Abstract
The gaseous plant hormone ethylene can trigger myriad physiological and morphological responses in plants. While many ethylene signaling pathway components have been identified and characterized, little is known about the function of the integral membrane protein ETHYLENE-INSENSITIVE2 (EIN2), a central regulator of all ethylene responses. Here, we demonstrate that Arabidopsis thaliana EIN2 is a protein with a short half-life that undergoes rapid proteasome-mediated protein turnover. Moreover, EIN2 protein accumulation is positively regulated by ethylene. We identified two F-box proteins, EIN2 TARGETING PROTEIN1 (ETP1) and EIN2 TARGETING PROTEIN2 (ETP2), that interact with the EIN2 C-terminal domain (EIN2-CEND), which is highly conserved and sufficient to activate most ethylene responses. Overexpression of ETP1 or ETP2 disrupts EIN2 protein accumulation, and these plants manifest a strong ethylene-insensitive phenotype. Furthermore, knocking down the levels of both ETP1 and ETP2 mRNAs using an artificial microRNA (amiRNA) leads to accumulation of EIN2 protein, resulting in plants that display constitutive ethylene response phenotypes. Finally, ethylene down-regulates ETP1 and ETP2 proteins, impairing their ability to interact with EIN2. Thus, these studies reveal that a complex interplay between ethylene, the regulation of ETP1/ETP2 F-box proteins, and subsequent targeting and degradation of EIN2 is essential for triggering ethylene responses in plants.
Keywords: Hormone, protein degradation, signal transduction, plant
In plants, ethylene (C2H4) is a regulator of various physiological and morphological responses, including inhibition of cell expansion, promotion of leaf and flower senescence, induction of fruit ripening and abscission, resistance to pathogen infection, and adaptation to stress conditions (Bleecker and Kende 2000; Guo and Ecker 2004). The molecular dissection of ethylene signal transduction began with genetic screens based on the well-documented triple response phenotype of ethylene-treated etiolated Arabidopsis seedlings. Through these screens, many ethylene mutants have been obtained, including the ethylene-insensitive mutants etr1, ein2, ein3, ein5 (Bleecker et al. 1988; Guzman and Ecker 1990; Roman et al. 1995); the ethylene-overproducing mutants eto1, eto2, eto3, and the ethylene constitutive response mutant ctr1 (Guzman and Ecker 1990; Kieber et al. 1993). Initial studies of these mutants have revealed a mostly linear framework for the ethylene-signaling pathway, leading from ethylene perception at the membrane to transcriptional activation in the nucleus (Stepanova and Ecker 2000; Chen et al. 2002; Guo and Ecker 2004).
Ethylene is perceived by a family of membrane-bound, endoplasmic reticulum-located receptors—ETHYLENE RESPONSE1 (ETR1), ETHYLENE RESPONSE SENSOR1 (ERS1), ETHYLENE RESPONSE2 (ETR2), ETHYLENE-INSENSITIVE4 (EIN4), and ETHYLENE RESPONSE SENSOR2 (ERS2)—that are similar in sequence and structure to bacterial two-component histidine kinases (Chang et al. 1993; Hua et al. 1998; Kendrick and Chang 2008). Each receptor has an N-terminal membrane-spanning domain that binds ethylene with a copper cofactor provided by the RESPONSIVE TO ANTAGONIST1 (RAN1) copper transporter (Hirayama et al. 1999). Briefly, in the absence of ethylene gas, the ethylene receptors repress downstream responses through interaction with CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) (Gao et al. 2003), which is a member of Raf kinase family that also acts as a negative regulator of the downstream ethylene signaling pathway (Kieber et al. 1993). In the presence of ethylene, the receptors stop repressing ethylene response through inactivation of CTR1. Additionally, ETHYLENE-INSENSITIVE2 (EIN2) is derepressed and positively regulates the levels of ETHYLENE-INSENSITIVE3 (EIN3) and ETHYLENE-INSENSITIVE3-LIKE1 (EIL1), the key transcription factors of ethylene signaling pathway, which results in the activation of transcription of ethylene-responsive genes (Chao et al. 1997; Solano et al. 1998). Recently, numerous studies have expanded the linear view of ethylene signaling pathway. For instance, a new protein, REVERSION-TO-ETHYLENE SENSITIVITY1 (RTE1), which is colocalized with the ethylene receptor ETR1, was identified as a positive regulator of ETR1 function, but the connection between RTE1 and ETR1 is still under investigation (Solano et al. 1998; Resnick et al. 2006; Dong et al. 2008). Additionally, a number of groups found that post-transcriptional regulation of protein levels is a key mechanism of modulating EIN3 activity by ethylene. Specifically, they found that ubiquitin/proteasome-mediated degradation negatively regulates ethylene responses by targeting EIN3 for turnover through two F-box proteins, EIN3-BINDING F-BOX PROTEIN1 (EBF1) and EIN3-BINDING F-BOX PROTEIN2 (EBF2) (Guo and Ecker 2003; Potuschak et al. 2003; Gagne et al. 2004). Interestingly, negative feedback regulation exists in this step of the ethylene signal-transduction pathway, in that EIN3 targets the promoter of EBF2 to control its expression level likely allowing fine-tuning of ethylene responses (Binder et al. 2007; Konishi and Yanagisawa 2008). Most recently, an alternative ethylene signaling pathway has been proposed that is based on studies of ethylene responses in Arabidopsis protoplasts (Varma Penmetsa et al. 2008; Yoo et al. 2008). Characterization of these genes/proteins has provided additional insight into the molecular mechanisms that may underlie the response of plants to ethylene gas.
EIN2 is an integral membrane protein with limited similarity in the N terminus to mammalian NRAMP metal transporters; the ∼850-amino-acid C terminus of EIN2 is conserved in all the known EIN2 homologs of angiosperms (Varma Penmetsa et al. 2008). Interestingly, expression of a portion of the C terminus (EIN2-CEND) is sufficient to constitutively activate ethylene and stress responses both in Arabidopsis (Alonso et al. 1999) and in Medicago (Mt) (Varma Penmetsa et al. 2008). Phenotypic, epistatic, and biochemical analyses place EIN2 in a central position in ethylene signaling pathway (Roman et al. 1995; Johnson and Ecker 1998; Guo and Ecker 2003). Identification of the signal perceived by EIN2, isolation of the EIN2-interacting proteins, and understanding its mode of action remain of major interest within plant hormone biology.
In this study, we demonstrate that EIN2, the key positive regulator of ethylene signal transduction, is a short half-life protein and it undergoes rapid proteasome-mediated turnover. We also identify two F-box proteins, EIN2 TARGETING PROTEIN1 (ETP1) and EIN2 TARGETING PROTEIN2 (ETP2), as the key regulators of EIN2 protein stability, and through the regulation of EIN2 they negatively affect ethylene signal transduction. Overall, our results suggest ethylene responses are specifically modulated in an EIN2 protein level-dependent manner, and reveal a complex interplay between ethylene, the regulation of ETP1/ETP2 F-box proteins, and subsequent targeting and degradation of EIN2 as essential for triggering appropriate ethylene responses in plants.
Results
EIN2 is a short half-life protein and is positively influenced by ethylene
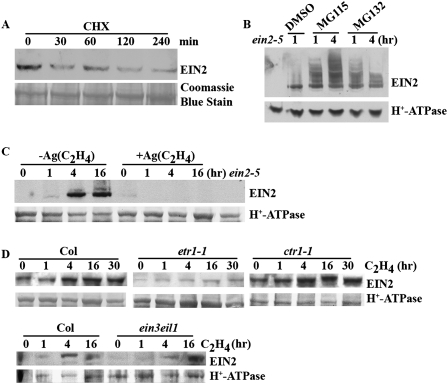
A previous study demonstrated that EIN2 mRNA is not altered in response to ethylene (Alonso et al. 1999). Here we examined whether EIN2 may be subject to post-transcriptional regulation. We first tested its stability by Western blotting after treatment with cycloheximide (CHX), which inhibits de novo protein biosynthesis. We found that EIN2 levels dramatically decreased after 30 min of CHX treatment and remained barely detectable for the subsequent 2 h (Fig. 1A). The rapid reduction of EIN2 protein levels indicates that EIN2 is a short-lived protein, with a half-life of ∼30 min or less. To further test whether the level of EIN2 is regulated by 26S proteasome-mediated protein turnover, we examined EIN2 protein levels from wild-type Col-0 etiolated seedlings treated with specific proteasome inhibitors MG115 or MG132 (Lee and Goldberg 1998). As shown in Figure 1B, after 1 h of MG132 or MG115 treatment, the accumulation of EIN2 protein markedly increased. Interestingly, this analysis also revealed the presence of higher molecular weight forms of the protein, suggesting that EIN2 might be modified, possibly by ubiquitinylation. These results suggested that EIN2 is a short half-life protein whose turnover is mediated by the proteosome.
Figure 1.
EIN2 is a short-lived protein whose accumulation is essential for ethylene responses. (A) EIN2 is a short half-life protein. Etiolated wild-type Col-0 seedlings grown in air supplied in the presence of 100 μM CHX for different amounts of time. Total protein lysates were subjected to immunoblotting with EIN2 antiserum. Coomassie blue staining of total membrane proteins was used as a lane loading control. (B) The protein level of EIN2 is stabilized by specific proteasome inhibitors. Total membrane protein extracts were derived from wild-type Col-0 etiolated seedlings treated with mock (1% DMSO), MG132 (50 μM), MG115 (50 μM) for 1 or 4 h, and used for immunoblot assays. (C) EIN2 accumulation is abolished by Ag+ treatment. Etiolated wild-type seedlings were grown on MS media without or with 100 μM AgNO3 for 3 d and treated with air or ethylene for the indicated amount of time. (D) EIN2 protein level is impaired in etr1-1 mutant seedlings and constitutively accumulates in ctr1-1 mutant seedlings. Wild-type Col-0, ctr1-1, etr1-1, and ein3-1eil1-1 mutant etiolated seedlings were grown on MS media in air for 3 d and subsequently treated with ethylene gas for the indicated amount of time. Western blotting using an anti-H+-ATPase antibody was used as a lane loading control.
Since EIN2 is a key regulator of the ethylene signaling pathway, we tested the effect of treatment with exogenous ethylene gas on EIN2 protein stability. To do this, we monitored the level of EIN2 protein in plants treated with ethylene (10 parts per million [ppm]) for increasing amounts of time with or without the presence of silver (Ag+), which is a potent inhibitor of ethylene action (Abeles et al. 1992). We found that the level of EIN2 protein rapidly increased in response to ethylene treatment, but in the presence of silver ion, EIN2 protein accumulation was completely inhibited (Fig. 1C). In addition, to test the effect of ethylene on EIN2 protein, we monitored EIN2 levels in wild-type Col-0 seedlings grown on MS medium supplemented with either 1-aminocyclopropane-1-carboxylic acid (ACC), an ethylene precursor, or aminoethoxyvinylglycine (AVG), an inhibitor of ethylene biosynthesis. EIN2 protein was not present at detectable levels in the presence of AVG. In contrast, the level of EIN2 was elevated in the presence of ACC (Supplemental Fig. S1), further suggesting that ethylene stabilizes EIN2 protein. To gain further insight into kinetics of EIN2 protein induction by ethylene, we examined EIN2 levels in different ethylene mutants. In wild-type Col-0 seedlings, the level of EIN2 protein increased in response to ethylene treatment (Fig. 1D). However, in etr1-1 mutant seedlings, EIN2 accumulation was not observed even with 30-h ethylene treatment. In contrast, the level of EIN2 was constitutively elevated in ctr1-1 mutant seedlings. Additionally, EIN2 levels were further increased in ctr1-1 mutant seedlings by ethylene treatment. In the ein3eil1 double mutant, EIN2 accumulation was similar to that seen for wild-type Col-0 seedlings (Fig. 1D). These results demonstrated that the accumulation of EIN2 protein is dramatically altered in the different ethylene response mutants. Specifically, the level of EIN2 protein is decreased in ethylene-insensitive mutants, but increased in the ethylene constitutive response mutants. Taken together, the results demonstrate that EIN2 protein is stabilized and accumulates by the presence of exogenous ethylene gas. This accumulation of EIN2 is dependent on an intact ethylene signaling pathway upstream of EIN3/EIL1. Overall, these results suggest that EIN2 protein levels are positively correlated with the ethylene response.
Two novel F-box proteins interact with the C-terminal end of EIN2
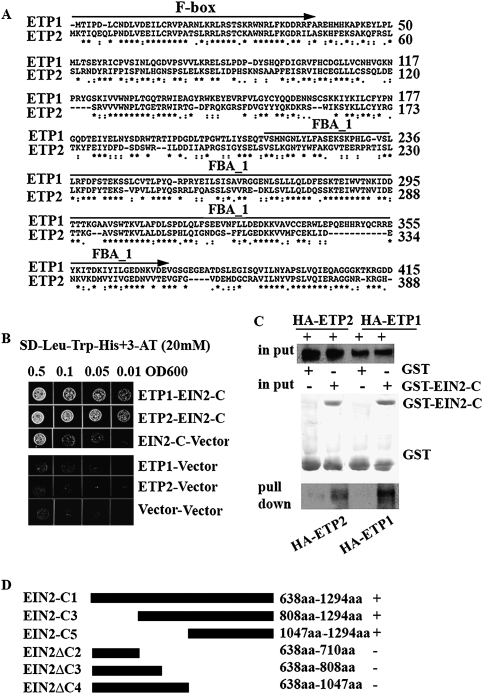
To further understand the post-transcriptional regulation of EIN2 levels, we performed a yeast two-hybrid screen to identify proteins that potentially interact with the EIN2-CEND. As a result, a novel F-box protein ETP1 (At3g18980, GB:Q9LJ68; GB:Q8LB99) was identified. Sequence analysis reveals the presence of a paralogous gene, ETP2 (At3g18910, GB:Q9LJ34), that is 50% identical to ETP1 at the amino acid sequence level (Fig. 2A). Therefore, we tested whether the ETP2 protein might also interact with EIN2-CEND. As shown in Figure 2, B and C, both directed yeast two-hybrid assays and pull-down experiments demonstrated that EIN2-CEND was able to interact with both F-box proteins, ETP1 and ETP2.
Figure 2.
Two novel F-box proteins, ETP1 and ETP2, interact with EIN2-CEND (EIN2-C). (A) Alignment of ETP1 and ETP2 amino acid sequences generated with the ClustalW program. The positions of amino acid residues are indicated with numbers; asterisks and dots indicate identical and conserved amino acids, respectively. The putative F-box motif and the FBA_1 (F-box protein-associated) domain are indicated by arrow above the sequences (B) EIN2-CEND interacts with ETP1 and ETP2 in yeast. Growth on selective plates lacking adenine, histidine, tryptophan with 20 mM 3-AT (−Leu, −Trp, −His, +3AT) and on control plates lacking only tryptophan and leucine (−Trp, −Leu) is shown. (C) EIN2 interacts with ETP1 and ETP2 in vitro. GST-EIN2-CEND (GST-EIN2-C) fusion protein and GST alone protein were purified from E. coli. These proteins, as well as GST alone, were assayed to pull-down with in vitro translated and HA-tagged ETP1 and ETP2 proteins. The same quantities of the GST fusion proteins (bottom panel) and the same amount of HA-tagged ETP1 or ETP2 (middle panel) were used as inputs. (Top panel) The HA-tagged ETP1 and ETP2 were detected by anti-HA antibody. A plus sign (+) indicates the addition of protein; a minus sign (−) indicates the protein was not added. (D) EIN2 highly conserved CEND (EIN2-C5) is sufficient to interact with ETP1 and ETP2 in yeast. The diagrams indicate different deletions of EIN2 CEND. A plus sign (+) indicates interaction; a minus sign (−) indicates no interaction. (WB) Western blot.
We further characterized the domain of EIN2-CEND that interacts with ETP1 and ETP2. Based on the alignment of EIN2 proteins from different plant species (Supplemental Fig. S3), a series of deletion mutants of EIN2-CEND was generated to map the region of EIN2 required for interaction with ETP1 and ETP2. As shown in Figure 2D and Supplemental Figure S2, A and B, EIN2-CEND1 (EIN2-C1), EIN2-CEND3 (EIN2-C3), and EIN2-CEND5 (EIN2-C5) interact with both ETP1 and ETP2. In contrast, we found that the EIN2ΔCEND2 (EIN2ΔC2), EIN2ΔCEND3 (EIN2ΔC3), and EIN2ΔCEND4 (EIN2ΔC4) completely lost the ability to interact with EIN2 (Supplemental Fig. S2A,B). These results demonstrated that the most highly conserved region of EIN2 (EIN2-C5), the last ∼250 amino acids (Supplemental Fig. S3), is both necessary and sufficient for the interaction of EIN2 with ETP1 and ETP2. Additionally, these results suggested that other portions of EIN2-CEND are not essential for these interactions but may enhance them.
ETP1 and ETP2 are the regulators of EIN2 stability
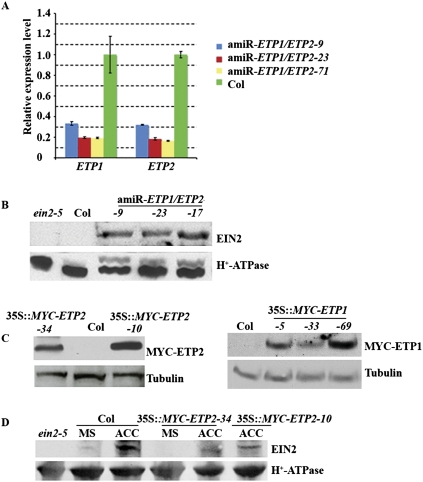
To test whether ETP1 and ETP2 are involved in the regulation of EIN2 protein turnover, we identified T-DNA insertion lines for mutants in both ETP1 and ETP2: etp1, etp2-1, etp2-2 (Supplemental Fig. S4A; Alonso et al. 2003). The response to ethylene of these mutant seedlings was tested. etp1 mutant seedlings manifested a slight ethylene hypersensitivity (Supplemental Fig. S4D), while the ethylene response of etp2 mutant seedlings was similar to that of wild-type Col-0 (Supplemental Fig. S4B,C). Because of their sequence similarity, it is possible that ETP1 and ETP2 may function redundantly. To test this hypothesis, we utilized an artificial microRNA (amiRNA) (Schwab et al. 2006) directed at knocking down the levels of both ETP1 and ETP2 mRNAs. A number of independent ETP1/ETP2 knockdown (amiR-ETP1/ETP2) transgenic lines were isolated. As shown in Figure 3A, the gene expression levels of ETP1 and ETP2 were significantly reduced in these knockdown lines to 15%–30% of the level in wild-type Col-0 plants as measured by quantitative PCR (qPCR) analysis. Since we found that ETP1 and ETP2 were able to interact directly with EIN2, we investigated whether ETP1 and ETP2 might be involved in EIN2 regulation. To do this, we examined the level of EIN2 protein in amiR-ETP1/ETP2 plants. We found that EIN2 protein accumulation was greatly increased in amiR-ETP1/ETP2 plants compared with wild-type Col-0 plants (Fig. 3B; Supplemental Fig. S5), suggesting that deficiency of ETP1 and ETP2 RNAs results in accumulation of EIN2 protein. To further illuminate the functions of ETP1 and ETP2 in the regulation EIN2 protein, we constructed transgenic plants containing either ETP1 or ETP2 under the control of the Cauliflower Mosaic Virus 35S promoter (35S), allowing constitutively high levels of expression for these two genes; the RNA levels of ETP1 and ETP2 were fivefold to 10-fold increased in comparison with wild-type plants (Supplemental Fig. S7). Compared with wild-type Col-0 plants, EIN2 protein levels were greatly reduced in plants overexpressing ETP1 or ETP2 (Fig. 3C). More interestingly, EIN2 accumulation was barely detectable in the ETP1 or ETP2 overexpressed seedlings even upon ACC treatment (Fig. 3D), suggesting that ethylene-dependent EIN2 accumulation is impaired by overexpression of ETP1 or ETP2. Taken together, these results demonstrate that ETP1 and ETP2 are negative regulators of EIN2 protein stability.
Figure 3.
ETP1 and ETP2 are regulators of EIN2 levels. (A) qPCR analysis of ETP1 and ETP2 transcript levels in amiR-ETP1/ETP2 mutant plants. Total RNA was extracted from the leaves of 3-wk-old light-grown plants. The data were normalized to the corresponding actin (input) controls. The data shown are the means ± SD of three independent experiments. (B) EIN2 protein accumulates in amiR-ETP1/ETP2 mutant plants. (C) MYC-ETP1 or ETP2 accumulates in 35S∷MYC-ETP1 or MYC-ETP2 transgenic plants, respectively. Wild-type Col-0 Arabidopsis plants were transformed with a binary vector carrying the MYC tag that fused with ETP1 or ETP2 open reading frame. Total proteins from 35S∷MYC-ETP1 or MYC-ETP2 transgenic plants were subjected to immunoblotting with anti-MYC antibody. The same membranes were stripped and subjected to immunoblotting with an anti-tubulin antibody as loading control. (D) Overexpression of ETP2 causes reduction of EIN2 protein. Wild-type Col-0 and ETP2 overexpression plants grown on soil for 3 wk and the total protein lysates from leaves were subjected to immunoblotting with EIN2 antiserum. The same membrane was stripped and subjected to immunoblotting with an anti-H+-ATPase antibody as a lane loading control.
Knockdown of ETP1 and ETP2 results in constitutive ethylene response
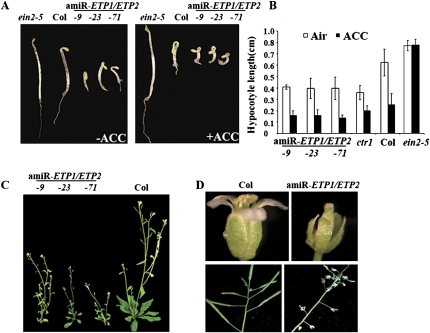
To better understand the molecular consequences of reduced ETP1/ETP2 RNA levels, we interrogated the transcriptome of wild-type Col-0 and amiR-ETP1/ETP2 plants before and after 4 h of treatment with ethylene gas using Affymetrix ATH1 arrays. Total RNA was prepared from 3-wk-old leaves of wild-type Col-0 and amiR-ETP1/ETP2 plants with or without 4 h of ethylene treatment. The microarray data revealed that ∼40% of genes with significant changes in expression of amiR-ETP1/ETP2 knockdown lines compared with wild-type Col-0 plants overlapped with genes whose expression levels change in wild-type Col-0 plants upon ethylene treatment (Supplemental Fig. S6; Supplemental Tables S1–S4), suggesting that ETP1 and ETP2 specifically affect numerous ethylene-responsive genes. Therefore, we examined the ethylene response of amiR-ETP1/ETP2 3-d-old etiolated seedlings. When grown on MS medium, the amiR-ETP1/ETP2 seedlings manifested a typical constitutive ethylene response phenotype (Fig. 4A–C). Interestingly, the amiR-ETP1/ETP2 seedlings still showed some response to exogenously added ethylene (Fig. 4B). This residual response may be a consequence of the remaining of ETP1 and ETP2 mRNA present in these knockdown lines or might also be due to redundancy of function for other members of this family of proteins.
Figure 4.
Constitutive ethylene response phenotypes ETP1 or ETP2 knockdown plants support a function in ethylene signaling. (A) Ethylene response phenotype of 3-d-old etiolated seedlings of amiR-ETP1/ETP2 plants. The plants were grown on MS media supplied with (left panel) or without (right panel) 10 μM ACC in the dark for 3 d. (B) Measurement of hypocotyl length of amiR-ETP1/ETP2 3-d-old etiolated seedlings. Each measurement is the average length (mean ± standard error) of >10 hypocotyls. (C) Phenotype of 5-wk-old amiR-ETP1/ETP2 plants grown on soil. (D) amiR-ETP1/ETP2 mutant plants flower and silique morphology. The plants were grown on soil with 16-h light and 8-h dark, the flowers and siliques were photographed for 8-wk-old plants or 10-wk-old plants.
As expected for plants that show a constitutive ethylene response phenotype, the adult amiR-ETP1/ETP2 plants had small rosettes and dwarfed growth habit, as well as displayed abnormal flowers whose gynoecium protrude from the unopened floral buds (Fig. 4C). The decreased level of ETP1 and ETP2 RNA in amiR-ETP1/ETP2 plants resulted in severe sterility, a phenotype that was also observed for plants that overexpress EIN2-CEND (Alonso et al. 1999). Under our experimental growth conditions wild-type plants typically produce ∼40 seeds per silique. However, in amiR-ETP1/ETP2 plants most siliques produce no seed although a few siliques do produce two to three seeds with an average of 20 seeds per plant. Overall, these results demonstrated that, in the absence of ETP1 or ETP2, plants manifest a constitutive ethylene-responsive phenotype, suggesting that ETP1 and ETP2 negatively regulate ethylene response through the degradation of EIN2.
Overexpression of ETP1 or ETP2 leads to a reduction in sensitivity to ethylene
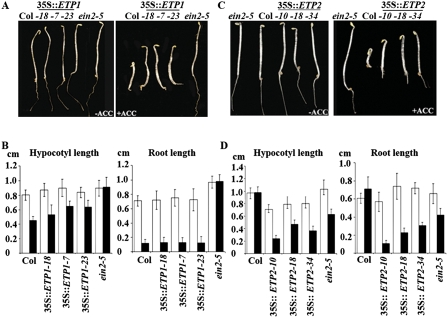
To further confirm the function of ETP1 and ETP2 in the ethylene signaling pathway, we examined the ethylene response phenotype of 3-d-old etiolated seedlings overexpressing ETP1 or ETP2. As demonstrated in Figure 5, A and B, overexpression of ETP1 resulted in a partial ethylene-insensitive phenotype of the hypocotyl, while the roots of these plants did not show any difference in ethylene response compared with wild-type Col-0 seedlings. Interestingly, overexpression of ETP2 resulted in a strong ethylene-insensitive phenotype both in roots and hypocotyls (Fig. 5C,D). The ethylene-insensitive phenotype displayed by plants overexpressing ETP2 is consistent with a greater reduction in EIN2 protein levels (Fig. 3D). These results demonstrate that overexpressing ETP1 or ETP2 results in significant ethylene-insensitive phenotypes, which further suggests that ETP1 and ETP2 negatively regulate ethylene response through their regulation of EIN2 levels.
Figure 5.
Ethylene-insensitive phenotypes ETP1 or ETP2 overexpression plants suggest a function in ethylene signaling. Phenotypes of 3-d-old etiolated seedlings overexpressing ETP1 (A) or ETP2 (C). Seedlings were grown on MS media supplemented without (left panel) or with 10 μM ACC (right panel) in dark for 3 d. Measurement of hypocotyl and root length for etiolated seedlings overexpressing ETP1 (B) or ETP2 (D). Three-day-old etiolated seedlings grown on the MS media supplemented with or without 10 μM ACC. Each measurement is the average length (mean ± standard error) of >20 hypocotyls or roots.
Ethylene negatively regulates ETP1 and ETP2 protein levels
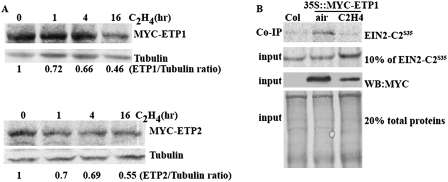
To better understand how ethylene gas regulates ETP1 and ETP2, the RNA expression of ETP1 and ETP2 was examined after different times of ethylene treatment. We found that the expression of ETP1 and ETP2 RNA was unaffected even with prolonged ethylene treatment (Supplemental Fig. S8). Therefore, we further examined ETP1 and ETP2 protein levels upon different times of ethylene treatment. As shown in Figure 6A, the protein levels of both ETP1 and ETP2 were down-regulated upon treatment with exogenous ethylene. In addition, a coimmunoprecipitation assay was used to examine the stability of the interaction between EIN2 and ETP1 upon ethylene treatment. An equivalent amount of S35-labeled EIN2-C2 from in vitro transcriptional/translational system was coimmunoprecipitated with anti-MYC antibody in the presence of total protein prepared from 35S∷MYC-ETP1 transgenic plants treated with or without ethylene. As shown in Figure 6B, the level of ETP1 protein was decreased by added ethylene, resulting in less EIN2 coimmunoprecipitating with ETP1. These findings suggest that ethylene may perturb the interaction between EIN2 and ETP1 through down-regulation of ETP1 protein levels, providing an additional layer of complexity of EIN2 regulation.
Figure 6.
Ethylene plays a negative role in ETP1 and ETP2 protein expression. (A) Protein levels of MYC-ETP1 (top panel) and MYC-ETP2 (bottom panel) after various times of ethylene treatment. The total protein extracts were subjected to immunoblotting with anti-MYC antibody. The same membranes were stripped and reprobed with an anti-tubulin antibody. (B) The interaction of ETP1 and EIN2 is affected by ethylene. Total protein extracts from 35S∷MYC-ETP1 transgenic plants with or without ethylene treatment were incubated with equal excessive amounts of 35S-methionine-labeled EIN2-C2 protein from in vitro transcriptional/translational system, and subsequently immunoprecipitated with an anti-MYC antibody. The EIN2-C235S was detected by autoradiography. (WB) Western blot.
Discussion
EIN2 protein level plays a key role in ethylene signaling
Our biochemical and genetic studies described here demonstrate ethylene responses in plants are facilitated by control of EIN2 protein turnover through the 26S proteasome pathway. Notably, the stability of EIN2 protein is decreased drastically in 30 min with CHX treatment. In contrast, EIN2 is stabilized and accumulates in the presence of specific proteasome inhibitors MG132 or MG115. The ubiquitin pathway has been shown to be important for degrading membrane proteins but, in many cases, the proteasome is not involved. Instead, the proteins are shuttled to the lysosome. However, this latter pathway is insensitive to MG132, indicating that EIN2 is a short half-life protein and the protein is subjected to proteasome-mediated protein degradation. In support of these findings, a recent study found that the EIN2 C terminus was capable of interacting with the COP1/signalsome (Christians et al. 2008); however, the biochemical consequence of this interaction was not reported.
The ein2-null mutant completely loses ethylene response (Alonso et al. 1999). In contrast, overexpression of EIN2-CEND causes many ethylene-responsive phenotypes in adult plants (Alonso et al. 1999). Furthermore, the accumulation of EIN3 protein is completely blocked in the ein2 mutant (Guo and Ecker 2003). All of these studies have demonstrated that the EIN2 plays an irreplaceable function in the ethylene response pathway.
Since EIN2 mRNA levels are unaffected by treatment with exogenous ethylene (Alonso et al. 1999), we examined EIN2 protein levels in plants treated with ethylene and in various ethylene response mutants. We found that the constitutive ethylene-responsive mutant ctr1 significantly overaccumulated EIN2 protein relative to wild type at all time points tested, while there was no detectable accumulation of EIN2 in the ethylene-insensitive mutants etr1. Moreover, protein accumulation of EIN2 in ein3eil1 plants was much reduced compared with wild-type plants upon the ethylene treatment. Additionally, we found that the level of EIN2 protein positively correlates with the ethylene response phenotypes, suggesting that plants respond rapidly to exogenous ethylene by adjusting the level of EIN2 protein through its turnover. According to the current model, when plants are grown in air (absence of ethylene), the negative regulator CTR1 actively represses ethylene responses. Genetics evidence demonstrated that EIN2 is the first positive regulatory factor downstream from CTR1 (Ecker 1995; Guo and Ecker 2004), and our findings now demonstrate that EIN2 protein dramatically accumulates in the constitutive ethylene response mutant, ctr1, suggesting that the accumulation of EIN2 results in the constitutive ethylene response phenotype of these plants. However, the connection between CTR1 and EIN2 is unknown. One possibility is that, in the absence of ethylene, CTR1 may function in concert with other, yet-to-be identified protein(s) to prevent EIN2 from accumulating. Previous studies have shown that accumulation of EIN3 protein fully depends on the presence of EIN2 protein (Guo and Ecker 2003). We found that EIN3 protein also accumulates in the amiR-ETP1/ETP2 knockdown plants where EIN2 accumulates (H. Qiao and J.R. Ecker, unpubl.), which offers additional evidence that the protein level of EIN2 is crucial for EIN3 stability and ethylene signaling. Interestingly, we found the protein level of EIN2 does not become saturated until after 4 h of ethylene treatment, while EIN3 protein level is saturated after 1 h of ethylene treatment (Guo and Ecker 2003). The molecular mechanism behind these differences is unclear. However, a question of major biological interest is to uncover the link between EIN2 and EIN3 in the ethylene signaling pathway, which may shed light on the differences in timing for accumulation of protein levels.
EIN2 C-terminal end is of crucial importance to ethylene response
EIN2 is a ubiquitous protein and exists in all the plant species examined. Interestingly, an EIN2 homolog is even present in the algae, Chlamydomonas reinhardtii (H. Qiao and J.R. Ecker, unpubl.). Through most of the plant species, EIN2-CEND is the most highly conserved domain (Supplemental Fig. S3), suggesting this domain may be of high importance for ethylene signal transduction. Interestingly, one of the ein2 mutant alleles isolated previously that carries a single substitution (A to C at 3943) mutation in the CEND of EIN2 still manifests a strong ethylene-insensitive phenotype (Alonso et al. 1999). This mutant allele of EIN2 suggests that an intact EIN2-CEND is essential for maintaining normal EIN2 function in ethylene signaling.
To identify EIN2-interacting proteins and potentially fill in gaps within the ethylene signaling pathway, we used EIN2-CEND to perform a yeast two-hybrid screen. Among the potential interactors, an F-box protein, ETP1, was identified. Furthermore, both directed yeast two-hybrid and pull-down experiments demonstrated that EIN2-CEND interacts with ETP1 and its Arabidopsis homolog ETP2. In addition, we demonstrated that the most highly conserved domain of the EIN2-CEND is necessary and sufficient for the interaction of EIN2 with ETP1 and ETP2, and these ∼250 amino acids of EIN2 may be essential for regulation of the ethylene response. Overall, these results suggest that the interaction between EIN2-CEND and ETP1 or ETP2 is crucial to a proper ethylene response in Arabidopsis, and this regulation through C-terminal interaction with F-box proteins mediating protein turnover is important for ethylene response in most if not all plant species.
The regulation of EIN2 by ETP1 and ETP2 is integral to ethylene signaling
In plants, there are ∼700 F-box proteins (Gagne et al. 2002), and they are involved in a plethora of biological processes including plant hormone responses (Xie et al. 1998; Gray et al. 2001; Guo and Ecker 2003; Potuschak et al. 2003; Fu et al. 2004; Dharmasiri et al. 2005; Kepinski and Leyser 2005), lateral root formation (Coates et al. 2006), light signaling and clock control (Mas et al. 2003; Imaizumi et al. 2003, 2005), pollen recognition and rejection (Qiao et al. 2004a,b; Sijacic et al. 2004), and plant–pathogen interactions (Kim and Delaney 2002; Duyvesteijn et al. 2005; van den Burg et al. 2008). Based on phylogenetic analysis, the superfamily of F-box proteins is divided into 20 groups (Gagne et al. 2002). ETP1 and ETP2 belong to a novel subfamily of F-box protein of which little is known (Gagne et al. 2002). Specifically, both ETP1 and ETP2 carry the F-box protein-associated (FBA_1) motif, which is different from typical protein–protein interaction domains carried by most well-known F-box proteins (Lechner et al. 2006). We found that the nontypical FBA_1 domain interacts directly with EIN2, indicating that this region may play a role in recognition of its substrates, which in our case is EIN2 (data not shown). However, at this point we cannot be certain that EIN2 is the only target of ETP1 or ETP2, and if other substrates might be recognized by different portions of the ETP proteins.
Our study provides compelling evidence that ETP1 and ETP2 are negative regulators of EIN2 protein levels, and the EIN2–ETP1/ETP2 interaction is central in controlling ethylene responses. First, ETP1 and ETP2 interact with EIN2 physically. Second, our genetic experiments of knocking down gene expression of ETP1 and ETP2 simultaneously (amiR-ETP1/ETP2) demonstrate that plants lacking these two proteins manifest constitutive ethylene response phenotypes both in etiolated seedlings and adult plants. This phenotype is extremely strong although the amiR-ETP1/ETP2 plants are slightly larger than their ctr1 counterpart (data not shown). Strikingly, knocking down ETP1 and ETP2 expression also stunted growth and renders the plants partially sterile, which is similar to the phenotypes observed in the etr1/etr2/ein4/ers2 quadruple mutant (Hua et al. 1998) and the ebf1 ebf2 double mutant (Guo and Ecker 2003; Potuschak et al. 2003). In contrast, overexpression of ETP1 and ETP2 results in plants manifesting an ethylene-insensitive phenotype. Third, our biochemical data demonstrate that the level of EIN2 is greatly increased in the ETP1/ETP2 knockdown (amiR-ETP1/ETP2) plants, but in ETP1 or ETP2 overexpression plants EIN2 does not accumulate upon treatment with ACC, suggesting that ETP1 and ETP2 suppress EIN2 protein accumulation. Fourth, genome-wide microarray analysis demonstrated that ethylene-inducible genes are significantly induced in amiR-ETP1/ETP2 plants compared with wild-type Col-0 plants. Finally, our study demonstrated that ethylene negatively regulates ETP1 protein level and this impairs the interaction between EIN2 and ETP1, which likely results in accumulation of EIN2 protein. Further biochemical studies of this highly conserved and essential integral membrane protein will be critical to uncover the mechanistic details governing its hormone signaling properties.
Materials and methods
Plant growth condition, ethylene, and drug treatment
The Columbia ecotype (Col-0) was the parent strain for all mutant and transgenic lines used in this study. Arabidopsis seeds were surface-sterilized and plated on the MS medium (4.3 g of MS salt, 10 g sucrose at pH 5.7, 8 g of bactoagar per liter). After 3–4 d of cold (4°C) treatment, the plates were wrapped in foil and kept in a 24°C incubator before the phenotypes of seedlings were analyzed. For propagation, seedlings were transferred from plates to soil (Pro-mix-HP) and grown to maturity at 22°C under a 16-h light/8-h dark cycle. Ethylene treatment of Arabidopsis seedlings grown on plates was performed in air-tight containers (AirGas) by flowing hydrocarbon-free air supplied with 10 ppm ethylene or treated with hydrocarbon-free air alone (Kieber et al. 1993). For drug treatment, Arabidopsis etiolated seedlings were incubate with liquid MS medium for 3 h in the dark at room temperature. Afterward, the seedlings were treated with MG132 (100 μm), MG115 (100 μm), or DMSO (0.1%) for various times prior to harvesting the tissue.
Antibody preparation, immunoblot assays, and pull-down
The coding region corresponding to residues 808 amino acids to 1294 amino acids of EIN2 protein was PCR-amplified, purified from Escherichia coli and used to raise polyclonal antibodies in rabbits. Immnunoblot assays were performed as described (Guo and Ecker 2003) with minor modifications. Membrane proteins were extracted according to Chen et al. (2002); protein samples were mixed with 2× SDS-PAGE sample buffer, heated for 3 min at 90°C, and cooled on ice for 2 min. The proteins were fractionated by 4%–12% gradient Bis-Tris Novex precast gels (Invitrogen), transferred onto nitrocellulose filter and the blot was probed with anti-EIN2 or anti-H+-ATPase antibody (kindly provided by Dr M.J. Chrispeels).
In vitro transcribed and translated S35-labeled EIN2-C2 proteins were generated according to the protocol of TNT-Coupled Wheat Germ Extract Systems (Promega). Total protein extracts were prepared from MYC-ETP1 plants treated with or without ethylene. The same amount of S35-labeled EIN2-C2 was incubated with total protein extract from MYC-ETP1 plants treated with or without ethylene, anti-MYC antibody, and IgG agarose overnight at 4°C. The IgG agarose beads were washed by PBST for six times, and the proteins were eluted by 2× SDS-PAGE sample buffer, heated for 3 min at 90°C, and cooled on ice for 2 min. The proteins were fractionated by 4%–12% gradient Bis-Tris Novex precast gels (Invitrogen). The presence of EIN2-C2S35 was detected by autoradiography.
Yeast two-hybrid interaction assay
The cDNA sequences of the EIN2, ETP1, and ETP2 (Yamada et al. 2003) and their derivatives were cloned into pAS2 LOXP or pACT2 LOXP vector (Clontech; H. Li and J.R. Ecker, unpubl.; http://signal.salk.edu/pHOST.html). Yeast transformation, growth conditions, and interaction assays were performed according to the manufacturer's instructions (Clontech).
Construction of transgenic Arabidopsis plants
Knockdown of ETP1 and ETP2 using an amiRNA was carried out as described (Schwab et al. 2006; http://wmd.weigelworld.org/cgi-bin/mirnatools.pl). The oligonucleotides used for amiRNA constructed are “TCTTTGAATAAACGGTCCCAT” and “TCTTTGAATAAACGGTGCCAT”. The binary vector used was pCHF3, and contained the amiRNA backbone miR 319. The binary vector pKYLX7 was modified by inserting a loxP site and MYC tag in the MCS region (H. Li and J.R. Ecker, unpubl.; http://signal.salk.edu/pHOST.html). The full-length ORFs of ETP1 and ETP2 were cloned into pUNI15 vector at the NcoI/SaiI site (a gift from Dr. Stephen Elledge). An in vitro plasmid recombination reaction, catalyzed by Cre recombinase, was carried out between pUNI15 (containing F-box cDNA sequence) and the modified pKYLX7 with Myc tag. The resulting plasmids that harbor ETP1 and ETP2 coding regions driven by CaMV 35S promoter were introduced into Agrobacterium strain GV3101 and subsequently transformed into Arabidopsis plants. Transgenic T1 plants were identified by selection for kanamycin resistance. The triple response phenotype was scored in T2 seedlings originated from individual transgenic T1 plants. Homozygous T3 seedlings were used for phenotype analysis and immunoblotting studies.
Acknowledgments
We thank Brian D. Gregory, Su-qiung Fang, Joanne Chory, and members of the Ecker and Chory laboratories for critical reading of the manuscript. The plasmid containing the amiRNA backbone was generously provided by Dr. Detlef Weigel (Max Planck Institute for Developmental Biology). We also thank the Salk Institute Genomic Analysis Laboratory for the full-length ETP1 and ETP2 cDNA clones and the T-DNA insertional mutants of etp1 and etp2. K.N.C. was supported by a National Science Foundation IGERT training grant (DGE0504645). This work was supported by a grant (MCB 0516888) from the National Science Foundation to J.R.E.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1765709.
Supplemental material is available at http://www.genesdev.org.
References
- Abeles F.B., Morgan P.W., Saltveit M.E., Jr . Ethylene in plant biology. 2nd ed. Academic Press; New York: 1992. [Google Scholar]
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Binder B.M., Walker J.M., Gagne J.M., Emborg T.J., Hemmann G., Bleecker A.B., Vierstra R.D. The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell. 2007;19:509–523. doi: 10.1105/tpc.106.048140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A.B., Kende H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- Bleecker A.B., Estelle M.A., Somerville C., Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Chang C., Kwok S.F., Bleecker A.B., Meyerowitz E.M. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chen Y.F., Randlett M.D., Findell J.L., Schaller G.E. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J. Biol. Chem. 2002;277:19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Christians M.J., Robles L.M., Zeller S.M., Larsen P.B. The eer5 mutation, which affects a novel proteasome-related subunit, indicates a prominent role for the COP9 signalosome in resetting the ethylene-signaling pathway in Arabidopsis. Plant J. 2008;55:467–477. doi: 10.1111/j.1365-313X.2008.03521.x. [DOI] [PubMed] [Google Scholar]
- Coates J.C., Laplaze L., Haseloff J. Armadillo-related proteins promote lateral root development in Arabidopsis. Proc. Natl. Acad. Sci. 2006;103:1621–1626. doi: 10.1073/pnas.0507575103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dong C.H., Rivarola M., Resnick J.S., Maggin B.D., Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signaling. Plant J. 2008;53:275–286. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyvesteijn R.G., van Wijk R., Boer Y., Rep M., Cornelissen B.J., Haring M.A. Frp1 is a Fusarium oxysporum F-box protein required for pathogenicity on tomato. Mol. Microbiol. 2005;57:1051–1063. doi: 10.1111/j.1365-2958.2005.04751.x. [DOI] [PubMed] [Google Scholar]
- Ecker J.R. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- Fu X., Richards D.E., Fleck B., Xie D., Burton N., Harberd N.P. The Arabidopsis mutant sleepy1gar2-1 protein promotes plant growth by increasing the affinity of the SCFSLY1 E3 ubiquitin ligase for DELLA protein substrates. Plant Cell. 2004;16:1406–1418. doi: 10.1105/tpc.021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Downes B.P., Shiu S.H., Durski A.M., Vierstra R.D. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Smalle J., Gingerich D.J., Walker J.M., Yoo S.D., Yanagisawa S., Vierstra R.D. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin–protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc. Natl. Acad. Sci. 2004;101:6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Chen Y.F., Randlett M.D., Zhao X.C., Findell J.L., Kieber J.J., Schaller G.E. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. J. Biol. Chem. 2003;278:34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Kepinski S., Rouse D., Leyser O., Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Guzman P., Ecker J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Kieber J.J., Hirayama N., Kogan M., Guzman P., Nourizadeh S., Alonso J.M., Dailey W.P., Dancis A., Ecker J.R. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell. 1999;97:383–393. doi: 10.1016/s0092-8674(00)80747-3. [DOI] [PubMed] [Google Scholar]
- Hua J., Sakai H., Nourizadeh S., Chen Q.G., Bleecker A.B., Ecker J.R., Meyerowitz E.M. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Tran H.G., Swartz T.E., Briggs W.R., Kay S.A. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Johnson P.R., Ecker J.R. The ethylene gas signal transduction pathway: A molecular perspective. Annu. Rev. Genet. 1998;32:227–254. doi: 10.1146/annurev.genet.32.1.227. [DOI] [PubMed] [Google Scholar]
- Kendrick M.D., Chang C. Ethylene signaling: New levels of complexity and regulation. Curr. Opin. Plant Biol. 2008;11:479–485. doi: 10.1016/j.pbi.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim H.S., Delaney T.P. Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell. 2002;14:1469–1482. doi: 10.1105/tpc.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. Plant J. 2008;55:821–831. doi: 10.1111/j.1365-313X.2008.03551.x. [DOI] [PubMed] [Google Scholar]
- Lechner E., Achard P., Vansiri A., Potuschak T., Genschik P. F-box proteins everywhere. Curr. Opin. Plant Biol. 2006;9:631–638. doi: 10.1016/j.pbi.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 1998;8:397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- Mas P., Kim W.Y., Somers D.E., Kay S.A. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Qiao H., Wang F., Zhao L., Zhou J., Lai Z., Zhang Y., Robbins T.P., Xue Y. The F-box protein AhSLF-S2 controls the pollen function of S-RNase-based self-incompatibility. Plant Cell. 2004a;16:2307–2322. doi: 10.1105/tpc.104.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Wang H., Zhao L., Zhou J., Huang J., Zhang Y., Xue Y. The F-box protein AhSLF-S2 physically interacts with S-RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell. 2004b;16:582–595. doi: 10.1105/tpc.017673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick J.S., Wen C.K., Shockey J.A., Chang C. REVERSION-TO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc. Natl. Acad. Sci. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman G., Lubarsky B., Kieber J.J., Rothenberg M., Ecker J.R. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic P., Wang X., Skirpan A.L., Wang Y., Dowd P.E., McCubbin A.G., Huang S., Kao T.H. Identification of the pollen determinant of S-RNase-mediated self-incompatibility. Nature. 2004;429:302–305. doi: 10.1038/nature02523. [DOI] [PubMed] [Google Scholar]
- Solano R., Stepanova A., Chao Q., Ecker J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes & Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Ecker J.R. Ethylene signaling: From mutants to molecules. Curr. Opin. Plant Biol. 2000;3:353–360. doi: 10.1016/s1369-5266(00)00096-0. [DOI] [PubMed] [Google Scholar]
- van den Burg H.A., Tsitsigiannis D.I., Rowland O., Lo J., Rallapalli G., Maclean D., Takken F.L., Jones J.D. The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell. 2008;20:697–719. doi: 10.1105/tpc.107.056978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma Penmetsa R., Uribe P., Anderson J., Lichtenzveig J., Gish J.C., Nam Y.W., Engstrom E., Xu K., Sckisel G., Pereira M., et al. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 2008;55:580–595. doi: 10.1111/j.1365-313X.2008.03531.x. [DOI] [PubMed] [Google Scholar]
- Xie D.X., Feys B.F., James S., Nieto-Rostro M., Turner J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yamada K., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Tena G., Xiong Y., Sheen J. Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature. 2008;451:789–795. doi: 10.1038/nature06543. [DOI] [PMC free article] [PubMed] [Google Scholar]