the total protein levels of these components in the H1-depleted cells. Indeed, immunoblotting of cell lysates showed that the cellular levels of total HP1 and H3K9Me2 proteins are actually increased upon the knockdown of H1 (Fig. 3D). However, qRT–PCR assays showed that the levels of the Su(var)2-5 (HP1) and Su(var)3-9 histone methyltransferase mRNAs were not altered in the H1-depleted cells (Fig. 2C). We conclude that H1 is required for proper polytene chromosome structure and for formation of the chromocenter, including localization of the major heterochromatin markers of the chromocenter.
Drosophila H1 depletion causes misalignment of sister chromatids in polytene chromosomes
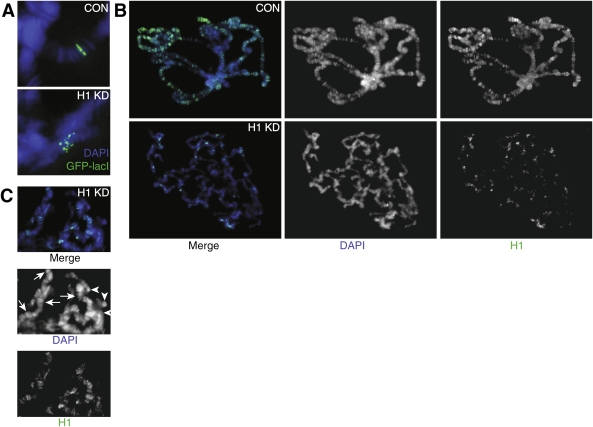
To further investigate the causes of the abnormalities seen in the polytene chromosome structure in the H1-depleted cells, we examined the alignment of chromatids in the polytene chromosomes by using a system in which a GFP-tagged lac repressor DNA-binding domain (GFP-lacI) associates with multiple copies of lac operator (lacO) sequences inserted into the 61F interband region on the third chromosome (Danzer and Wallrath 2004). The GFP-lacI transgene is driven by the hsp70 promoter. When its expression is induced, the binding of the fusion protein to the lacO repeats can be detected with an anti-GFP antibody. This approach has been used previously to detect disruption of the normal parallel alignment of the chromatids in polytene chromosomes (Deng et al. 2005). In control polytene chromosomes squashes, the GFP signal was observed as a tight, straight band perpendicular to the chromosome axis (Fig. 4A, top panel). However, in H1-depleted larvae, the GFP signal was dispersed and in many chromosomes consisted of clearly separated spots (Fig. 4A, bottom panel). Nevertheless, these spots were localized together, suggesting that even though the normal morphology of the chromosomes is severely altered by H1 depletion, partial alignment of the individual fibers along the longitudinal chromosome axis is maintained to some extent.
Figure 4.
Effects of H1 depletion on alignment of sister chromatids in polytene chromosomes. (A) H1 depletion results in misalignment of chromatin fibrils in an interband region in 61F. The genomic position of a P-element insertion containing 256 lacO sites was visualized by indirect immunofluorescence (IF) through binding of an ectopically expressed GFP-lacI protein to the locus in polytene chromosomes. In control squashes (pINT-1-Nau + Tubulin-GAL4, CON), the GFP signal was observed as a single straight band, indicative of an alignment of endoreplicated chromatin fibrils. In H1-depleted polytene chromosomes (pINT-1-H15M + Tubulin-GAL4, H1 KD), the GFP signal was dispersed into multiple isolated spots, indicating the loss of perfect alignment. The animals were incubated at 29°C throughout their life cycle. (Blue) DAPI; (green) GFP-lacI. (B) Residual H1 protein in H1-depleted polytene chromosomes is unevenly distributed and correlates with regions of persistent polytene band–interband structure. Polytene chromosome squashes from control (pINT-1-Nau + Tubulin-GAL4, CON) and H1-depleted (pINT-1-H15M + Tubulin-GAL4, H1 KD) salivary glands were stained with the H1 antibody. In control chromosomes, H1 localizes primarily to bands in euchromatic arms and to the pericentric region. In H1-depleted larvae, the H1 signal is dramatically reduced, and the residual protein is distributed over a limited number of loci. The majority of these loci also have a partially conserved polytene band–interband structure, as evidenced by DAPI staining. In contrast, chromosome regions that do not contain detectable H1 typically exhibit amorphous, clumped morphology. The animals were incubated at 29°C throughout their life cycle. (Blue) DAPI; (green) H1. (C) In H1 knockdown larvae, polytene band–interband structure is partially preserved in loci with elevated residual H1. Whereas DAPI stains equally brightly the residual structured bands (arrows) and unstructured chromatin clumps (arrowheads), H1 staining predominantly has an appearance of discrete bands. No diffuse signal can be observed for the H1 staining. Furthermore, there is a substantial overlap between the residual DAPI and H1 bands.
When H1 is depleted by RNAi, the residual H1 (<5% of the wild-type level) is not distributed uniformly in polytene chromosomes but, instead, is mostly localized to a small number of specific H1-enriched loci (Fig. 4B). Very consistently, these loci of higher residual H1 content also maintain a partially conserved band–interband polytene structure. In contrast, many chromosome regions that do not contain detectable H1, although occasionally brightly stained with DAPI, have no discernable banding pattern and exhibit unstructured, clumped morphology (Fig. 4C). Thus, Drosophila histone H1 helps to maintain alignment of sister chromatids during endoreplication in larval cells and plays an important role in the formation of the normal banding pattern of polytene chromosomes.
Discussion
Drosophila H1 depletion by RNAi
Our work provides evidence that maintaining the level of histone H1 expression is essential for proper Drosophila development. We used in vivo transcription of an H1-specific dsRNA “hairpin” as a means to induce post-transcriptional gene silencing in Drosophila. We observed that lethality caused by abrogation of histone H1 synthesis is temperature-dependent. In our system, the transcription of the H1-specific hairpin RNA is activated ubiquitously by the yeast transactivator protein GAL4, which is known to exert stronger effects at elevated temperatures (Brand et al. 1994). Indeed, the depletion of H1 protein and penetrance of the RNAi-induced lethality in our transgenic strains both directly correlated with the temperature (Table 1; Supplemental Fig. 1; Supplemental Table 1). Thus, the temperature dependence of GAL4 transcriptional activity allows temporal control over the post-transcriptional silencing of H1; that is, by transferring developing animals from the permissive (18°C) to the restrictive (29°C) temperatures, or vice versa, one can target the RNAi effect to a specific developmental time period. For instance, we found that activating the synthesis of the H1-specific RNAi during late stages of Drosophila development (in pupae and adults) did not cause an appreciable effect on viability, in contrast to H1 abrogation in embryos and larvae. Thus, there may be a less stringent requirement for maintaining H1 expression after metamorphosis. Alternatively, the endogenous H1 protein that accumulates in larvae prior to metamorphosis may be sufficient for proper cell function throughout the rest of the life cycle in Drosophila.
Our previous studies with single and compound H1 subtype-specific knockout mice also revealed a direct correlation between the levels of H1 expression and survival (Fan et al. 2003). Mice lacking only one or two H1 subtypes, but containing a normal H1 to nucleosome ratio, survive and appear normal. On the other hand, mice lacking five H1 alleles, with a reduction from 20% to up to 50% in the H1-to-nucleosome ratios in different tissues, were small and born at a significantly lower rate than the single and double H1 knockout mice. Embryos lacking six alleles (three H1 subtypes) and containing approximately half of the normal H1 levels developed multiple abnormalities and died in midgestation, an indication that a minimum threshold level of H1 protein is required for normal mammalian embryonic development (Fan et al. 2003). Our data in Drosophila parallel these findings, since at subpermissive temperatures (26°C or lower), intermediate reduction of H1 expression to ∼70% of the wild-type larval level resulted in partial survival of affected animals. Thus, in contrast to simpler eukaryotes, in which the linker histone is not essential, metazoans require maintenance of a certain level of H1 expression for normal development.
The roles of Drosophila H1 in heterochromatin formation
Pericentric heterochromatin has been implicated in gene silencing that occurs when euchromatic genes are placed adjacent to heterochromatin by chromosome rearrangement or transposition—a phenomenon that was initially described in Drosophila as PEV (Henikoff 1990). Through genetic screening, many important chromatin regulators have been identified, which, when mutated, act as modifiers (suppressors or enhancers) of PEV (Reuter and Spierer 1992). Thus, PEV in Drosophila represents a valuable assay for identification and molecular study of evolutionarily conserved functions controlling epigenetic programming in eukaryotes (Reuter and Spierer 1992; Wallrath 1998). We observed that the linker histone H1 stimulates silencing in pericentric heterochromatin. Although it was not feasible to make a classical mutant of the H1 genes, dose reduction of H1 by ∼15% (Supplemental Fig. 1; Supplemental Table 1) resulted in PEV suppression. In that respect, H1 resembles other dominant suppressors of PEV, such as Su(var)2-5, which encodes HP1. Dose reduction of HP1 in Su(var)2-5 heterozygotes results in strong PEV suppression (Eissenberg et al. 1990). Our data indicate that H1 is an essential structural component of pericentric heterochromatin, or it is necessary for recruitment of another such essential biochemical component(s) to heterochromatin. In fact, we found that the level of H1 does affect the localization of two major markers of pericentric heterochromatin, HP1 and H3K9Me2.
HP1 is an abundant nonhistone chromosomal protein first discovered in Drosophila because of its association with heterochromatin. HP1 is conserved in many eukaryotes, including fission yeast, insects, and mammals; involved in gene silencing; and consistently associated with pericentric heterochromatin and telomeres (Fanti and Pimpinelli 2008). In Drosophila polytene chromosomes, HP1 is diagnostic of heterochromatin, and the vast majority of HP1 protein concentrates at the chromocenter. Indirect immunofluorescence staining of polytene chromosomes indicates that histone H1 is abundant in pericentric heterochromatin (Fig. 4B). Furthermore, the chromocenter is severely disrupted in polytene chromosomes of salivary gland cells with depleted H1, and H1 abrogation also results in a delocalization of HP1 (Fig. 3B). The dispersion of the chromocenter is not produced by mechanical stress during squashing, since it is similarly observed in whole-mount salivary gland cells (Fig. 3C). Thus, H1 plays important roles in the establishment and/or maintenance of the structure as well as in the biochemical composition of proximal heterochromatin in Drosophila larvae. It remains to be seen whether H1 is directly required for faithful deposition/recruitment of HP1 to its cognate loci in pericentric heterochromatin, or mislocalization of HP1 in chromosomes of H1-depleted cells is a secondary effect mediated by disruption of other nuclear processes that are regulated by the abundance of H1 (e.g., transcription). The former explanation is certainly possible since there are several reports that HP1 interacts directly with H1 (Nielsen et al. 2001; Daujat et al. 2005; Hale et al. 2006).
Methylation of histone H3 Lys 9 (H3K9) has a well-established role in heterochromatin formation in metazoans, and H3K9Me3 (H3K9Me2 in Drosophila) is highly enriched in condensed heterochromatin (Lachner et al. 2001; Rice et al. 2003). The chromodomain of HP1 specifically recognizes methylated H3K9, which facilitates its recruitment and leads to an overlapping distribution of HP1 and the H3K9 methylation mark in the genome (James et al. 1989; Lachner et al. 2001; Schotta et al. 2002). Upon H1 abrogation, however, very little or no H3K9Me2 is detected in the loci where HP1 remains present. We conclude that in polytene chromosomes of H1-depleted larvae, HP1 is deposited by a mechanism that does not require histone H3 dimethylation. The persistence of HP1 in proximal heterochromatin in the absence of dimethylated H3K9 is consistent with reports indicating that HP1 can bind nonspecifically to nucleosome core particles and even to naked DNA (Zhao et al. 2001). It is also consistent with the findings of Danzer and Wallrath (2004), who, by using a tethering system to recruit HP1 to euchromatic sites, showed that HP1-mediated silencing can operate in a Su(var)3-9-independent manner. Our findings strengthen the view that, whereas HP1 may normally cooperate with Su(var)3-9 and K9-methylated H3 in heterochromatin formation and gene silencing at pericentric chromosome sites, it can be deposited in these regions independently of these other components, and even without the presence of H1.
The Su(var)3-9-null mutants, although also lacking an appreciable level of H3K9Me2 signal in immunofluorescence-stained polytene chromosomes, do not exhibit the same spectrum of phenotypes as H1-depleted animals. For instance, the single polytene chromocenter is not disrupted in Su(var)3-9-null mutants (Schotta et al. 2002). Thus, the observed phenotypes and defects in chromatin structure upon abrogation of H1 cannot be explained exclusively by the loss of H3K9 dimethylation, and H1 is therefore predicted to play a separate and unique role in the establishment and/or maintenance of pericentric heterochromatin. In the future, it will be interesting to see whether in addition to the reversal of heterochromatic silencing, similar to other suppressors of variegation, H1 depletion also affects other properties of heterochromatin, such as the reduced rates of meiotic recombination normally observed in these regions (Westphal and Reuter 2002).
It is an intriguing observation that H3K9Me2 is not detectable in chromatin of H1-depleted salivary glands by indirect immunofluorescence, although total protein levels in cell lysates are elevated rather than reduced. Thus, H1 may be required for H3K9Me2 deposition in chromatin. Alternatively, if histone H3 Lys 9 is dimethylated by Su(var)3-9 predominantly in the context of a nucleosome, H1 depletion may result in specific expulsion of the K9-dimethylated form of H3 from pericentric regions and potentially other H3K9Me2-enriched loci. We analyzed the presence of other repressive, heterochromatin-specific histone marks, such as H4K20Me2, H3K9Me1, and H3K9Me3, in polytene chromosomes of H1 knockdown larvae by IF microscopy and discovered that they were all largely absent in pericentric heterochromatin (Supplemental Figs. 5, 6). In contrast, there was no substantial effect on the active H3K4Me2 mark, which remained widely distributed in polytene chromosomes (Supplemental Fig. 5). Thus, H1 appears to be required for global maintenance of repressive marks in heterochromatin, rather than stimulation of particular programs/enzymes that affect specific histone modification states. This function of H1 might be linked to its role in the transcriptional activity of heterochromatin. Indeed, our studies of heterochromatic gene expression in H1-depleted larvae showed that low levels of H1 cause altered transcriptional activity in heterochromatin. Further studies of the dynamics of formation and maintenance of H3K9Me2 and other repressive marks in H1-depleted chromatin may allow us to better understand this relationship.
The roles of Drosophila H1 in transcriptional repression and activation
H1 depletion has a dramatic effect on the distribution of H3K9Me2-containing nucleosomes in the genome. It is possible that H1 is similarly involved in maintenance of other repressive histone marks in Drosophila. However, it is unlikely that H1 is involved in Polycomb silencing (Schwartz and Pirrotta 2008), since we did not observe homeiotic phenotypes in adult escapers that survive partial H1 depletion (at 26°C and below).
Our previous work with H1-depleted mouse ES cells, as well as studies in other species, suggested that H1 may participate in both transcriptional activation as well as repression in vivo (Shen and Gorovsky 1996; Hellauer et al. 2001; Fan et al. 2005; Ni et al. 2006). Likewise, our studies with H1-depleted Drosophila larvae support dual roles for H1 in transcriptional regulation. Similar to other suppressors of PEV, H1 stimulates silencing of genes that are brought into juxtaposition with heterochromatin. On the other hand, certain Drosophila genes that are embedded in heterochromatin (e.g., concertina, light, and rolled) are dependent on their genomic localization for proper transcriptional regulation, as their expression is reduced when their genomic loci are rearranged to lie next to a euchromatic breakpoint (Wakimoto and Hearn 1990; Hearn et al. 1991; Eberl et al. 1993; Howe et al. 1995) or when heterochromatin component genes are mutated (Lu et al. 2000). By qRT–PCR assay, we demonstrated that concertina, light, and rolled are repressed in third instar larval salivary glands upon reduction of H1 levels (Fig. 2C). Thus, H1 is also required for activation of heterochromatic genes within the context of pericentric heterochromatin.
Wakimoto and Hearn (1990) proposed that heterochromatin-associated proteins function to support normal transcription of heterochromatic genes when those genes are at their normal chromosomal sites and that position effects result when these genes are deprived of such essential proteins by displacement away from heterochromatin “compartments.” Similarly, H1 may contribute to the formation of a particular chromatin structure that interferes with activation of euchromatic genes but to which heterochromatic genes have become adapted. The loss of H1 would deplete the nucleus of this particular chromatin conformation, releasing silenced genes from repression while simultaneously depriving the resident heterochromatin genes of their functional context. Interestingly, mutations of rolled, similar to H1 depletion, lead to late larval or early pupal lethality and defective imaginal disc formation (Hilliker 1976; Dimitri 1991). It remains to be seen whether one of the effects contributing to the lethality of H1-depleted animals is down-regulation of specific heterochromatic genes.
As a control, we performed a limited analysis of possible effects of H1 abrogation on expression of several euchromatic genes. So far, we have not discovered a euchromatic in vivo transcriptional target for H1 in Drosophila larvae. However, this lack of apparent effect can be explained by our limited sample size (four genes) and the choice of targets. We assayed only abundant, ubiquitous genes, whose transcription units in the wild-type animals (without H1 abrogation) may be positioned within chromatin that already contains little or no H1. In the future, it will be important to extend this analysis to tissue-specific, tightly regulated genes and to perform this experiment in an unbiased, genome-wide (microarray) format.
The roles of Drosophila H1 in sister chromatid adhesion
Although the Drosophila polytene chromosome has served as a model to study chromatin structure, remarkably little is known about its spatial organization or the molecular mechanisms that maintain the alignment of sister chromatids. Previous studies suggested that interchromatid cohesion is generated and maintained in the banded regions (Ananiev and Barsky 1985; Urata et al. 1995). H1 is widely distributed in euchromatic arms of polytene chromosomes; however, it localizes predominantly to bands of compacted chromatin (Fig. 4B). H1 depletion disrupts the normal band–interband structure of polytene chromosomes. Thus, H1 functions to establish or maintain the parallel alignment of band chromosome fibrils. When depleted by RNAi, residual H1 protein is not distributed uniformly in polytene chromosomes. Remarkably, the residual H1 maxima correlate with the persistent band–interband structure over short fragments of the H1-depleted polytene chromosomes (Fig. 4C). This result emphasizes the requirement for H1 in polytene chromatid alignment/adhesion. Similarly, the dissociation of the normal single chromocenter in polytene chromosomes into several foci of HP1 localization in the H1 knockdown larvae may also be related to the loss of adhesion.
Linker histone H1 is an abundant protein component of chromatin. It binds to DNA outside the core particle region, and its function in internucleosomal interactions and chromatin condensation is widely accepted (Robinson and Rhodes 2006; Woodcock et al. 2006). It is possible that internucleosomal interactions directly mediated by H1 can occur in trans between two distinct chromatin fibrils and, thus, play a role in adhesion of sister chromatids in polytene chromosomes. In that case, genomic regions of intrinsically higher H1 density (bands) would then cluster (“align”) in polytene chromosomes. This direct mechanism is consistent with the partial conservation of the polytene chromosome banding structure of H1-depleted salivary gland cells in regions that contain elevated levels of residual H1 (Fig. 4C). However, we cannot exclude a possibility that H1 activity in chromatid alignment is mediated through interactions with other molecules important for chromatin structure maintenance, such as H3S10 kinase JIL-1 (Deng et al. 2005).
Although JIL-1 hypomorphic or null alleles exhibit a defect in polytene chromosome alignment comparable with that observed in H1 knockdown alleles, other functions of these proteins are remarkably dissimilar. Unlike H1, JIL-1 localizes to gene-active interbands and counteracts the function of Su(var)3-9 (Deng et al. 2007). JIL-1 is also an enhancer of PEV (Bao et al. 2007). Furthermore, in JIL-1 alleles, polytene chromosome arms are highly condensed and interband regions are missing, with the male X chromosome affected the most severely (Wang et al. 2001). None of these phenotypes are observed in H1 knockdown animals. On the contrary, H1-depleted polytene chromosomes are rather extended, probably due to the dispersal of normally compacted band regions. However, both H1 and JIL-1 appear to contribute to polytene fibril alignment. It is possible that the polytene chromosome structure is established through interplay between antagonistic effects mediated by several effectors, such as H1 and JIL-1 (or its substrates). In the future, it will be interesting to elucidate fine details of these putative functional interactions between H1 and JIL-1.
Although H1 is clearly required for chromatid alignment in endoreplicating cells, it is likely dispensable or less critical for sister chromatid alignment in G2–M of proliferating cells. Mutations that affect Drosophila genes coding for the Rad21 subunit of cohesin, CAP-G subunit of condensin, and Orc2 and Orc5 subunits of the origin recognition complex have been shown previously to affect sister chromatid alignment and segregation in vivo (Pflumm and Botchan 2001; Dej et al. 2004; Pauli et al. 2008). Mutations in these genes result in massive missegregation of chromosomes during mitosis, which was not observed in H1-depleted animals. On the other hand, these mutations do not cause any abnormalities in polytene chromosome structure. Thus, adhesion of replicating chromatin in dividing and endoreplicating cells in Drosophila is likely to be maintained through distinct mechanisms.
In conclusion, we demonstrated that the linker histone H1 is essential for normal development in Drosophila and required for proper chromosome structure and function. Specifically, H1 is involved in the establishment of repressive pericentric heterochromatin and deposition/maintenance of the several histone modification marks that are localized in proximal heterochromatin. Furthermore, reduced H1 expression results in defective polytene chromosome structure with dissociation of the chromocenter and an almost complete loss of the banding pattern in the chromosome arms. Thus, linker histone H1 plays an essential role in the architecture and activity of metazoan chromosomes.
Materials and methods
Fly strains and genetics
Flies were grown on standard corn meal, sugar, and yeast medium with Tegosept. Stocks and crosses were maintained in an environmental chamber at 18°C, or in vials placed in water baths at other indicated temperatures. The pINT-1-H1 and pUAST-H1 constructs were prepared by PCR and standard cloning techniques and sequenced. Transgenic fly strains were generated by Bestgene in the y w background. Six different pINT-1-H1 insertion lines—pINT-1-H11M, pINT-1-H12M, pINT-1-H13M, pINT-1-H14M, pINT-1-H15M, and pINT-1-H16F—contained P-element insertions in the second (2M–5M and 6F) or third (1M) chromosomes and differed in their potency in H1 depletion. Three different pUAST-H1 insertion lines—DH1-6, DH1-7, and DH1-10—contained P-element insertions in the second (DH1-6 and DH1-7) or third (DH1-10) chromosomes. Trans-heterozygous flies containing the pINT-1-H1 + Tubulin-GAL4 transgenes were produced by crossing homozygous pINT-1-H1 flies with Tub∷GAL4/TM3, Sb flies. The crosses were performed at the indicated temperatures (Table 1), and offspring were analyzed for the presence of pINT-1-H1 + Tubulin-GAL4 adults using chromosome markers. To determine the developmental stage of H1 RNAi-induced lethality, the foregoing described crosses were initiated at 18°C, and after a certain time transferred to 29°C. Cytologic experiments were performed with the pINT-1-H15M insertion, which produces the highest degree of H1 depletion when combined in trans-heterozygotes with the Tubulin-GAL4 driver at 29°C. For rescue experiments with the histone cluster duplications and pUAST-H1 cDNA transgenes, the pINT-1-H12M insertion line was combined with the Tubulin-GAL4 transgenic line, and the double-balanced trans-heterozygous stock was maintained at 18°C. Viability of pINT-1-H12M + Tubulin-GAL4 in combination with various duplications and pUAST-H1 insertions was analyzed in crosses at 29°C (Tables 2, 3). The GFP-lacI transgenic stock 128.1 and the lacO repeat transgenic stock (61F) were generous gifts of Dr. L. Wallrath (University of Iowa). The pINT-1-Nau transgenic line was generously provided by Dr. B. Paterson (NIH). The histone gene cluster duplication and GAL4 driver alleles as well as balancer stocks were obtained from the Bloomington Stock Center.
For details of cloning, genetic crosses, and viability calculations, see the Supplemental Material.
Micrococcal nuclease digestion of chromatin
y w or pINT-1-H15M + Tubulin-GAL4 animals were incubated at 29°C from egg deposition to the third instar (L3) larvae. Micrococcal nuclease digestion assay and analyses were performed as described previously (Cartwright et al. 1999; Fyodorov et al. 2004) on nuclei from whole L3 larvae. The digested DNA was resolved on a 1.2% agarose (Fig. 1C) or 6% native polyacrylamide gel (Fig. 1D) in 1× TBE and stained with ethidium bromide.
Immunohistochemistry
Salivary glands and wing discs of the wandering third instar larvae were dissected in PBS + 0.1% Triton X-100. For whole-mount staining, they were fixed for 15 min in PBS containing 3.7% formaldehyde, washed in PBS + 0.1% Triton X-100, and permeabilized for 1 h with PBS + 1% Triton X-100. Alternatively, to prepare polytene chromosomes, they were fixed in 3.7% paraformaldehyde for 30 sec, squashed in 45% acetic acid + 3.7% formaldehyde, and frozen in liquid nitrogen. The glands or polytene spreads were incubated overnight in PBS + 10% fat-free milk + 0.1% Triton X-100 with primary antibodies at the indicated dilutions: affinity-purified rabbit anti-Drosophila H1 (1:5000), affinity-purified rabbit anti-H3K9Me2, and anti-H3K9Me3 (1:100; Upstate Biotechnologies), affinity-purified rabbit anti-H3K4Me2, anti-H3K9Me1, and anti-H4K20Me2 (1:100; Abcam), monoclonal mouse anti-Drosophila HP1, C1A9 (Developmental Studies Hybridoma Bank; 1:50), and monoclonal mouse anti-GFP antibody (1:100; BD Bioscience). The preparations were washed twice in PBS + 400 mM NaCl + 0.2% NP-40 for 30 min. Goat anti-rabbit Alexa Fluor 488 and goat anti-mouse Alexa Fluor 568 (Molecular Probes) were used at 1:200 dilution in PBS + 0.1% Triton X-100. The final preparations were mounted in Vectashield mounting solution (Vector) and stained with DAPI (0.5 μg/mL) for DNA visualization. The preparations were examined using epifluorescence optics on an Olympus IX81 microscope, and images were captured and digitized using a high-resolution 12-bit Cooke Sensicam QE cooled CCD camera.
Immunoblot analyses of H1, HP1, and H3K9Me2 in Drosophila salivary gland lysates
Five pairs of salivary glands from larvae of each genotype were dissected and homogenized in 30 μL of Laemmli loading buffer. The crude lysates were boiled for 5 min and centrifuged. Total protein concentrations were determined by the Bradford assay, and ∼10 μg of protein was loaded per well on a 12% SDS-PAGE gel. Proteins were transferred to nitrocellulose membranes using an electro-blot apparatus at 120 mA for 2 h. The membranes were blocked for 1 h in the blocking buffer (LI-COR Bioscience). Rabbit anti-H1 (1:50,000), monoclonal anti-HP1 (1:3000), or rabbit anti-H3K9Me2 (1:1000) antibodies were incubated with the membranes for 3 h. Subsequently, the blots were washed in PBS + 0.1% Tween 20 and incubated with infrared dye-labeled secondary antibodies (1:15,000; LI-COR Bioscience). The blots were reprobed with mouse monoclonal anti-tubulin antibody, E7 (University of Iowa Hybridoma Bank; 1:500) and affinity-purified goat anti-histone H3 antibody (1:1000; Santa Cruz Biotechnologies), which were used as loading controls. Images were obtained and quantitated using the LI-COR Odyssey Infrared Imaging System. To verify that the measured Western signals were within the liner range of the instrument, we performed a titration with 4–24 μg of total protein from wild-type salivary glands loaded in each lane (Supplemental Fig. 7).
Quantitative real-time PCR
Total RNA from 10 pairs of salivary glands from larvae of each genotype was isolated by Trizol extraction (Invitrogen) and quantitated with a NanoDrop 1000 Spectrophotometer (Thermo Scientific). One microgram of total RNA was treated with RNAse-free DNase I (Promega), and random-primed cDNA was prepared using the SuperScript II kit (Invitrogen). Real-time quantitative PCR amplification reactions were carried out in an ABI Prism 7700 sequence detection system (Applied Biosystems). One-step RT–PCR was done using a SYBR Green Quantitative RT–PCR kit as per the manufacturer's instructions. Primer sequences are available in the Supplemental Material. To quantitate the expression levels, CT values of an endogenous reference gene, rp49, were included. All reactions were carried out in triplicate, along with no-template controls.
Acknowledgments
We are grateful to Jim Kadonaga (University of California at San Diego), Thomas Kornberg (University of California at San Francisco), Bruce Paterson (NIH), and Lori Wallrath (University of Iowa) for fly stocks, DNA constructs, and anibodies; and to Barbara Wakimoto (University of Washington) for primer sequences. We thank Elena Vershilova for technical assistance, and Konstantin Beirit, Slava Elagin, Michael Keogh, Alexandra Lusser, and members of the Fyodorov and Skoultchi laboratories for critical reading of the manuscript. We thank Laura Norwood for discussions and advice. This work was supported by grants from the NIH to D.V.F. (GM074233) and A.I.S. (CA079057). D.V.F. was a Scholar of the Sidney Kimmel Foundation for Cancer Research.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1749309.
Supplemental material is available at http://www.genesdev.org.
References
- Ananiev E.V., Barsky V.E. Elementary structures in polytene chromosomes of Drosophila melanogaster. Chromosoma. 1985;93:104–112. doi: 10.1007/BF00285853. [DOI] [PubMed] [Google Scholar]
- Bao X., Deng H., Johansen J., Girton J., Johansen K.M. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect variegation at pericentric sites in Drosophila heterochromatin. Genetics. 2007;176:1355–1358. doi: 10.1534/genetics.107.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H., Manoukian A.S., Perrimon N. Ectopic expression in Drosophila. Methods Cell Biol. 1994;44:635–654. doi: 10.1016/s0091-679x(08)60936-x. [DOI] [PubMed] [Google Scholar]
- Brown D.T., Alexander B.T., Sittman D.B. Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 1996;24:486–493. doi: 10.1093/nar/24.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I.L., Cryderman D.E., Gilmour D.S., Pile L.A., Wallrath L.L., Weber J.A., Elgin S.C. Analysis of Drosophila chromatin structure in vivo. Methods Enzymol. 1999;304:462–496. doi: 10.1016/s0076-6879(99)04028-8. [DOI] [PubMed] [Google Scholar]
- Corona D.F., Siriaco G., Armstrong J.A., Snarskaya N., McClymont S.A., Scott M.P., Tamkun J.W. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:2011–2021. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink A.K., Linsk R., Birchler J.A. The Lighten up (Lip) gene of Drosophila melanogaster, a modifier of retroelement expression, position effect variegation and white locus insertion alleles. Genetics. 1994;138:153–163. doi: 10.1093/genetics/138.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer J.R., Wallrath L.L. Mechanisms of HP1-mediated gene silencing in Drosophila. Development. 2004;131:3571–3580. doi: 10.1242/dev.01223. [DOI] [PubMed] [Google Scholar]
- Daujat S., Zeissler U., Waldmann T., Happel N., Schneider R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J. Biol. Chem. 2005;280:38090–38095. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- Dej K.J., Ahn C., Orr-Weaver T.L. Mutations in the Drosophila condensin subunit dCAP-G: Defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics. 2004;168:895–906. doi: 10.1534/genetics.104.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Zhang W., Bao X., Martin J.N., Girton J., Johansen J., Johansen K.M. The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma. 2005;114:173–182. doi: 10.1007/s00412-005-0006-8. [DOI] [PubMed] [Google Scholar]
- Deng H., Bao X., Zhang W., Girton J., Johansen J., Johansen K.M. Reduced levels of Su(var)3-9 but not Su(var)2-5 (HP1) counteract the effects on chromatin structure and viability in loss-of-function mutants of the JIL-1 histone H3S10 kinase. Genetics. 2007;177:79–87. doi: 10.1534/genetics.107.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitri P. Cytogenetic analysis of the second chromosome heterochromatin of Drosophila melanogaster. Genetics. 1991;127:553–564. doi: 10.1093/genetics/127.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J.A., Kosmidou E., Morgan A., Jackson S.P. Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol. Cell. 2003;11:1685–1692. doi: 10.1016/s1097-2765(03)00197-7. [DOI] [PubMed] [Google Scholar]
- Eberl D.F., Duyf B.J., Hilliker A.J. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster. Genetics. 1993;134:277–292. doi: 10.1093/genetics/134.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A., Lein S., Schotta G., Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C., James T.C., Foster-Hartnett D.M., Hartnett T., Ngan V., Elgin S.C. Mutation in a heterochromatin-specific chromosomal protein is associated with suppression of position-effect variegation in Drosophila melanogaster. Proc. Natl. Acad. Sci. 1990;87:9923–9927. doi: 10.1073/pnas.87.24.9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Nikitina T., Morin-Kensicki E.M., Zhao J., Magnuson T.R., Woodcock C.L., Skoultchi A.I. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol. 2003;23:4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- Fanti L., Pimpinelli S. HP1: A functionally multifaceted protein. Curr. Opin. Genet. Dev. 2008;18:169–174. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Fyodorov D.V., Blower M.D., Karpen G.H., Kadonaga J.T. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes & Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale T.K., Contreras A., Morrison A.J., Herrera R.E. Phosphorylation of the linker histone H1 by CDK regulates its binding to HP1α. Mol. Cell. 2006;22:693–699. doi: 10.1016/j.molcel.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Hearn M.G., Hedrick A., Grigliatti T.A., Wakimoto B.T. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics. 1991;128:785–797. doi: 10.1093/genetics/128.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellauer K., Sirard E., Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1. J. Biol. Chem. 2001;276:13587–13592. doi: 10.1074/jbc.M011196200. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Position-effect variegation after 60 years. Trends Genet. 1990;6:422–426. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- Hilliker A.J. Genetic analysis of the centromeric heterochromatin of chromosome 2 of Drosophila melanogaster: Deficiency mapping of EMS-induced lethal complementation groups. Genetics. 1976;83:765–782. doi: 10.1093/genetics/83.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A.J., Holm D.G. Genetic analysis of the proximal region of chromosome 2 of Drosophila melanogaster. I. Detachment products of compound autosomes. Genetics. 1975;81:705–721. doi: 10.1093/genetics/81.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe M., Dimitri P., Berloco M., Wakimoto B.T. Cis-effects of heterochromatin on heterochromatic and euchromatic gene activity in Drosophila melanogaster. Genetics. 1995;140:1033–1045. doi: 10.1093/genetics/140.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Keles S., Prestel M., Hochheimer A., Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes & Dev. 2007;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T.C., Eissenberg J.C., Craig C., Dietrich V., Hobson A., Elgin S.C. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- Konev A.Y., Yan C.M., Acevedo D., Kennedy C., Ward E., Lim A., Tickoo S., Karpen G.H. Genetics of P-element transposition into Drosophila melanogaster centric heterochromatin. Genetics. 2003;165:2039–2053. doi: 10.1093/genetics/165.4.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Laybourn P.J., Kadonaga J.T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lifton R.P., Goldberg M.L., Karp R.W., Hogness D.S. The organization of the histone genes in Drosophila melanogaster: Functional and evolutionary implications. Cold Spring Harb. Symp. Quant. Biol. 1978;42:1047–1051. doi: 10.1101/sqb.1978.042.01.105. [DOI] [PubMed] [Google Scholar]
- Lin Q., Inselman A., Han X., Zhang W., Handel M.A., Skoultchi A.I. Reductions in linker histone levels are tolerated in developing spermatocytes but cause changes in specific gene expression. J. Biol. Chem. 2004;279:23525–23535. doi: 10.1074/jbc.M400925200. [DOI] [PubMed] [Google Scholar]
- Lu B.Y., Emtage P.C., Duyf B.J., Hilliker A.J., Eissenberg J.C. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.Q., Liu L.P., Hess D., Rietdorf J., Sun F.L. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes & Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.L., Oulad-Abdelghani M., Ortiz J.A., Remboutsika E., Chambon P., Losson R. Heterochromatin formation in mammalian cells: Interaction between histones and HP1 proteins. Mol. Cell. 2001;7:729–739. doi: 10.1016/s1097-2765(01)00218-0. [DOI] [PubMed] [Google Scholar]
- Patterton H.G., Landel C.C., Landsman D., Peterson C.L., Simpson R.T. The biochemical and phenotypic characterization of Hho1p, the putative linker histone H1 of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:7268–7276. doi: 10.1074/jbc.273.13.7268. [DOI] [PubMed] [Google Scholar]
- Pauli A., Althoff F., Oliveira R.A., Heidmann S., Schuldiner O., Lehner C.F., Dickson B.J., Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev. Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflumm M.F., Botchan M.R. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 1997;26:83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- Ramon A., Muro-Pastor M.I., Scazzocchio C., Gonzalez R. Deletion of the unique gene encoding a typical histone H1 has no apparent phenotype in Aspergillus nidulans. Mol. Microbiol. 2000;35:223–233. doi: 10.1046/j.1365-2958.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- Reuter G., Spierer P. Position effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- Rice J.C., Briggs S.D., Ueberheide B., Barber C.M., Shabanowitz J., Hunt D.F., Shinkai Y., Allis C.D. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Richards E.J., Elgin S.C. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Robinson P.J., Rhodes D. Structure of the ‘30 nm’ chromatin fibre: A key role for the linker histone. Curr. Opin. Struct. Biol. 2006;16:336–343. doi: 10.1016/j.sbi.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., Rea S., Jenuwein T., Dorn R., Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y.B., Pirrotta V. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Shen X., Gorovsky M.A. Linker histone H1 regulates specific gene expression but not global transcription in vivo. Cell. 1996;86:475–483. doi: 10.1016/s0092-8674(00)80120-8. [DOI] [PubMed] [Google Scholar]
- Shen X., Yu L., Weir J.W., Gorovsky M.A. Linker histones are not essential and affect chromatin condensation in vivo. Cell. 1995;82:47–56. doi: 10.1016/0092-8674(95)90051-9. [DOI] [PubMed] [Google Scholar]
- Urata Y., Parmelee S.J., Agard D.A., Sedat J.W. A three-dimensional structural dissection of Drosophila polytene chromosomes. J. Cell Biol. 1995;131:279–295. doi: 10.1083/jcb.131.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushinsky S.C., Bussey H., Ahmed A.A., Wang Y., Friesen J., Williams B.A., Storms R.K. Histone H1 in Saccharomyces cerevisiae. Yeast. 1997;13:151–161. doi: 10.1002/(SICI)1097-0061(199702)13:2<151::AID-YEA94>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Van Holde K.E. Chromatin. Springer Verlag; New York: 1988. [Google Scholar]
- Wakimoto B.T., Hearn M.G. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics. 1990;125:141–154. doi: 10.1093/genetics/125.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L.L. Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Jin Y., Johansen J., Johansen K.M. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell. 2001;105:433–443. doi: 10.1016/s0092-8674(01)00325-7. [DOI] [PubMed] [Google Scholar]
- Wei Q., Rong Y., Paterson B.M. Stereotypic founder cell patterning and embryonic muscle formation in Drosophila require nautilus (MyoD) gene function. Proc. Natl. Acad. Sci. 2007;104:5461–5466. doi: 10.1073/pnas.0608739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal T., Reuter G. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics. 2002;160:609–621. doi: 10.1093/genetics/160.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A.P. Histone H1. Int. J. Biochem. Cell Biol. 1997;29:1463–1466. doi: 10.1016/s1357-2725(97)00026-5. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. Chromatin: Structure and function. Academic Press; New York: 1998. [Google Scholar]
- Woodcock C.L., Skoultchi A.I., Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- Zhao T., Heyduk T., Eissenberg J.C. Phosphorylation site mutations in heterochromatin protein 1 (HP1) reduce or eliminate silencing activity. J. Biol. Chem. 2001;276:9512–9518. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]