Abstract
Rictor is a component of the target of rapamycin complex 2 (TORC2). While TORC2 has been implicated in insulin and other growth factor signaling pathways, the key inputs and outputs of this kinase complex remain unknown. We identified mutations in the Caenorhabditis elegans homolog of rictor in a forward genetic screen for increased body fat. Despite high body fat, rictor mutants are developmentally delayed, small in body size, lay an attenuated brood, and are short-lived, indicating that Rictor plays a critical role in appropriately partitioning calories between long-term energy stores and vital organismal processes. Rictor is also necessary to maintain normal feeding on nutrient-rich food sources. In contrast to wild-type animals, which grow more rapidly on nutrient-rich bacterial strains, rictor mutants display even slower growth, a further reduced body size, decreased energy expenditure, and a dramatically extended life span, apparently through inappropriate, decreased consumption of nutrient-rich food. Rictor acts directly in the intestine to regulate fat mass and whole-animal growth. Further, the high-fat phenotype of rictor mutants is genetically dependent on akt-1, akt-2, and serum and glucocorticoid-induced kinase-1 (sgk-1). Alternatively, the life span, growth, and reproductive phenotypes of rictor mutants are mediated predominantly by sgk-1. These data indicate that Rictor/TORC2 is a nutrient-sensitive complex with outputs to AKT and SGK to modulate the assessment of food quality and signal to fat metabolism, growth, feeding behavior, reproduction, and life span.
Keywords: C. elegans, fat metabolism, life span, insulin/IGF, AKT
The target of rapamycin (tor) gene is conserved from slime molds to mammals. TOR exists in two structurally and functionally distinct multiprotein complexes whose components are highly conserved across phylogeny, TOR complex 1 (TORC1) and TORC2 (Sarbassov et al. 2005a; Bhaskar and Hay 2007). TORC1 regulates cell growth, protein synthesis, and autophagy, whereas TORC2 has been implicated in cytoskeletal reorganization in cell lines (Sarbassov et al. 2005b). Because of the embryonic lethality of TORC2 component knockout mice (Guertin et al. 2006; Shiota et al. 2006; Yang et al. 2006), its in vivo role can only be assessed using tissue-specific inactivation in mammals (Bhaskar and Hay 2007).
In multicellular eukaryotes, TORC2 is composed of TOR, mLST8 Rictor, Sin-1, and Protor (Sarbassov et al. 2004; Frias et al. 2006; Yang et al. 2006; Pearce et al. 2007). TORC2 was recently identified as the hydrophobic motif (HM) kinase of AKT and may also regulate PDK1-mediated phosphorylation of AKT (Hresko and Mueckler 2005; Sarbassov et al. 2005b). Phosphorylation of the HM is important for AKT to phosphorylate downstream targets, such as mFOXO3, in an insulin-dependent manner (Guertin et al. 2006). However, TORC2 may also phosphorylate other AGC family kinases, such as protein kinase C (PKC) and SGK (Sarbassov et al. 2004; Guertin et al. 2006; Garcia-Martinez and Alessi 2008).
In Saccharomyces cerevisiae, Schizosaccharomyces pombe, and mammals, genetic inactivation of TORC2 is lethal (Schmidt et al. 1996; Kamada et al. 2005). The lethality in yeast is due to defects in cytoskeletal arrangement and ceramide synthesis (Schmidt et al. 1996; Tabuchi et al. 2006; Aronova et al. 2008). Genetic screens identified gain-of-function alleles of a yeast homolog of SGK, Ypk2, as suppressors of the lethality of TORC2 mutations (Kamada et al. 2005). Furthermore, Ypk2 gain-of-function mutations fully rescue the ceramide synthesis defects of TORC2 mutants (Aronova et al. 2008), identifying the SGK homolog as a critical downstream effector of TORC2 in yeast.
In mammals, TOR, Rictor, Sin-1, and mLST8 are all necessary for the assembly, full activity, and substrate specificity of the TORC2 complex. Activation of TORC2 kinase activity by growth factors or insulin is abrogated by elimination of any component in mouse embryonic fibroblasts (MEFs) (Frias et al. 2006; Guertin et al. 2006; Shiota et al. 2006; Yang et al. 2006). Studies of TORC2 component knockout mice have been limited due to defects in placentogenesis and embryogenesis that result in lethality by embryonic day 10.5 (E10.5) to E11.5 (Guertin et al. 2006; Shiota et al. 2006), although muscle-specific inactivation of rictor produces glucose intolerance and increased basal glycogen synthesis (Bentzinger et al. 2008; Kumar et al. 2008), presumably via decreased AKT HM phosphorylation.
In Caenorhabditis elegans, as in mammals, insulin-like signaling through PI3 Kinase regulates fat mass, life span, and whole-organism metabolism (Morris et al. 1996; Ogg et al. 1997; Ogg and Ruvkun 1998; Paradis and Ruvkun 1998; Paradis et al. 1999; Wolkow et al. 2000; Porte et al. 2005). While TORC2 has been firmly implicated in insulin signaling (Bhaskar and Hay 2007), its role in whole-organism fat mass and life-span regulation has yet to be determined. Here we present evidence that TORC2 functions in vivo to regulate the metabolic state of C. elegans. Mutations in C. elegans rictor show elevated body fat content, defective feeding behavior on different bacterial diets, and shortened life span. Furthermore, for all phenotypes except for triglyceride accumulation, sgk-1 phenocopies rictor, implicating SGK-1 as the critical downstream kinase of TORC2 for these phenotypes. For the full high-fat phenotype of rictor mutants, the AKT homologs akt-1 and akt-2 are necessary in addition to sgk-1.
Results
Identification and expression of the C. elegans homolog of Rictor, RICT-1
We identified two mutantations in the C. elegans homolog of rictor (F29C12.3, subsequently referred to as rict-1) based on a classical, forward genetic screen for substantially increased fat mass in animals fed the standard laboratory bacteria, Escherichia coli OP50. While C. elegans was not initially predicted to contain a homolog of Rictor, this was likely due to misannotation of the gene to two separate mRNAs, F29C12.2 and F29C12.3, separated by an intronic gene in the antisense direction, pqn-32 (F29C12.1), and that rictor is divergent among eukaryotes, sharing only 44% similarity in a conserved region of 200 amino acids (Sarbassov et al. 2004). C. elegans RICT-1 is approximately as similar to mammalian Rictor as Drosophila Rictor (16% identity and 50% similarity and 22% identity and 57% similarity, respectively, over the entire coding region). In addition, BLASTP comparison of C. elegans RICT-1 versus the human genome database detects Rictor as the top hit with an e value of e-33, and and the converse search detects C. elegans RICT-1 as the only homolog, with an e value of e-35.
The two alleles of rict-1 that emerged from the high-fat genetic screen contain early stop mutations in the coding region of rict-1, removing 97 (allele mg450) or 211 (allele mg451) of the 1580 amino acids encoded by the major mRNA species (Supplemental Fig. S1). mg450 and mg451 are predicted to be loss-of-function mutations based on the removal of a highly conserved C-terminal region (Supplemental Fig. S1), recessive inheritance, and noncomplementation of mg450 and mg451. Furthermore, the high-fat phenotype is phenocopied by RNAi of rict-1 (Supplemental Fig. S2A), and a rict-1 promoter-driven rict-1 cDNA transgene rescues the high-fat, small body size, and life span phenotypes (Supplemental Fig. S3A). RNAi of rict-1 does not further enhance the high-fat phenotype of rict-1 mutants, supporting the probable null nonsense mutations (Supplemental Fig. S2B). rict-1 is expressed in the nervous system (head neurons, ventral nerve cord), intestine, body wall muscle, pharynx, and spermatheca as assessed by a promoter-driven RFP reporter (Supplemental Fig. S3B). In adult animals, the most prominent sites of expression are head neurons and the intestine (Supplementary Fig. S3B). In support of rict-1 mutations acting to increase fat mass through TORC2 and not through associations with other cellular signaling components, RNAi of sinh-1, the C. elegans homolog of the TORC2 complex component mSin1, also produces a high-fat phenotype (Supplemental Fig. S2A).
RICT-1 regulates fat mass and the response to different diets
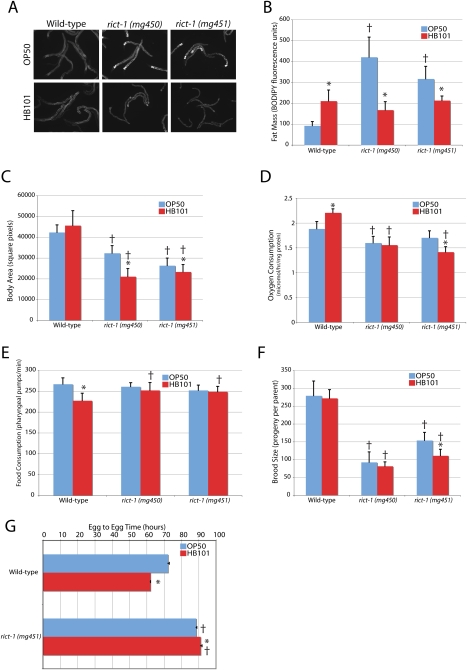
While wild-type animals increase fat mass, as assessed by labeling with the fluorescent fatty acid analog C1C12-BODIPY, when fed the nutrient-rich bacterial strain E. coli HB101 as compared with the relatively nutrient-poor OP50 (Fig. 1A). HB101 is considered to be nutrient-rich based on its superior ability to support wild-type C. elegans growth when compared with an OP50-derived strain (Avery and Shtonda 2003; Shtonda and Avery 2006). Compared with OP50, wild-type C. elegans fed HB101 develop from egg to reproductive adult >10 h faster (Fig. 1G; Avery and Shtonda 2003), demonstrate increased intestinal BODIPY fat staining, and increase oxygen consumption (Fig. 1A,B,D).
Figure 1.
The metabolic defects of rict-1 mutants. (A,B) BODIPY-labeled fatty acid vital dye revealed a threefold to fourfold increase in fat content in both rict-1 alleles (mg450 and mg451) relative to wild type (N2) as day 3 adults when fed E. coli OP50 bacteria. This phenotype was not evident when rict-1 mutant animals were fed E. coli HB101. (C) Body size is 24%–38% reduced in rict-1 mutants and further reduced by feeding HB101. (D) Energy expenditure is reduced slightly in rict-1 mutants, increased in wild type by feeding HB101 versus OP50, and decreased by HB101 versus OP50 in rict-1 (mg451). (E) Food intake as measured by pharyngeal pumping rate when animals are on food is equivalent in wild-type and rict-1 mutant worms on OP50. On HB101, wild-type worms decrease pumping rate as day 1 adults, but rict-1 mutants maintain a higher pumping rate on HB101. However, rict-1 mutants spend less time on HB101 and therefore consume less of this bacteria. (F) Brood size is reduced 45%–67% in rict-1 mutants and in the mg451 allele is reduced further by HB101 feeding. (G) Egg-to-egg time is delayed 16 h in rict-1 mutants. Feeding HB101 accelerates egg-to-egg time by 10 h in wild-type worms and paradoxically slows this interval by 2.5 h in rict-1 mutants. (†) P < 0.05 versus wild-type; (*) P < 0.05 versus same strain on OP50 by two-tailed, equal variance t-test. Error bars indicate ±SD, except for G, where they indicate ±SEM.
When fed OP50, rict-1 mutants display a 200%–300% increase in BODIPY fat content as day 3 adults (Fig. 1A,B). A 30%–50% increase is seen in rict-1 mutants as day 1 adults (Fig. 3A, below), indicating that BODIPY accumulation is preferentially activated post-developmentally during the reproductive period. Despite having dramatically high lipid levels, rict-1 mutants are delayed 16 h in reaching reproductive maturity (Fig. 1G), and mature adults matched for developmental stage are smaller (Fig. 1C). This highlights a conserved role for TORC2 in growth as rictor also regulates body size in Drosophila (Lee and Chung 2007), and the murine embryos of TORC2 knockout components are small (Guertin et al. 2006).
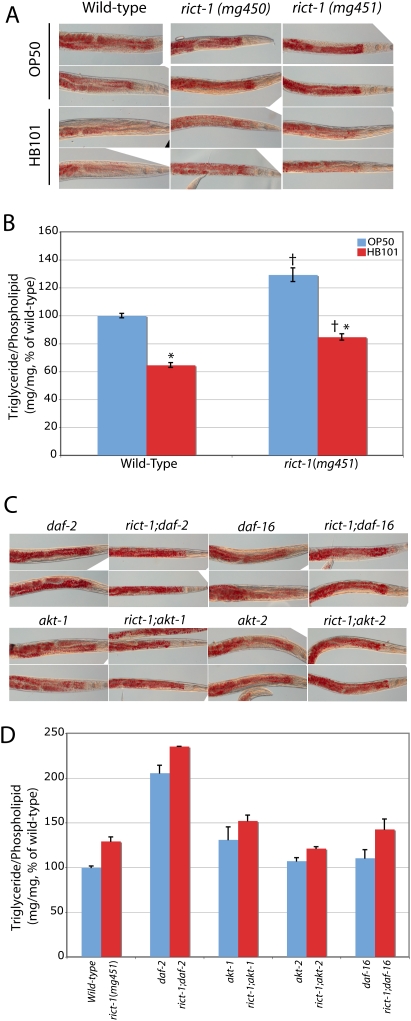
Figure 3.
Oil-Red-O staining and quantitative lipid biochemistry in rict-1 mutants. (A) Oil-Red-O staining confirms elevated fat mass in rict-1 mutants fed E. coli OP50. Lipid is evident in red droplets. Fat mass in worms fed E. coli HB101, unlike in BODIPY fatty acid labeling, is lower in wild-type worms and rict-1 mutants, but rict-1 mutants have relatively elevated fat mass on either diet. (B) Quantitation of lipid species by solid-phase extraction followed by gas chromatography and mass spectroscopy in day 1 adult rict-1 mutants revealed a 30% increase in triglyceride mass when normalized to total phospholipid content. In both wild-type and rict-1 mutants, lipid mass is lower when animals are fed HB101, supporting the Oil-Red-O staining observations, in contrast to what is seen with BODIPY staining. (†) P < 0.05 versus wild type; (*) P < 0.05 versus same strain on OP50 by two-tailed, equal variance t-test. Mean ± SEM for four biological replicates. (C,D) Oil-Red-O staining and quantitative lipid biochemistry of rict-1 double mutants with daf-2/InsulinR, akt-1, akt-2, and daf-16/FOXO. Fat mass of rict-1 mutants is additive with daf-2, partially additive with akt-1, partially blunted by akt-2, and unaffected by daf-16. These methods confirm the previous observations of daf-2 mutants having elevated triglyceride mass. Mean ± SEM for paired biological replicates.
Increased food consumption is not responsible for the observed fat-level differences in high-fat rict-1 animals (Fig. 1E). A nonsignificant trend toward decreased oxygen consumption was seen in rict-1(mg451) animals [significant in rict-1(mg450) animals], and thus decreased energy expenditure may play some role in the development of the high-fat phenotype of rict-1 mutants. rict-1 mutants display an attenuated brood size of 30%–50% as compared with wild type (Fig. 1F). Therefore, shunting of calories away from reproduction or growth or inadequate utilization of stored fat could also provide an explanation for the elevated fat content of rict-1 mutants.
Surprisingly, rict-1 mutants displayed a paradoxical, detrimental response to HB101 feeding in all metabolic phenotypes examined: The BODIPY high-fat phenotype of the rict-1 animals fed OP50 was not evident when feeding HB101 bacteria (Fig. 1A,B), body size decreased (Fig. 1C), oxygen consumption did not increase (Fig. 1D), total brood size decreased (in the stronger mg451 allele) (Fig. 1F), and time to reproductive maturity was further delayed by >2.5 h (Fig. 1G). This suggests either that rict-1 animals eat less when fed HB101 or that rict-1 mutations prevent utilization of nutrients particular to the HB101 strain. A decreased pumping rate was not evident in rict-1 animals found on the lawn of bacteria (Fig. 1E). However, when put on a plate with HB101, rict-1 mutant animals displayed decreased residence time on the lawn of food (day 1 adult wild-type worms off HB101 lawn 3.1% ± 0.9% vs. rict-1 mutants off HB101 lawn 10.8% ± 1.5%; P < 0.01), suggesting that over time they may consume less HB101. We also observed decreased lawn residence when RNAi was used to knock down rict-1, leading to 26.7% ± 7.3% and 36.9% ± 2.1% of animals being off the lawn for two different RNAi clones targeting rict-1 (vs. 0% ± 0% for vector RNAi control). This indicates that Rictor/TORC2 is necessary for proper feeding behavior as wild-type worms normally do not decrease residence on the lawn of food when fed HB101 or the E. coli strain HT115 used to conduct RNAi. Of note, both HB101 and HT115 are closely related to each other and to the E. coli strain K12, versus OP50, which is much more closely related to the E. coli B strain (B. Samuel and G. Ruvkin, unpubl.).
In insulin/IGF signaling pathway mutants, BODIPY and Nile red do not accurately indicate body fat mass
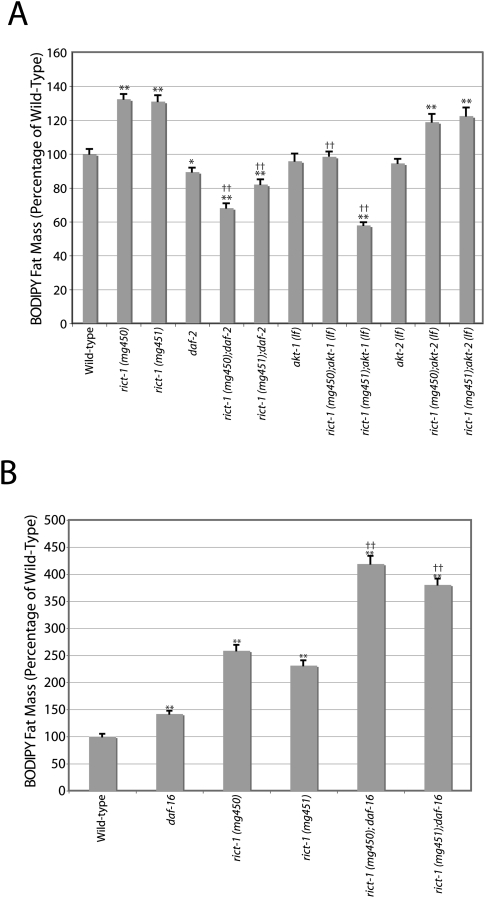
Insulin-like signaling in many animals, including C. elegans, is a major endocrine signal of nutrients and fat (Porte et al. 2005). Since TORC2 had been reported to activate AKT via HM phosphorylation, we examined the relationship between the BODIPY high-fat phenotype of rict-1 mutants and the insulin/IGF/AKT signaling pathway (Supplementary Fig. S4). When assessed by BODIPY-labeled fatty acids, inactivation of daf-2/insulin-like signaling or akt-1 loss-of-function mutations (lf) in combination with rict-1 reduced fat mass to wild-type or lower levels, suggesting that insulin-like signaling through akt-1 is required for the development of the high-fat phenotype of rict-1 mutants (Fig. 2A). Additionally, daf-16 animals accumulated more BODIPY, and rict-1;daf-16 double mutants showed an even greater increase than either single mutant (Fig. 2B). However, the BODIPY phenotype of daf-2 mutant animals was puzzlingly contradictory to the previously reported high-fat phenotypes determined by Sudan black fixative-based staining (Kimura et al. 1997; Ogg et al. 1997) and biochemical fat quantitation using solid-phase chromatography or thin-layer chromatography followed by gas-chromatography/mass spectroscopy (Ashrafi et al. 2003; Perez and Van Gilst 2008). As BODIPY fat mass assessment requires normal feeding, uptake, and accumulation, this raises the possibility that BODIPY fat mass assessment may not be accurate when mutants consume more or less food or uptake and accumulate the dye with different efficiency.
Figure 2.
BODIPY-labeled fatty acid assessed fat mass interaction of rict-1 with insulin/IGF signaling components. (A) rict-1 BODIPY fat mass accumulation is reduced by combination with the insulin/IGF-1 receptor hypomorphic mutation daf-2 (e1370) or akt-1 (mg306)-null mutation. rict-1 fat accumulation is not affected by the akt-2 (ok393)-null mutation. BODIPY measurements were inconsistent with previous examinations of daf-2 mutant worms demonstrating increased fat mass by fixative-based lipid staining with Sudan black or biochemical measurement of triglyceride content. Mean of two experiments ±SEM. (*) P < 0.01 versus wild type; (**) P < 0.001 versus wild type; (††) P < 1e−6 versus rict-1 single mutant. (B) Fat mass of rict-1; daf-16/FOXO(mgDf47) double mutants is significantly elevated above rict-1 single mutants on OP50. A small but significant elevation was also seen in daf-16/FOXO single mutants that did not phenocopy the magnitude of fat elevation in rict-1 mutants. Mean ± SEM. (**) P < 1e−6 versus wild type; (††) P < 1e−6 versus rict-1 single mutant.
RICT-1 partially regulates fat via insulin-like signaling through AKT
To more accurately assess fat storage using histochemistry, we fixed adult animals of various genotypes and assayed stored neutral lipid using the standard assay for adipocyte fat storage, the dye Oil-Red-O (Soukas et al. 2001). Oil-Red-O staining of rict-1 mutants recapitulated the high-fat phenotype of rict-1 mutants fed BODIPY-labeled fatty acids (Fig. 3A). Oil-Red-O, unlike Sudan black staining, is much more consistent across animals of a given genotype and correlated well with biochemical measurements of triglyceride mass (see below), and was thus used preferentially to assess fat mass. The rict-1 allele mg451 demonstrated stronger staining with Oil-Red-O than the mg450 allele and was used for all subsequent analyses. Biochemical assessment of triglyceride mass using solid-phase chromatography followed by gas chromatography/mass spectroscopy (Perez and Van Gilst 2008) in day 1 adult rict-1 mutants was ∼30% elevated (Fig. 3B), confirming the results obtained with neutral lipid staining with Oil-Red-O. However, the increase in BODIPY fat mass in wild-type worms fed HB101 when compared with OP50 was no longer evident when worms were stained with Oil-Red-O; in fact, HB101 feeding led to decreases in fat mass in both wild-type worms and in rict-1 mutants as assessed by Oil-Red-O and by biochemical quantitation of triglyceride content (Fig. 3A,B). rict-1 mutants, despite having lower fat mass on HB101 than on OP50, still demonstrated elevated triglyceride accumulation above wild-type HB101 levels (Fig. 3A,B). This indicates that despite decreased lawn residence time on HB101, rict-1 animals still accumulate greater amounts of triglyceride.
Oil-Red-O staining of daf-2 mutants showed elevated fat mass (Fig. 3C), as previously observed with other fixative stains (Kimura et al. 1997; Ogg et al. 1997). Additionally, akt-1 mutants demonstrated an increase in Oil-Red-O staining, consistent with AKT-1 participating in metabolic signaling (Fig. 3C). These results were confirmed by quantitative biochemical analysis of triglycerides and phospholipids in daf-2 and akt-1 mutants (Fig. 3D). Interestingly, we found rict-1;daf-2 and rict-1;akt-1 double mutants to have fat mass above either single mutant, indicating that it is unlikely that rict-1 regulates fat mass exclusively through insulin-like signaling via akt-1. The increase in fat mass evident in rict-1 mutants was partially blunted in the rict-1;akt-2 double mutant (Fig. 3C,D), suggesting that akt-2 plays a positive role in the generation of the high-fat phenotype of rict-1 mutants, or that increased akt-1 signaling in an akt-2 mutant may partially suppress the rict-1 phenotype.
To explore interactions with insulin-like signaling further, we examined interactions of rict-1 with the single C. elegans FOXO homolog daf-16. daf-16 is epistatic to the long-lived, dauer, and high-fat phenotypes of insulin-signaling mutant inactivations (e.g., insulin/IGF receptor mutant daf-2, PI3K homolog mutants age-1, or akt-1;akt-2 double mutants) (Kimura et al. 1997; Ogg et al. 1997; Finch and Ruvkun 2001). If mutations in rict-1 increase fat by loss of AKT-mediated phosphorylation and inhibition of FOXO, FOXO loss of function should be epistatic to rict-1 (i.e., the high-fat phenotype should be mitigated). This was not the case, as rict-1;daf-16 double mutants fed OP50 show an identical increase in fat mass when compared with rict-1 single mutants when assessed by Oil-Red-O and quantitative lipid biochemistry (Fig. 3C,D). This supports the conclusion that rict-1 also regulates fat in parallel to insulin-like signaling through daf-2, akt-1, and daf-16.
RICT-1 regulates life span in a diet-dependent manner
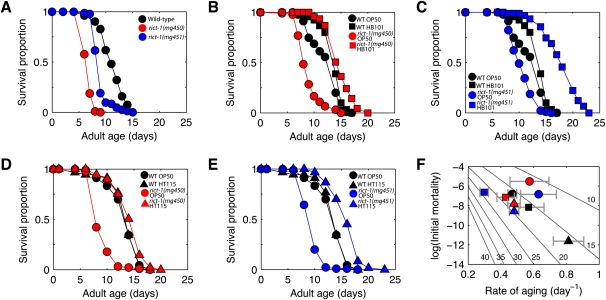
Because nutrient availability is a critical determinant of life span and TORC1 component mutants CeTor(let-363) and CeRaptor(daf-15) have been reported to be long-lived on OP50 (Vellai et al. 2003; Jia et al. 2004; Long et al. 2004; Meissner et al. 2004), we assayed the longevity of rict-1 mutant animals. Unlike tor and CeRaptor mutants, rict-1 mutants display an increased rate of aging and were short-lived on OP50 (Figs. 2F, 4A; Supplemental Table 1). This was unexpected, given that TORC2 has been reported to activate the insulin/IGF pathway signaling component AKT and reduction of insulin/IGF signaling in C. elegans and higher organisms increases life span (Finch and Ruvkun 2001; Wolff and Dillin 2006). This suggests that TORC2 is unlikely to regulate life span by activating AKT in worms fed OP50. Unlike wild-type animals, which do not appreciably change (Fig. 4B,C) or slightly decrease life span (Supplemental Fig. S5A,B) in response to nutrient-rich food (HB101), rict-1 mutants feeding on HB101 have a profoundly extended life span (Figs. 2F, 4B,C; Supplemental Fig. S6; Supplemental Table 1). A similar observation was made when mutants were fed the E. coli strain HT115 (Fig. 4D,E). This is consistent with decreased consumption of nutrient-rich food by rict-1 mutants, as decreased caloric intake is well known to prolong life span in C. elegans and other organisms (Wolff and Dillin 2006).
Figure 4.
The life span of rict-1 mutants is highly responsive to diet, unlike wild type. (A) Median life span of rict-1 mutants is reduced 24%–43% on OP50 relative to wild type (N2). (B) While HB101 versus OP50 had marginal (13%) impact on wild-type life span, HB101 extended rict-1 (mg450) life span by 75%. In a repeat experiment (Supplemental Fig. S5A), life span impacts were −13% and 54%, respectively. (C) HB101 versus OP50 diet had a similar impact on rict-1(mg451), increasing life span by 75%, and in a repeat experiment (Supplemental Fig. S5B) by 74%. (D,E) Similar to HB101, feeding the K12 derived strain E. coli HT115 had insignificant (2%) impact on wild-type life span versus OP50, whereas for rict-1 (mg450) (D) and rict-1 (mg451) (E), life span increased by 71% and 76%, respectively. (F) Changes in diet affect Gompertz parameters for wild type but not life span, because a higher rate of aging is coupled with lower initial mortality; in contrast, rict-1 mutants have higher rates of aging at comparable initial mortality on OP50 versus HB101 and HT115. Error bars ±SD. (Gray diagonals) Lines of constant life span calculated assuming zero accident rate. Legend for F as in B–E.
To formally test the possibility that rict-1 mutants have life-span extension on HB101 due to decreased caloric intake, life span was studied in rict-1;skn-1 double mutants fed HB101, since skn-1 is required for dietary restriction-induced longevity (Bishop and Guarente 2007). No life-span extension was seen in rict-1;skn-1 double mutants fed HB101, supporting the conclusion that decreased consumption of HB101 is responsible for life-span extension in rict-1 worms (Supplemental Fig. S5C). In contrast, tor and raptor C. elegans mutants show increased fat content but arrest before adulthood in an incomplete dauer-like state (Long et al. 2002; Jia et al. 2004) and are thus fundamentally different from rict-1.
Insulin-like signaling through AKT is necessary but not sufficient for the short life span of rict-1 mutants
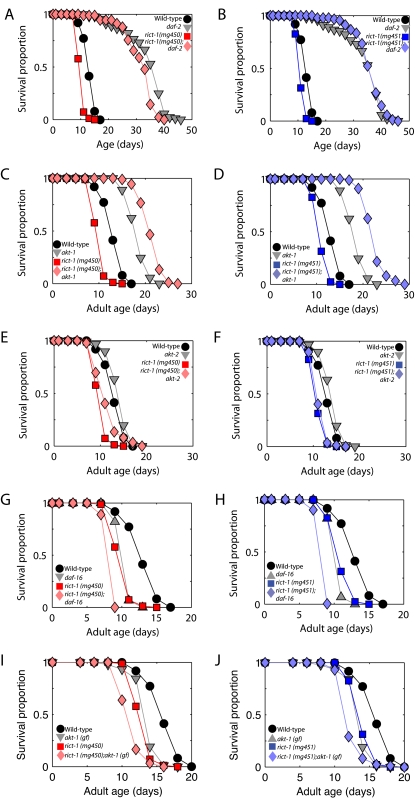
The short-life-span phenotype of rict-1 mutants requires daf-2 and akt-1 signaling as rict-1;akt-1 double mutants shift to being long-lived, rather than short-lived, and rict-1;daf-2 double mutants show a life span nearly identical to daf-2 mutants (Fig. 5A–D). rict-1;akt-2 double mutants have life spans similar to rict-1 single mutants (Fig. 5E,F). These data suggest that a high level of insulin-like signaling via the daf-2 and akt-1 pathway is necessary for the short life span of the rict-1 mutant. However, rict-1;daf-16 double mutants demonstrate shorter life span than either single mutant, indicating that rict-1 does not shorten life span only via regulation of the DAF-16/FOXO transcriptional outputs of insulin signaling (Fig. 5G,H; Supplemental Table S2). Furthermore, rict-1;akt-1(gf) double mutants also demonstrate a significantly shorter life span than either single mutant (Fig. 5I,J). Thus, rict-1 generates these phenotypes by also signaling in parallel to akt-1 and FOXO.
Figure 5.
Life-span regulation by insulin/IGF signaling in rict-1 mutants. (A–D) Life span is extended in rict-1;daf-2 and rict-1;akt-1(lf) double mutants. This is in contrast to the normal short life span of rict-1 mutants, which requires intact insulin-like signaling through akt-1 to shorten life span. (E,F) rict-1;akt-2(lf) double mutants do not show alteration in life span, indicating that the primary interaction is between rict-1 and akt-1. (G–J) Life span of rict-1;daf-16/FOXO (mgDf47) and rict-1;akt-1(gain of function) on OP50 is shortened significantly relative to single mutants, indicating that insulin signaling regulates life span in parallel to TORC2, and that lack of akt-1 activation, leading to decreased inhibitory phosphorylation (thus, activation) of daf-16/FOXO, is not responsible for life-span shortening in rict-1 mutants. (Tabular data is available in Supplemental Table 2.)
Mutations in sgk-1 phenocopy and do not enhance rict-1 growth, reproductive, and life-span defects
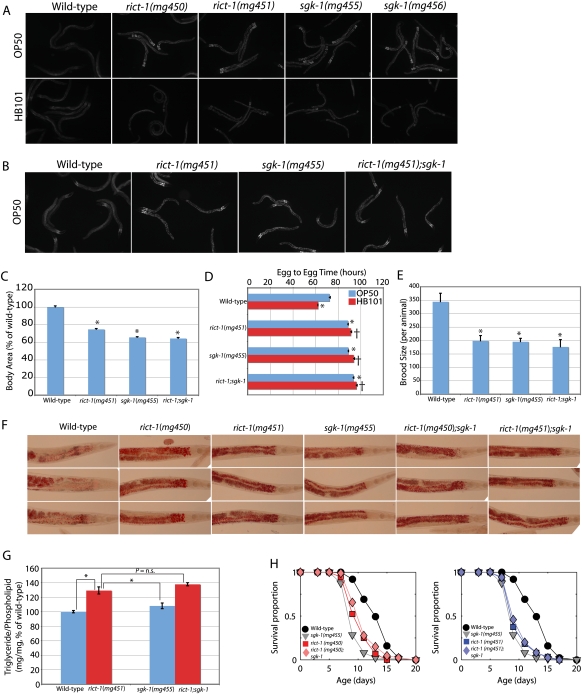
Because akt-1 and akt-2 signaling do not explain the growth, high-fat, reproductive, life-span, and feeding defects in rict-1 mutants, we returned to the mutant collection generated in the forward genetic screen for elevated fat mass. Examining these mutants for high fat and small body size indicated an additional complementation group defined by two mutants that mapped to a region on the long arm of chromosome X containing the AGC family kinase sgk-1. The Ashrafi laboratory had informed us previously that sgk-1 acts in the same genetic pathway as rict-1 by their analyses using the vital dye Nile red (K. Jones and K. Ashrafi, pers. comm.) and it had been shown previously that the sgk-1(ok538) allele also affects body size (Hertweck et al. 2004), as did our two mutants mapping to this 1.0-cM genetic region, so this gene was likely to be affected in these two alleles. Sequencing of sgk-1 exons in these two mutants identified two mutations: a nonsense mutation that removes 178 residues including critical components of the kinase domain and the HM phosphorylation site (mg455), and a missense mutation that changed a nonconserved proline residue to serine (mg456) (Supplemental Fig. S7).
Examination of sgk-1 mutants indicated an identical BODIPY phenotype to rict-1 when fed OP50 and HB101 (Fig. 6A); i.e., elevated BODIPY staining on OP50 and normal staining when fed HB101. Additionally, sgk-1 mutants have decreased body size, are slow growing, and have decreased brood size (Fig. 6C–E). These phenotypes were nearly identical when examined in a rict-1;sgk-1 double mutant, indicating that these genes are likely to be in the same genetic pathway (Fig. 6C–E). The rict-1;sgk-1 double mutant was slightly slower in reaching reproductive maturity (Fig. 6D), which could indicate divergence of action of rict-1 and sgk-1 in regulating developmental rate. However, the fat mass as assessed by Oil-Red-O staining in sgk-1(mg455), which encodes a predicted null allele, was not increased as greatly as in rict-1 mutants (Fig. 6F). Quantitative biochemical analysis confirmed this observation and indicated greater fat mass in the rict-1;sgk-1 double mutant than the sgk-1 single mutant (Fig. 6G). This suggests that outputs of TORC2 other than sgk-1 may be necessary for the full extent of the high-fat phenotype.
Figure 6.
sgk-1 and rict-1;sgk-1 double-mutant analysis. (A) BODIPY-labeled fatty acid fat mass imaging of two alleles of sgk-1 that have elevated fat mass indicate a similar phenotype to rict-1 mutants: high BODIPY signal when fed OP50 and blunting of BODIPY staining and smaller body size when fed HB101. (B–E) Analysis of rict-1;sgk-1 double mutants indicates that sgk-1 phenocopies rict-1 for BODIPY fat mass, body size, and egg-to-egg time, and that none of these phenotypes is enhanced in the rict-1;sgk-1 double mutant. Mean ± SEM. (*) P < 0.05 versus wild type; (†) P < 0.05 versus same strain on OP50. (F,G) Oil-Red-O staining and quantitative lipid biochemistry of rict-1, sgk-1, and rict-1;sgk-1 show that rict-1 produces a higher-fat phenotype than is explained by sgk-1 alone, indicating that TORC2 has additional outputs other than sgk-1 to regulate fat mass. Mean ± SEM. (H) Unlike fat mass, sgk-1 completely phenocopies rict-1 for life span and the short life span of sgk-1 mutants is not enhanced in the rict-1;sgk-1 double mutant. rict-1 and sgk-1 are thus in the same genetic pathway shortening life span when animals are fed OP50.
In contrast, sgk-1 mutants fed OP50 display an identically shortened life span when compared with rict-1 and rict-1;sgk-1 double mutants (Fig. 6H), indicating that these genes are likely in the same genetic pathway regulating life span. Thus, the life span, growth, reproductive, and BODIPY or Nile red increase phenotypes of rict-1 may be explained by sgk-1, but not the full extent of triglyceride accumulation assessed biochemically and by Oil-Red-O staining.
RICT-1/TORC2 regulates fat mass through AKT and SGK
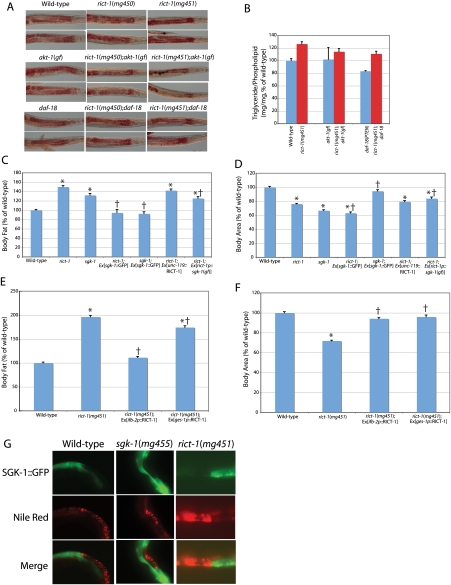
To further examine the mechanism by which rict-1 produces an increase in fat mass, we looked at fat mass in akt-1 gain-of-function genetic mutants, in transgenic animals overexpressing wild-type sgk-1 under its own promoter, and in transgenic animals expressing activated sgk-1 [sgk-1(gf)] under the control of the rict-1 promoter. For the latter, the same rict-1 promoter was used that was able to rescue rict-1 mutant life span and fat mass by driving rict-1 cDNA because sgk-1 would be predicted to act in the same cells as rict-1 for the phenotype observed since TORC2 is an upstream kinase of sgk-1 in cell lines (Garcia-Martinez and Alessi 2008). We found that rict-1;akt-1(gf) double mutants partially suppressed the rict-1 elevated fat mass as assessed by Oil-Red-O and quantitative lipid biochemistry (Fig. 7A,B). Expression of activated sgk-1(gf) under the rict-1 promoter only partially suppressed the high-fat phenotype of rict-1, as assessed with Nile red (Fig. 7C). rict-1;sgk-1(gf) transgenic animals did not rescue body size (Fig. 7D). Failure to rescue could be due to lack of appropriate expression of the transgene in necessary tissues, lack of sufficient expression level, or failure of gain-of-function mutation of the HM threonine to aspartate to activate SGK-1 as has been demonstrated previously (Park et al. 1999). Surprisingly, however, high gene dosage of wild-type sgk-1 under its own promoter was able to completely suppress Nile red fat mass of rict-1 mutants, demonstrating that sgk-1 can act downstream from rict-1 to regulate fat (Fig. 7C). This same rescuing transgene also completely rescued the high-fat phenotype of sgk-1 mutants (Fig. 7D). However, high gene dosage of wild-type sgk-1 did not rescue the small body size of rict-1 animals (but did rescue the small body size of sgk-1 mutants), suggesting that functional TORC2 is necessary for growth regulation (Fig. 7D) or that there are additional outputs of TORC2 that regulate body size. Finally, activation of the PI3 kinase cascade by a daf-18 (PTEN homolog)-null mutation reduced fat mass in a wild-type background, but rict-1;daf-18 double mutants still increased fat mass above daf-18 single mutants by 33%, indicating regulation of fat mass in rict-1 in parallel to PI3K (Fig. 7A,B).
Figure 7.
Rescue of the rict-1 mutant high-fat and growth phenotypes. (A,B) Oil-Red-O staining and quantitative lipid biochemistry indicate that rict-1 high fat is partially suppressed by a chromosomally located akt-1 gain-of-function (gf) mutation. In contrast, while wild-type fat mass is decreased by a daf-18/PTEN inactivating mutation, rict-1 still increases fat in a daf-18 background, indicating that rict-1 increases fat mass in parallel to PI3K signaling. (C,D) Neuron-specific transgene rescue of rict-1 under the unc-119 promoter (rict-1;Ex[unc-119p∷RICT-1]) fails to rescue the small body size or high-fat phenotype of rict-1 mutants as assessed by Nile red. Transgenic expression of an activated sgk-1 under the rict-1 promoter partially rescues the small body size and high-fat phenotypes of rict-1 {rict-1;Ex[rict-1p∷sgk-1(gf)]}. In contrast, high gene dosage of wild-type sgk-1 in rict-1 mutants fully suppresses the high-fat phenotype but does not rescue the small body size (rict-1;Ex[sgk-1∷GFP]). However, since this is very high copy overexpression, it does not mimic physiologic regulation of sgk-1 or rule out that other genes play a role in the high-fat phenotype of rict-1. (*) P < 1e−6 versus wild type; (†) P < 1e−6 versus nonrescued mutant. (E,F) Intestine-specific transgene rescue of RICT-1 under either the ifb-2 and ges-1 promoters (rict-1;Ex[ifb-2p∷RICT-1] and rict-1;Ex[ges-1p∷RICT-1], respectively) fully rescues the small-body-size phenotype. The ifb-2 promoter rescuing transgene completely rescues the high-fat phenotype of rict-1, whereas a partial rescue is seen in the case of the ges-1 promoter-rescuing transgene. (*) P < 1e−6 versus wild type; (†) P < 1e−6 versus nonrescued mutant. (G) Mosaic analysis of a SGK-1 translational fusion with GFP, which allows expression to be followed visually, indicates that in intestinal cells expressing the SGK-1∷GFP transgene, Nile red fat mass is reduced substantially, whereas in those cells lacking the transgene, Nile red fat mass is substantially higher. This suppression is seen in wild-type, sgk-1, and rict-1 animals.
RICT-1 regulates fat mass and whole organism growth via the intestine
The mechanism of action of rict-1 was further dissected by tissue-specific rescue experiments in the two main sites of expression (Supplemental Fig. S3B), the nervous system and the intestine, which are also known to play a key role in fat storage and life span (Wolkow et al. 2000; Ashrafi et al. 2003; Libina et al. 2003). Transgenic expression of rict-1 in neurons did not rescue the high-fat or growth defects of rict-1 mutants (Fig. 7C,D). Additionally, neuron-specific rescue failed to rescue normal life span in rict-1 mutants (Supplemental Fig. S8). Expression of rict-1 in the intestine of rict-1 mutants under two different intestine-specific promoters, the ges-1 promoter and the ifb-2 promoter, rescues the small-body-size phenotype in transgenic progeny in the F1 generation (Fig. 7F). Examination of rict-1;Ex[ifb-2p∷RICT-1] F1 animals rescued for rict-1 in the intestine also revealed normalization of Nile red fat mass (Fig. 7E). Examination of the Nile red phenotype of transgenic rict-1;Ex[ges-1p∷RICT-1] rescued for body size indicated on average a 12%, significant reduction in fat mass for the whole population (Fig. 7E). This was an incomplete rescue because only ∼20% of size-rescued mutants showed wild-type Nile red fat levels. We were unable to obtain a sufficient number of transmitting lines of rict-1 mutants expressing rict-1 in the intestine; this may be due to rict-1 gene dosage or to imbalances between tissues in rict-1 signaling. (In the single transgenic line obtained carrying an intestine-specific rescue construct, neither body size, nor Nile red fat mass, nor life span was rescued [Supplemental Fig. 8], perhaps due to insufficient rict-1 gene dosage to rescue.)
We also examined animals mosaic for sgk-1 overexpression in the intestine. These studies indicated that sgk-1 suppresses Nile red fat mass in the intestine in a cell-autonomous manner (Fig. 7G). In wild-type, sgk-1(mg455), and rict-1(mg451) mutants, an extrachromosomal transgene overexpressing an SGK-1∷GFP translational fusion suppressed Nile red fat mass in the intestine (Fig. 7G). In the surrounding intestinal cells lacking the SGK-1∷GFP fusion protein, Nile red fat mass was higher than in transgene-containing intestinal cells. Since fat mass is also suppressed in wild-type worms by overexpression of SGK-1, these data do not allow the conclusion that sgk-1 is the sole output of TORC2-regulating fat mass. These data in aggregate suggest that sgk-1 acts directly within intestinal cells, the same site of rict-1 action, likely downstream from TORC2 to regulate fat mass.
Discussion
The data presented here highlight a new role for Rictor/TORC2 in energy metabolism, feeding behavior, life-span modulation, growth, and reproduction. We suggest that rict-1 acts as a critical sensor of organismal energetic status and is a master regulator of metabolic processes such as growth, energy storage, reproduction, and maintenance of life span. When this process is disabled, the organism is no longer able to appropriately channel calories into essential metabolic processes and inappropriately activates long-term energy storage in the form of triglycerides. These data also show that the site of action of rict-1 in regulating growth and fat mass is the intestine, which is the key site of fat storage in C. elegans (Ashrafi et al. 2003; Mak et al. 2006). Given the evolutionary conservation of Rictor, components of the TORC2 complex, AKT and SGK, these results suggest that these genes may be involved in fat storage and life span across phylogeny.
RICT-1 acts through SGK-1 to regulate life span, growth, reproduction, and both SGK-1 and AKT to regulate fat mass
Many phenotypes in rict-1 mutants are shared by the AGC kinase sgk-1, the sole C. elegans homolog of the serum and glucocorticoid-induced kinase gene family. While the life-span, growth, and reproductive phenotypes of rict-1 mutants can be explained by sgk-1, the full extent of excess triglyceride accumulation is dependent on the akt kinases in addition to sgk-1. Even though fat mass in rict-1 mutants as assessed by Nile red can be suppressed by overexpression of wild-type SGK-1, based on loss-of-function data, it is unlikely that sgk-1 is the only output of TORC2 affecting fat mass. Furthermore, it is difficult to say whether SGK-1 is the only downstream effector of TORC2 for fat mass because overexpression of SGK-1 suppresses fat mass in wild-type worms. Additionally, a chromosomally located gain-of-function mutation in akt-1 (Paradis and Ruvkun 1998) partially suppresses rict-1 fat mass as assessed biochemically and by Oil-Red-O, implying that TORC2 signals to regulate fat both through SGK and AKT. Thus, a model is proposed whereby RICT-1/TORC2 modulates fat mass, aging, brood delivery, and growth through both SGK and AKT (Supplemental Fig. S9).
There are emerging data to suggest that SGK is an important, downstream effector of TORC2 in yeast and in mammals (Kamada et al. 2005; Aronova et al. 2008; Garcia-Martinez and Alessi 2008). The SGK family has been extensively implicated in sodium and potassium homeostasis in mammals (Lang and Cohen 2001; Lang et al. 2006). In mice and in humans, SGK1 and SGK3 have been linked to glucose homeostasis, salt balance, and blood pressure, and the SGK3 knockout mouse has a subtle, transient growth defect (Busjahn et al. 2002; McCormick et al. 2004). The SGK1 knockout mouse has no life-span phenotype (Lang et al. 2006). Our data show that SGK is a critical metabolic output of TORC2 in regulating growth and metabolism. It remains to be shown whether mammalian SGK1, SGK2, or SGK3 share these roles. It is possible that SGK2 plays this role, the knockout for which has not yet been reported, or that functional redundancy in the SGK isoforms in mammals prevents development of growth phenotypes in SGK1 and SGK3 knockouts and SGK1 and SGK3 double knockout mice.
RICT-1 and SGK-1 modulate life span in a nutrient-dependent manner
Based on the results we present here in rict-1 mutants and in rict-1;sgk-1 double mutants, we conclude that the defects evident in sgk-1 mutants are due to failure to activate the HM by TORC2 phosphorylation. Null, deletion mutations in C. elegans sgk-1(ok538) had been previously reported to show defects in growth, reproduction, and life span (Hertweck et al. 2004). Our loss-of-function allele sgk-1(mg455) shares all the defects in brood size, growth rate, and body size evident in the deletion allele. However, we observed a shortened life span in sgk-1 mutants that was identical to and not enhanced by rict-1 loss of function when animals were fed OP50. This is in contrast to previously published life-span analysis demonstrating dramatic life-span extension in sgk-1 knockdown by RNAi (Hertweck et al. 2004). In contrast to our studies, these previous studies were done without fluorodeoxyuridine (to inhibit progeny production) and by RNAi against sgk-1 (not the genetic mutant) using the E. coli strain HT115. These studies used RNAi because the genetic mutant life-span analysis was not possible without fluorodeoxyuridine due to an egg-laying defect and causing progeny to hatch within the parent (bag of worms phenotype). We also could not assess the life span of sgk-1(mg455) or rict-1 mutants without FUDR due to a highly penetrant bag of worms phenotype. Here, using genetic mutants and fluorodeoxyuridine, we demonstrate that rict-1 animals and sgk-1 animals are short-lived when fed the standard laboratory diet, E. coli OP50. We also showed that when rict-1 animals are fed either E. coli HB101 or HT115, they shift from being short-lived to being long-lived and consume less bacteria than wild type. Finally, RNAi of rict-1 in our hands under the conditions reported previously (i.e. without FUDR on HT115) also leads to life-span extension (data not shown). Thus, we conclude that rather than strictly extending or shortening life span, rict-1 and likely also sgk-1 modulate life span by affecting feeding and metabolism on different diets.
Feeding behavior is modulated by rict-1 such that mutants feed paradoxically on diets of different nutritional value. Our observations suggest that rict-1 mutants feed more robustly on E. coli OP50 and consume less E. coli HB101 by residing less on the lawn of bacteria. This behavior is not observed for wild-type worms, which rarely leave a lawn of HB101. Thus, rict-1 participates in the genetic pathway that regulates feeding behavior in response to foods of different quality. It remains to be shown what signal from the diet is sensed either directly or indirectly by RICT-1/TORC2 and how this modulates feeding behavior. There is a strong precedent for TOR complex 1 to be nutrient- and growth factor-regulated (Sarbassov et al. 2005a; Bhaskar and Hay 2007; Sancak et al. 2008). In cell lines and MEFs, TORC2-mediated phosphorylation of the HM of AKT is insulin-sensitive (Hresko and Mueckler 2005; Guertin et al. 2006), but TORC2 itself is not known to directly sense nutrients. It remains to be seen whether the normal response of C. elegans to different diets depends on direct nutrient-dependent Rictor/TORC2 modulation versus descending input to Rictor/TORC2 from other signaling pathways. However, our data indicate that rict-1 and sgk-1 are critical links between food and metabolic outputs, including feeding behavior, fat mass, growth, reproduction, and life span.
Fixative-based fat staining differs from labeling with BODIPY-labeled fatty acids in the case of the insulin-like signaling pathway mutants
A protocol to stain neutral lipid using the dye Oil-Red-O was developed in order to bypass the need for animals to uptake or accumulate vital dyes or fluorescently labeled fatty acids to indicate fat mass. While for the case of the rict-1 mutant, an increase in BODIPY and Nile red agreed with the increase in Oil-Red-O staining, in the case of insulin-like signaling mutants, particularly daf-2 and akt-1, BODIPY staining was not representative of triglyceride mass as assessed by Oil-Red-O or quantitative lipid biochemistry. This indicates, in some (and likely the minority of) cases, that Nile red or BODIPY fat mass may not be indicative of the true lipid stores within the animal. We do not yet know why the vital dyes or BODIPY behave differently in insulin-like signaling mutants than they do in most other animals, but one possible explanation is that there is decreased dye absorption via the hypodermis or the intestine, as insulin signaling pathway mutants have a decreased gut surface area (Albert and Riddle 1988; Hu 2007).
Genetic pathways upstream of and downstream from RICT-1/TORC2
Our results indicate that RICT-1 and the AGC kinases AKT-1, AKT-2, and SGK-1 participate in a pathway regulating fat metabolism, feeding, growth, reproduction, and life span. However, based on the data, the upstream inputs and downstream effectors of rict-1, akt, and sgk-1 are not known. In mammals and in C. elegans, it has been shown previously that SGK-1 could be activated by PI3K signaling through the upstream kinase PDK-1, much like AKT (Park et al. 1999; Lang and Cohen 2001; Hertweck et al. 2004). It is likely that PI3K/PDK-mediated activation of AKT and SGK regulate fat differently from proposed HM phosphorylation of these kinases by TORC2. Furthermore, here we demonstrate that activation of canonical PI3K signaling by genetic inactivation of daf-18 (PTEN) regulates fat mass in parallel to rict-1/TORC2. Thus, rict-1 does not appear to be downstream from daf-18 (PTEN) or the sole insulin-like receptor daf-2 in its regulation of fat mass. Thus, we suggest that pathways other than insulin-like or PIP3 signaling are upstream of rict-1 for fat regulation. For rict-1 life-span regulation, insulin-like signaling through akt-1 is required for the short life span. However, rict-1 short life span is modulated by akt-1 and not strictly caused by alterations in insulin-like or akt signaling (Supplemental Fig. S9). Thus, for rict-1 modulation of life span, the upstream signals also remain unclear.
In C. elegans, the only genetically and biochemically defined target of the AKT and SGK kinases had been the FOXO transcription factor DAF-16 (Hertweck et al. 2004). More recently, AKT-1, AKT-2, and SGK-1 have been shown to phosphorylate the transcription factor SKN-1 (Tullet et al. 2008). From our analysis of life span and of fat mass, the phenotypes of rict-1 animals are not mediated by daf-16, while interactions of rict-1 with skn-1 have yet to be explored in more detail. Thus, other than daf-16, additional downstream targets of AKT and SGK must be responsible for mediating rict-1 phenotypes. Along these lines, in mammals, AKT and SGK phosphorylate several targets in addition to the daf-16 homolog FOXO3a (Brunet et al. 2001; Lang et al. 2006). Finally, TORC2 also phosphorylates the protein kinase C family (Sarbassov et al. 2004; Guertin et al. 2006; Facchinetti et al. 2008). We have just begun to understand the role of rict-1 and sgk-1 in metabolic regulation in C. elegans, and further work is needed to elucidate the upstream and downstream signaling molecules.
Overall, these observations indicate that TORC2 regulates metabolism, feeding, growth, and reproduction, and modulates life span in C. elegans. The dominant site of TORC2 and sgk-1 action is the intestine, and the role of these kinases in other tissues remains to be more fully explored. Since the intestine is the major site of fat storage and is an endocrine organ regulating life span (Libina et al. 2003), it is not surprising that signals generated within the intestine could have such pleiotropic effects in C. elegans. Given the evolutionary conservation of Rictor, the components of TORC2, and the SGK family of kinases, it will be interesting to see if conditional, tissue-specific inactivation of rictor, sgk isoforms, or other TORC2 components in mice produces alterations in body fat, growth, and life span, or protects them from obesity induced by nutrient-rich food, as do rict-1 mutations in C. elegans.
Materials and methods
Strains used
N2 Bristol was used as the wild-type strain. The following mutant strains were used: daf-2(e1370), daf-16(mgDf47), akt-1(mg306), akt-2(ok393), daf-18(mg198), skn-1(zu169). For tissue distribution of rict-1, mgEx744, mgEx745: Ex[F29C12.3p∷mRFP;myo-2p∷GFP], 1.4 kb of upstream rict-1 promoter sequence was used. For rescue experiments, mgEx747: Ex[F29C12.3p∷RICT-1 cDNA∷SL2∷GFP; myo-2p∷mCherry]. For neuronal rescue, mgEx754: Ex[unc119p∷RICT-1 cDNA∷SL2∷GFP; myo-2p∷GFP], 2.2 kb of upstream promoter was used. For intestinal rescue, Ex[ges-1p∷RICT-1∷SL2∷GFP;myo-2p∷GFP], 2.1 kb of upstream promoter was used. For Ex[ifb-2p∷RICT-1∷SL2∷GFP;myo-2p∷GFP], 3 kb of upstream promoter was used. For sgk-1 gain of function, mgEx757[rict-1p∷SGK-1(Δ453T → K) cDNA∷SL2∷GFP;myo-2p∷GFP]. For sgk-1 rescue, BR2773(byEx[sgk-1∷GFP]) (Hertweck et al. 2004) was crossed into rict-1(mg451) and sgk-1(mg455). rict-1 cDNA was amplified obtaining a dominant PCR product from wild-type worms and cloned, and several clones were sequenced to confirm the intact reading frame (Supplemental Fig. S1).
Isolation of rict-1(F29C12.3) and sgk-1 mutants
C. elegans N2 Bristol strain was mutagenized with EMS, and the resultant F2 generation was screened for increased staining with the vital dye Nile red (Ashrafi et al. 2003; Mak et al. 2006). Individual isolates were back-crossed to N2 Bristol and positionally cloned based on polymorphisms between N2 and the multiply polymorphic C. elegans strain CB4586.
Body fat assessment
Body fat analysis was conducted as described previously (Mak et al. 2006). Briefly, wild-type or mutant C. elegans was seeded on NGM plates containing either E. coli OP50 or HB101 supplemented with 1 μM the fluorescent fatty acid analog C1-BODIPY 500/510-C12 (Invitrogen) or Nile red (Invitrogen) from a 2-h synchronous egg lay or as L1 following overnight hatching and synchronization at 20°C in minimal media. Imaging and quantitation was conducted after growth at 20°C as day 3 adults (Figs. 1, 3B) or day 1–2 adults (Fig. 3A) using an Axioplan microscope and Axiovision software (Zeiss). At least 30 animals were imaged on at least three separate occasions, and results were consistent between experiments. The vital dye Nile red (Invitrogen) was used to confirm phenotypes of rict-1(mg450) and rict-1(mg451) observed by BODIPY staining, as described (Mak et al. 2006). All body fat analyses were carried out on animals grown at 20°C.
Oil-Red-O staining was conducted by washing 200–300 day 1 adult animals from plates synchronized at the time of the first progeny hatching (12 h after the first egg is laid). The worms were resuspended and washed twice with PBS and then suspended in 120 μL of PBS to which an equal volume of 2× MRWB buffer containing 2% paraformaldehyde was added (composition: 160 mM KCl, 40 mM NaCl, 14 mM Na2EGTA, 1 mM Spermidine HCl, 0.4 mM Spermine, 30 mM Na PIPES at pH 7.4, 0.2% BME). The worms were taken through three freeze–thaw cycles between dry-ice/ethanol and warm running tap water, followed by spinning at 14,000g, washing once in PBS to remove PFA, resuspension in 60% isopropanol to dehydrate, and addition of 60% Oil-Red-O stain (prepared as follows: from 0.5 g/100 mL isopropanol stock solution equilibrated for several days, freshly dilute with 40% water, 60% stock, and allow to sit 10 min and filter 0.2 to 0.4 μm). Oil-Red-O analysis in mosaic animals could not be completed due to the disappearance of GFP with the fixation necessary to complete Oil-Red-O staining. Although it is possible to quantitate the amount of Oil-Red-O staining in individual animals, for the purposes of this study we relied on quantitative lipid biochemistry and not Oil-Red-O quantitation to determine the amount of stored triglyceride in a given genotype. When quantitated, Oil-Red-O correlated well with triglyceride mass determined by quantitative lipid biochemistry (data not shown).
Quantitative lipid biochemistry was conducted as published previously (Perez and Van Gilst 2008) with the following modifications: Briefly, 7500 worms at day 1 of adulthood were washed from three 10-cm plates containing the indicated food, and washed over a 20-μm mesh (Small Parts, Inc.) with an excess of S-Basal medium to remove progeny, eggs, and bacteria. These were allowed to sit on an empty plate for 20 min to empty bacteria from the gut. Afterward, worms were reharvested and frozen on liquid nitrogen. The total worm pellet was sonicated in 0.25 mL of PBS, after which we added 50 μL of triglyceride and phospholipids standards dissolved in chloroform:methanol 2:1 (16.7 nmol of tritridecanoin, Nu-Chek Prep, and 25 nmol of 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine, Matreya, respectively), and immediately added to 1.5 mL of 2:1 chloroform:methanol to extract lipids. This mixture was spun down (1000g for 1 min), and the lower organic phase was recovered without debris. This was washed with 3 mL of 0.9% NaCl and spun at 1000g to separate phases. The lower organic phase was evaporated under nitrogen, and lipids were resuspended in 100% chloroform to conduct solid-phase chromatography using prepacked silica gel columns (Thermo Scientific) followed by GCMS as reported previously (Perez and Van Gilst 2008). For all measurements, at least two biological replicates were performed, with data shown as mean ± SEM. Where a minimum of four measurements were conducted, statistical analysis was carried out using pairwise Student's t-tests.
Pharyngeal pumping rates
Pumping rate was determined using a Sony camera attached to a Zeiss microscope for between 10 and 12 synchronized, well-fed day 1 adult animals per genotype grown on the same food that pumping assays were determined on. Pharyngeal contractions in 20- to 30-sec time periods were counted using iMovie (Apple), and pumping rates per minute were calculated.
Oxygen consumption
Synchronized, day 1 adult worms grown on either OP50 or HB101 were washed repeatedly over a 35-μm nylon mesh (Small Parts, Inc., for wild type) or a 20-μm nylon mesh (for rict-1 mutants) to remove bacteria, eggs, and larvae. Oxygen consumption of 2500 worms was then measured immediately in 1.5 mL of S-basal medium without cholesterol using a Strathkelvin 928 6-channel Clark electrode respirometer. Six to 10 independent measurements were conducted and averaged per strain, per food source and normalized to total protein content, determined using a BCA assay (Pierce).
Brood size determination
Ten to 12 synchronized animals per strain, per food source were transferred daily to fresh NGM plates containing OP50 or HB101, and hatched progeny were counted 3 d later. Total brood size was determined by adding progeny produced across all days.
Body size determination
Maximal, longitudinal cross-sectional area was obtained by quantitative microscopy using Zeiss axiovision software at identical time points that images were taken for fat mass determination.
Egg-to-egg time
Eggs were synchronized by 2 h synchronous egg lay. At the L4 stage, 20 animals per strain per food source were placed on an individual plate and monitored every 1–2 h until the first egg was laid. The data were expressed as the mean time until first egg laid ±SEM.
Feeding RNAi
RNAi clones were isolated from a genome-wide E. coli feeding RNAi library and fed to C. elegans as described previously (Kamath and Ahringer 2003; Kamath et al. 2003). E. coli clones expressing dsRNA corresponding to control vector L4440, rict-1, or sinh-1 were supplemented with C1-BODIPY 500/510 C12 or Nile red, to determine fat content as above, or put on unsupplemented plates for Oil-Red-O staining. Synchronous populations of worms were obtained by bleach treatment of gravid adults, and after one generation of RNAi feeding, animals were examined and imaged as 2-d adults for fat content.
Longevity assay
Animals from the parental generation were grown on the same food source that life span was conducted on. Synchronous animals were obtained by bleach treatment of gravid adults and seeded onto NGM OP50 or HB101 plates. As day 1 adults, 30 animals per genotype were transferred in triplicate (90 total) to fresh NGM OP50 or HB101 plates supplemented with 100 μM 5-fluorodeoxyuridine (FUDR) solution to suppress progeny production. Three independent assays were carried out using strains on different diets: (1) OP50 diet only, (2) OP50 or HB101, and (3) OP50, HB101, or HT115 diets. daf-2(e1370), daf-16 (mgDf47), akt-1 (mg306), akt-1(mg144gf), akt-2 (ok393), and sgk-1(mg455) epistasis analysis was carried out in triplicate using strains grown on OP50. skn-1(zu169) epistasis was carried out in triplicate on OP50 and HB101. All life-span analyses reported were conducted at 25°C.
Gompertz-Makeham survival analysis
Prior to analysis, data were pooled across independent experiments to give unbiased estimates of survival at each assayed age. The Gompertz-Makeham model for age-specific mortality M = M0 exp(Gx) + M∞, with initial mortality M0, rate of aging G, age x, and age-independent mortality M∞, can be shown to give the survival proportion S(x) = exp[(M0/G)(1 − exp(Gx) − M∞x)]. We performed population-weighted nonlinear least-squares Gompertz model fits using a Trust-Region Reflective Newton algorithm implemented by the MATLAB fit() function (The MathWorks). Median and maximum life span were calculated from each fit as the age of 50% and 25% estimated survival, respectively.
Acknowledgments
We thank Sean Curran, Eyleen O'Rourke, Susana Garcia, Meng Wang, Chi Zhang, and Buck Samuel for creative input, sharing materials, and reading of the manuscript. Thanks to Eyleen O'Rourke for help in developing Oil-Red-O staining and methods for quantitative lipid biochemistry. Thanks to Joseph Avruch for critical reading of the manucript and help with interpretation of the data. We are indebted to Mason Freeman for the use of his GCMS for lipid analysis. We also thank members of the Kaplan and Ausubel laboratories for creative exchanges, and Diane Sacchetti for administrative assistance. Thank you to Kaveh Ashrafi and Kevin Jones for sharing their findings on Rictor with us. Thanks to the Caenorhabditis Genome Center and Ralf Baumeister for providing strains, and the C. elegans Knockout Consortium for generating ok alleles. This work was supported by Award numbers R01DK070147 (to G.R.) and F32DK080607 (to A.S.) from the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1775409.
Supplemental material is available at http://www.genesdev.org.
References
- Albert P.S., Riddle D.L. Mutants of Caenorhabditis elegans that form dauer-like larvae. Dev. Biol. 1988;126:270–293. doi: 10.1016/0012-1606(88)90138-8. [DOI] [PubMed] [Google Scholar]
- Aronova S., Wedaman K., Aronov P.A., Fontes K., Ramos K., Hammock B.D., Powers T. Regulation of ceramide biosynthesis by TOR complex 2. Cell Metab. 2008;7:148–158. doi: 10.1016/j.cmet.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K., Chang F.Y., Watts J.L., Fraser A.G., Kamath R.S., Ahringer J., Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003;421:268–272. doi: 10.1038/nature01279. [DOI] [PubMed] [Google Scholar]
- Avery L., Shtonda B.B. Food transport in the C. elegans pharynx. J. Exp. Biol. 2003;206:2441–2457. doi: 10.1242/jeb.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger C.F., Romanino K., Cloetta D., Lin S., Mascarenhas J.B., Oliveri F., Xia J., Casanova E., Costa C.F., Brink M., et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Bhaskar P.T., Hay N. The two TORCs and Akt. Dev. Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Bishop N.A., Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans . Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Brunet A., Park J., Tran H., Hu L.S., Hemmings B.A., Greenberg M.E. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol. Cell. Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busjahn A., Aydin A., Uhlmann R., Krasko C., Bahring S., Szelestei T., Feng Y., Dahm S., Sharma A.M., Luft F.C., et al. Serum- and glucocorticoid-regulated kinase (SGK1) gene and blood pressure. Hypertension. 2002;40:256–260. doi: 10.1161/01.hyp.0000030153.19366.26. [DOI] [PubMed] [Google Scholar]
- Facchinetti V., Ouyang W., Wei H., Soto N., Lazorchak A., Gould C., Lowry C., Newton A.C., Mao Y., Miao R.Q., et al. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 2008;27:1932–1943. doi: 10.1038/emboj.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch C.E., Ruvkun G. The genetics of aging. Annu. Rev. Genomics Hum. Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- Frias M.A., Thoreen C.C., Jaffe J.D., Schroder W., Sculley T., Carr S.A., Sabatini D.M. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr. Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Garcia-Martinez J.M., Alessi D.R. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem. J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- Guertin D.A., Stevens D.M., Thoreen C.C., Burds A.A., Kalaany N.Y., Moffat J., Brown M., Fitzgerald K.J., Sabatini D.M. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCα, but not S6K1. Dev. Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Hertweck M., Gobel C., Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev. Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Hresko R.C., Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J. Biol. Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- Hu P.J. Dauer. The C. elegans Research Community, editor. WormBook. 2007 doi: 10.1895/wormbook.1.144.1. WormBook. http://www.wormbook.org. [DOI]
- Jia K., Chen D., Riddle D.L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Kamada Y., Fujioka Y., Suzuki N.N., Inagaki F., Wullschleger S., Loewith R., Hall M.N., Ohsumi Y. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol. 2005;25:7239–7248. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R.S., Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans . Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kimura K.D., Tissenbaum H.A., Liu Y., Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans . Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Kumar A., Harris T.E., Keller S.R., Choi K.M., Magnuson M.A., Lawrence J.C., Jr Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances basal glycogen synthase activity. Mol. Cell. Biol. 2008;28:61–70. doi: 10.1128/MCB.01405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F., Cohen P. Regulation and physiological roles of serum- and glucocorticoid-induced protein kinase isoforms. Sci. STKE. 2001;2001:RE17. doi: 10.1126/stke.2001.108.re17. [DOI] [PubMed] [Google Scholar]
- Lang F., Bohmer C., Palmada M., Seebohm G., Strutz-Seebohm N., Vallon V. (Patho)physiological significance of the serum- and glucocorticoid-inducible kinase isoforms. Physiol. Rev. 2006;86:1151–1178. doi: 10.1152/physrev.00050.2005. [DOI] [PubMed] [Google Scholar]
- Lee G., Chung J. Discrete functions of rictor and raptor in cell growth regulation in Drosophila . Biochem. Biophys. Res. Commun. 2007;357:1154–1159. doi: 10.1016/j.bbrc.2007.04.086. [DOI] [PubMed] [Google Scholar]
- Libina N., Berman J.R., Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Long X., Spycher C., Han Z.S., Rose A.M., Muller F., Avruch J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 2002;12:1448–1461. doi: 10.1016/s0960-9822(02)01091-6. [DOI] [PubMed] [Google Scholar]
- Long X., Muller F., Avruch J. TOR action in mammalian cells and in Caenorhabditis elegans . Curr. Top. Microbiol. Immunol. 2004;279:115–138. doi: 10.1007/978-3-642-18930-2_8. [DOI] [PubMed] [Google Scholar]
- Mak H.Y., Nelson L.S., Basson M., Johnson C.D., Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- McCormick J.A., Feng Y., Dawson K., Behne M.J., Yu B., Wang J., Wyatt A.W., Henke G., Grahammer F., Mauro T.M., et al. Targeted disruption of the protein kinase SGK3/CISK impairs postnatal hair follicle development. Mol. Biol. Cell. 2004;15:4278–4288. doi: 10.1091/mbc.E04-01-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner B., Boll M., Daniel H., Baumeister R. Deletion of the intestinal peptide transporter affects insulin and TOR signaling in Caenorhabditis elegans . J. Biol. Chem. 2004;279:36739–36745. doi: 10.1074/jbc.M403415200. [DOI] [PubMed] [Google Scholar]
- Morris J.Z., Tissenbaum H.A., Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans . Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Ogg S., Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol. Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G.I., Lee L., Tissenbaum H.A., Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans . Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Paradis S., Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes & Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S., Ailion M., Toker A., Thomas J.H., Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans . Genes & Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Leong M.L., Buse P., Maiyar A.C., Firestone G.L., Hemmings B.A. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. EMBO J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce L.R., Huang X., Boudeau J., Pawlowski R., Wullschleger S., Deak M., Ibrahim A.F., Gourlay R., Magnuson M.A., Alessi D.R. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem. J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C.L., Van Gilst M.R. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans . Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Porte D., Jr, Baskin D.G., Schwartz M.W. Insulin signaling in the central nervous system: A critical role in metabolic homeostasis and disease from C. elegans to humans. Diabetes. 2005;54:1264–1276. doi: 10.2337/diabetes.54.5.1264. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Kim D.H., Guertin D.A., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sabatini D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005a;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005b;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Kunz J., Hall M.N. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc. Natl. Acad. Sci. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiota C., Woo J.T., Lindner J., Shelton K.D., Magnuson M.A. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell. 2006;11:583–589. doi: 10.1016/j.devcel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Shtonda B.B., Avery L. Dietary choice behavior in Caenorhabditis elegans . J. Exp. Biol. 2006;209:89–102. doi: 10.1242/jeb.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas A., Socci N.D., Saatkamp B.D., Novelli S., Friedman J.M. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J. Biol. Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- Tabuchi M., Audhya A., Parsons A.B., Boone C., Emr S.D. The phosphatidylinositol 4,5-biphosphate and TORC2 binding proteins Slm1 and Slm2 function in sphingolipid regulation. Mol. Cell. Biol. 2006;26:5861–5875. doi: 10.1128/MCB.02403-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet J.M., Hertweck M., An J.H., Baker J., Hwang J.Y., Liu S., Oliveira R.P., Baumeister R., Blackwell T.K. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans . Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A.L., Orosz L., Muller F. Genetics: Influence of TOR kinase on lifespan in C. elegans . Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Wolff S., Dillin A. The trifecta of aging in Caenorhabditis elegans . Exp. Gerontol. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Wolkow C.A., Kimura K.D., Lee M.S., Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Yang Q., Inoki K., Ikenoue T., Guan K.L. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes & Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]