Abstract
Phosphorylation is essential for the SR family of splicing factors/regulators to function in constitutive and regulated pre-mRNA splicing; yet both hypo- and hyperphosphorylation of SR proteins are known to inhibit splicing, indicating that SR protein phosphorylation must be tightly regulated in the cell. However, little is known how SR protein phosphorylation might be regulated during development or in response to specific signaling events. Here, we report that SRPK1, a ubiquitously expressed SR protein-specific kinase, directly binds to the cochaperones Hsp40/DNAjc8 and Aha1, which mediate dynamic interactions of the kinase with the major molecular chaperones Hsp70 and Hsp90 in mammalian cells. Inhibition of the Hsp90 ATPase activity induces dissociation of SRPK1 from the chaperone complexes, which can also be triggered by a stress signal (osmotic shock), resulting in translocation of the kinase from the cytoplasm to the nucleus, differential phosphorylation of SR proteins, and alteration of splice site selection. These findings connect the SRPK to the molecular chaperone system that has been implicated in numerous signal transduction pathways and provide mechanistic insights into complex regulation of SR protein phosphorylation and alternative splicing in response to developmental cues and cellular signaling.
Keywords: SR protein-specific kinase, molecular chaperones, stress signaling, SR protein phosphorylation, alternative splicing
Pre-mRNA processing consists of a highly regulated cascade of events that are critical for gene expression in higher eukaryotic cells. SR proteins are an important class of splicing factors or regulators because of their involvement in both constitutive and regulated splicing (Lin and Fu 2007). More recent studies reveal an even broader role of SR proteins in gene expression from transcriptional elongation to protein synthesis (Sanford et al. 2004; Lin et al. 2008; Michlewski et al. 2008). These fundamentally important functions render SR proteins essential for viability of proliferating cells (Wang et al. 1996; Lin et al. 2005; Xiao et al. 2007). SR proteins are regulated in development, which are translocated from the cytoplasm to the nucleus during zygotic activation of gene expression (Sanford and Bruzik 1999). Individual SR proteins are also autoregulated, suggesting critical importance in maintaining their homeostasis in somatic cells (Lareau et al. 2007). Indeed, some SR proteins exhibit altered expression in human cancers (Ghigna et al. 1998; Stickeler et al. 1999; Pind and Watson 2003), and a recent study demonstrated that overexpression of a specific SR protein, SF2/ASF, is sufficient to trigger cellular transformation (Karni et al. 2007). These observations indicate that SR proteins must be tightly regulated and alteration of such regulation can have a profound impact on the physiological state of the cell.
Typical SR proteins contain one or two RNA recognition motifs (RRMs) followed by a signature serine/arginine-rich sequence known as the RS domain at the C terminus. The RS domains are extensively phosphorylated. While SR protein phosphorylation is essential for nuclear import of SR proteins as well as for their functions in mediating spliceosome assembly (Roscigno and Garcia-Blanco 1995; Xiao and Manley 1997; Yeakley et al. 1999; Yun and Fu 2000; Lai et al. 2001), partial dephosphorylation of SR proteins is also critical for progression of the assembled spliceosome to catalysis (Mermoud et al. 1994; Cao et al. 1997) and for a series of post-splicing events from the interaction with the mRNA transport machinery to SR protein-mediated translational control (Huang et al. 2004; Lai and Tarn 2004; Lin et al. 2005; Sanford et al. 2005). Consequently, it may not be surprising that experimental induction of both SR protein hypo- and hyperphosphorylation inhibits splicing (Prasad et al. 1999). While these observations clearly suggest that SR protein phosphorylation is under precise control, little is known about the mechanism of such regulation in the cell.
Multiple protein kinases have been implicated in SR protein phosphorylation. Among the growing list of SR protein kinases, the SRPK and Clk/Sty families are best characterized. Mammalian cells express two SRPKs and four members of the Clk/Sty family of kinases. Interestingly, SRPK1 and SRPK2 were shown recently to differentially associate with U1 and tri-snRNP particles, respectively, indicating that these kinases have both overlapping and unique functions in mammalian cells (Mathew et al. 2008). Enzymatic analysis reveals that SRPKs use a highly processive mechanism to phosphorylate a defined region in the RS domain in each SR protein (Aubol et al. 2003; Ngo et al. 2008), and Clk/Sty can further phosphorylate the remaining sites in the RS domain (Ngo et al. 2005; Velazquez-Dones et al. 2005), suggesting the possibility that these kinases may catalyze a cooperative phosphorylation relay to modulate SR protein function at different biochemical steps and/or in various cellular locations (Ngo et al. 2005; Hagopian et al. 2008). This idea is consistent with their cellular distributions: While members of the Clk/Sty family of kinases are predominately localized in the nucleus (Colwill et al. 1996; Nayler et al. 1998), the SRPK family of kinases are detected in both the cytoplasm and the nucleus (Wang et al. 1998; Ding et al. 2006).
Despite its importance, little is known about how SR protein phosphorylation might be regulated in the cell and how a specific signal might be transduced to control RNA processing via modulation of SR protein phosphorylation. Recent discoveries from the Manley laboratory shed a critical light on these important questions by revealing dramatic dephosphorylation of a specific SR protein (SRp38) in response to heat shock (Shin et al. 2004; Shi and Manley 2007). The regulation is achieved by an increased exposure of the protein to the activated protein phosphatase PP1 in combination with limited accessibility of the protein to SR protein kinases under heat-shock conditions, underscoring the importance of a balanced action between SR protein kinases and phosphatases in controlling the phosphorylation state of SR proteins.
In the present study, we focused on understanding how SR protein kinases are regulated. Our previous work showed that SRPK1 is a constitutively active kinase (Nolen et al. 2001; Lukasiewicz et al. 2007) and that an accessory domain (a spacer sequence that splits conserved kinase domains into two blocks) is involved in partitioning of the kinase between the cytoplasm and nucleus, suggesting that the cellular distribution of the kinase, rather than activity, is subject to regulation (Ding et al. 2006). We now show that SRPK1 directly interacts with two specific cochaperones for major heat-shock proteins and that the ATPase activity of Hsp90 plays a critical role in regulating the cellular distribution of the kinase in the cell. We further demonstrate that osmotic stress can induce SRPK1 nuclear translocation by modulating the dynamic interaction of SRPK1 with the chaperone complexes, thereby inducing differential SR protein phosphorylation and alternative splice site selection. These findings reveal a novel strategy by which to regulate SR protein phosphorylation and alternative splicing in higher eukaryotic cells.
Results
Identification of specific heat-shock cochaperones as SRPK1-interacting proteins
The SRPK family of kinases is highly conserved from budding yeast to humans. Each family member contains a unique spacer sequence that splits the conserved kinase domains into two halves (Fig. 1A), a feature characteristic of many receptor tyrosine kinases where the spacer sequences are frequently involved in signal transduction (Pawson and Scott 1997). Previous studies showed that removal of the spacer in SRPK1 family members had little effect on their kinase activities (Nolen et al. 2001; Aubol et al. 2003), but dramatically altered the cellular distribution of the kinases from largely cytoplasmic to exclusively nuclear (Takeuchi and Yanagida 1993; Siebel et al. 1999; Ding et al. 2006). These observations led us to hypothesize that the accessory spacer domain may function as an effector domain in response to certain cellular signals to control the cellular distribution of the kinase, thereby regulating SR protein phosphorylation and alternative splicing in the nucleus.
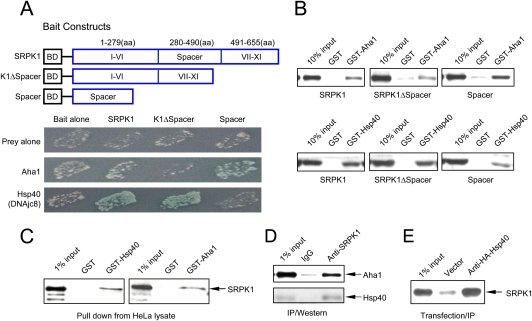
Figure 1.
Identification and validation of SRPK1-interacting proteins. (A) The bait constructs used in the two-hybrid screening. In full-length SRPK1, conserved kinase domains are split by a unique spacer sequence. The spacer was also constructed in the bait vector and used in a separate screen. The mutant SRPK1 deleted of the spacer (K1ΔSpacer) is toxic when stably expressed in yeast. This construct was only used for validation in transient assays. Pairwise two-hybrid interactions are indicated by both growth and the activity of the MEL1 reporter. (B) GST pull-down assay to confirm direct interactions between SRPK1 and specific chaperones in vitro. GST or GST fusion proteins as indicated were used to pull down recombinant SRPK1, K1ΔSpacer, or Spacer expressed and purified from bacteria. (C) GST or GST fusion proteins were also used to pull down SRPK1 from HeLa lysate. (D) Interaction of SRPK1 with Aha1 and Hsp40 in vivo. SRPK1 was immunoprecipitated from HeLa lysate followed by Western blotting analysis using anti-Aha1 and anti-pan-Hsp40 antibodies, the latter of which recognize multiple members of the Hsp40 family. (E) Specific interaction between SRPK1 and DNAjc8/Hsp40 in vivo. The HA-tagged DNAjc8/Hsp40 was transfected into HEK293 cells followed by anti-HA IP. SRPK1 in the complex was detected with anti-SRPK1. An empty vector was transfected and analyzed in parallel as a background control.
To test this hypothesis, we performed two-hybrid screens using both full-length SRPK1 and the spacer sequence from SRPK1 as baits (Fig. 1A). The spacer deletion mutant was not used in the initial screen because sustained expression of the mutant kinase was deleterious to yeast (Siebel et al. 1999). However, the mutant could still be used to test candidate targets in transient assays. Targets identified by the full-length SRPK1 bait include a large number of RS domain-containing splicing factors, consistent with our previous findings and confirming the specificity of the kinase for RS domain-containing proteins (Tronchere et al. 1997). In the present study, we focused on proteins outside this group, which led to the identification of a Hsp70 cochaperone known as DNAjc8, which belongs to the large Hsp40 family of heat-shock proteins (Ohtsuka and Hata 2000). Interestingly, DNAjc8 had been detected previously in purified spliceosomes, suggesting a potential role of this cochaperone in RNA processing-related processes (Zhou et al. 2002). For clarity, we use DNAjc8 and Hsp40 interchangeably in this report. A parallel screen with the spacer revealed another cochaperone called Aha1 for the major heat-shock protein Hsp90 (Panaretou et al. 2002). As indicated by the induced α-gal activity (blue), pairwise two-hybrid tests with different SRPK1 bait constructs confirmed that DNAjc8/Hsp40 preferentially interacted with SRPK1 outside the spacer, whereas Aha1 targeted the spacer (Fig. 1A).
The observation that SRPK1 interacts with specific cochaperones for major heat-shock proteins raised the possibility that the kinase may be regulated by heat-shock complexes, which have been implicated in numerous signal transduction pathways (Pratt and Toft 2003). In general, the assembly of chaperone complexes is initiated by Hsp40/Hsp70 binding to a specific client followed by the joining of Hsp90 and its cochaperones, which together are critical for proper folding of the client into its active conformation. The chaperone complexes also protect the client from degradation by the proteosome and the ATPase activity of Hsp90 is known to modulate dynamic client/chaperone interactions, thereby controlling the cellular localization and intracellular trafficking of the client. This functional profile of the molecular chaperone system is consistent with a potential role of molecular chaperones in regulating the cellular partitioning of SRPK1 and related kinases.
Direct interaction of SRPK1 with specific cochaperones
To biochemically characterize the candidates from the two-hybrid screens and determine whether SRPK1 directly interacts with the identified cochaperones, we prepared recombinant proteins for GST pull-down assays (Fig. 1B). We found that GST-Hsp40 interacts more robustly with full-length SRPK1 and the spacer-deleted kinase, compared with the spacer alone. Conversely, GST-Aha1 interacted more robustly with the spacer, compared with full-length and spacer-deleted SRPK1. The reduced affinity of Aha1 for the full-length kinase relative to the spacer alone suggests that, while the cochaperone targets the spacer, there may be some steric constraints for this domain in the full-length kinase (Fig. 1A,B). Taken together, these results suggest that SRPK1 is engaged in multiple and potentially cooperative interactions with specific cochaperones for major heat-shock proteins in the cell.
To substantiate the interaction between SRPK1 and the cochaperons in mammalian cells, we next used the GST pull-down assay to show that the interactions of Aha1 and Hsp40 with SRPK1 took place in HeLa lysate in the presence of numerous other cellular proteins (Fig. 1C). We also performed the coimmunoprecipitation (co-IP)/Western experiments to demonstrate that both cochaperones formed complexes with SRPK1 in vivo using an anti-Aha1 antibody and a pan-Hsp40 antiserum (Fig. 1D). To further confirm the specific interactions between SRPK1 and DNAjc8/Hsp40 in vivo, we transfected a HA-tagged DNAjc8 cDNA into 293 cells and showed its specific interactions with endogenous SRPK1 (Fig. 1E). Taken together, these results demonstrate the direct interactions of SRPK1 with the heat-shock cochaperones both in vitro and in vivo.
Requirement for cochaperones to link SRPK1 to major heat-shock proteins
The interactions between SRPK1 and specific cochaperones for heat-shock proteins strongly suggest that the kinase may interact with major heat-shock complexes in the cell. To test this possibility, we performed co-IP experiments with anti-SRPK1, and indeed, detected both Hsp70 and Hsp90 in SRPK1-containing complexes (Fig. 2A). This finding is consistent with the existing information on Hsp40 as cochaperone for Hsp70 and Aha1 as cochaperone for Hsp90 (Zylicz et al. 2001; Panaretou et al. 2002; Pratt and Toft 2003). To determine whether these cochaperones are required for bridging SRPK1 to major heat-shock proteins in the cell, we carried out RNAi knockdown experiments against HSP40 and AHA1. Quantitative RT–PCR confirmed specific down-regulation of respective transcripts in specific RNAi-treated, but not control siRNA-treated cells (Fig. 2B). We further confirmed by Western blotting a major reduction of the Aha1 protein in both single and double siRNA-treated cells (Fig. 2C). Because of the lack of specific antibody against DNAjc8/Hsp40, we could not directly verify its reduction at the protein level; however, the qPCR result strongly suggested a successful knockdown.
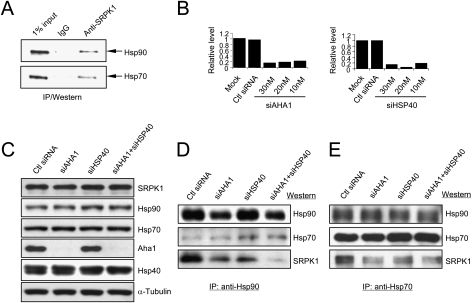
Figure 2.
Cochaperone-mediated association of SRPK1 with major heat-shock proteins. (A) Detection of Hsp70 and Hsp90 in SRPK1 complexes. (B) siRNA-mediated knockdown of Aha1 and DNAjc8/Hsp40 transcripts, which was quantified by real time RT–PCR. Scrambled siRNA was tested as control. (C) Knockdown of the Aha1 protein in single and double siRNA-treated HeLa cells. We could not directly validate DNAjc8/Hsp40 knockdown because of the lack of a specific antibody. (D,E) Reduced association of SRPK1 with major chaperones in HeLa cells deleted of Aha1, DNAjc8/Hsp40, or both. The chaperone complex was immunoprecipitated by using anti-Hsp90 (D) and anti-Hsp70 (E). The amount of associated SRPK1 was dramatically reduced in the absence of both Aha1 (cochaperone for Hsp90) and DNAjc8/Hsp40 (cochaperone for Hsp70). A similar, but less reduction of SRPK1 in Hsp70-containing complexes was observed.
Co-IP analysis of lysates from siRNA-treated cells revealed that RNAi knockdown of either AHA1 or HSP40 attenuated the association of SRPK1 with Hsp90 and simultaneous knockdown of both cochaperones practically eliminated the interaction (Fig. 2D). A similar, but smaller effect was also detected with anti-Hsp70 (Fig. 2E), perhaps reflecting a degree of functional overlap among members of the large Hsp40 family. We also confirmed attenuated association of SRPK1 with Hsp70 and Hsp90 in Hsp40 and Aha 1 knockdown cells by anti-SRPK1 IP followed by Western blotting against the major heat-shock proteins (Supplemental Fig. S1A). These experiments ruled out the possibility that the detected association of SRPK1 with Hsp70 and Hsp90 was due to the abundant nature of these heat-shock proteins in the cell. Importantly, despite specific partnership between Hsp40 and Hsp70 and between Aha1 and Hsp90, down-regulation of either cochaperone clearly affected the stability of SRPK1 in interacting with both Hsp70 and Hsp90. Although both the Hsp70 and Hsp90 chaperone systems have been implicated in many signal transduction processes (Pratt and Toft 2003), to our knowledge, our results show for the first time a crosstalk between the two chaperone systems, which likely also harbor other cofactors or connectors for the chaperone machineries.
Requirement of the Hsp90 ATPase activity for dynamic SRPK1/molecular chaperone interactions
Molecular chaperones are known to facilitate protein folding, and in many cases, they also serve to anchor folded proteins before releasing them to their sites of actions in the cell (Donze et al. 2001). Consistently, in Aha1 or Hsp40 knockdown cells, we detected by anti-SRPK1 IP the same amount of SRPK1, but less kinase activity toward the SR protein SF2/ASF relative to control siRNA-treated cells, indicating that SRPK1 folding was impaired in Aha1 and Hsp40 down-regulated cells (Supplemental Fig. S1B). Because we showed previously that the kinase activity is required for SRPK1 to enter the nucleus (Ding et al. 2006), we detected some minor alteration in the cellular distribution of SRPK1 in siAHA1-treated cells relative to control siRNA-treated cells and no difference in siHSP40-treated cells (Supplemental Fig. S1C).
Because of the protein folding problem in cells that had been subjected to relatively long-term (2–3 d) treatment with siRNAs, we explored an alternative strategy to modulate the association of SRPK1 with the chaperone system by using specific inhibitors, such as Geldanamycin or its derivative 17-AAG, to transiently inhibit the ATPase activity of Hsp90, which has been documented for its essential function in many documented cellular pathways (e.g., Goetz et al. 2003). To first determine whether the association of SRPK1 with heat-shock proteins depends on the ATPase activity of Hsp90, we treated HeLa cells with 17-AAG for 16 h, which is well known to induce the expression of multiple heat-shock proteins, including Hsp70 and Hsp90 (Fig. 3A). We next performed anti-SRPK1 IP followed by Western blotting analysis of Hsp90 in the complex and observed that the level of Hsp90 associated with SRPK1 was dramatically reduced, while Hsp70 appeared little altered, if not modestly increased, in response to the 17-AAG treatment (Fig. 3B). No signal was detected with control IgG side by side performed with anti-SRPK1 (data not shown). Note that, in light of the observation that the association of Hsp70 with SRPK1 is partially dependent on Aha1/Hsp90 as shown in Fig. 2E and Supplemental Fig. S1A, we suspect that Hsp70 overexpression in the presence of functional Hsp40 may have compensated for the loss of Aha1/Hsp90 from the SRPK1-containing complex in 17-AGG-treated cells. The precipitated kinase was equally, if not more, active from 17-AAG-treated cells compared with that from mock-treated cells, which is in contrast to the situation in Aha1 and Hsp40 knockdown cells (Supplemental Fig. S1B). This result indicates that SRPK1 was released from Hsp90 upon inhibition of the ATPase activity of the heat-shock protein and the released SRPK1 was an active SR protein kinase.
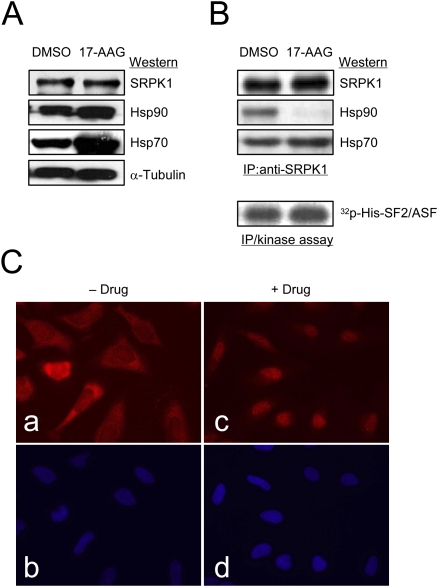
Figure 3.
The Hsp90 ATPase activity is required for dynamic interaction of SRPK1 with the Hsp70/Hsp90 chaperone complex. (A) Total protein detected by Western blotting as indicated from HeLa cells treated with vehicle (DMSO) or with 17-demthoxygeldanamycin (17-AAG). Both Hsp70 and Hsp90 are known to be induced by the treatment with 17-AAG. (B) Release of SRPK1 from the Hsp90-containing complex upon the inhibition of the Hsp90 ATPase activity. The association of the kinase with Hsp70 seemed unaffected. SRPK1 released from the Hsp90-containg complex was equally, if not more, active as an SR protein kinase when tested by in vitro phosphorylation using the SR protein SF2/ASF as a substrate. (C) Induction of SRPK1 nuclear translocation by the Hsp90 ATPase inhibitor (Geldanamycin used in this experiment; similar results also obtained with 17-AAG.) (Panels a,c) Endogenous SRPK1 as detected by anti-SRPK1 before and after the drug treatment. (Panels b,d) Corresponding nuclei were stained by DAPI. The cellular distribution of SRPK1 was shifted from the cytoplasm to the nucleus in response to the drug treatment.
Given the fact that a fraction of active SRPK1 could be released from the chaperone complexes by perturbation of the Hsp90 ATPase activity, we next determined whether the released kinase was capable of translocating to the nucleus by performing immunocytochemistry of SRPK1 before and after the drug treatment (Fig. 3C). As previously documented, we observed that SRPK1 was largely cytoplasmic with a fraction detectable in the nucleus (Ding et al. 2006). In contrast, the kinase was partially shifted to the nucleus after the treatment with the Hsp90 ATPase inhibitor. These data suggest that a fraction of released kinase can indeed translocate to the nucleus upon inhibitor-induced release from the chaperone complexes.
Induction of SRPK1 nuclear translocation in response to a stress signal
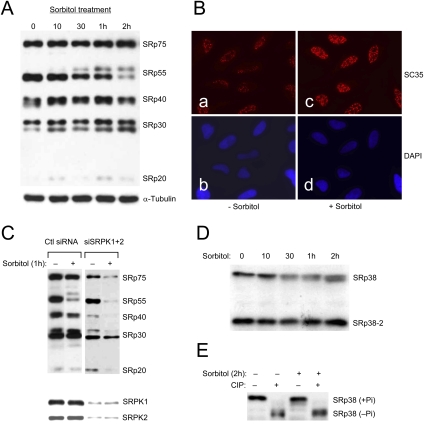
To determine whether SRPK1 nuclear translocation regulates SR protein phosphorylation and alternative splicing, we designed a series of experiments to identify potential signaling events that might dynamically modulate the interactions between SRPK1 and molecular chaperones. It has been previously reported that sorbitol-induced osmotic stress could trigger alternative splicing by redistributing a splicing suppressor hnRNP A1 from the nucleus to the cytoplasm (van der Houven van Oordt et al. 2000), which was similarly observed in our hands as shown in Figure 4A. To determine whether SRPK1 also contributed to osmotic stress-induced reprogramming of pre-mRNA splicing, we performed a time course experiment to determine the association of SRPK1 with key chaperone components. As shown in Figure 4B, the overall level of the chaperones and cochaperones was not affected by the sorbitol treatment up to 2 h, with the exception of a minor increase of Hsp70 (note that the sorbitol treatment represents a more transient permutation of the system related to that with the Hsp90 ATPase inhibitor). However, by IP/Western, we detected a dramatic rearrangement of these chaperone proteins with respect to their association with SRPK1 in the cell. After exposing HeLa cells to 0.6 M sorbitol from 30 min to 1 h, SRPK1 was practically free of Hsp70 and Hsp90.
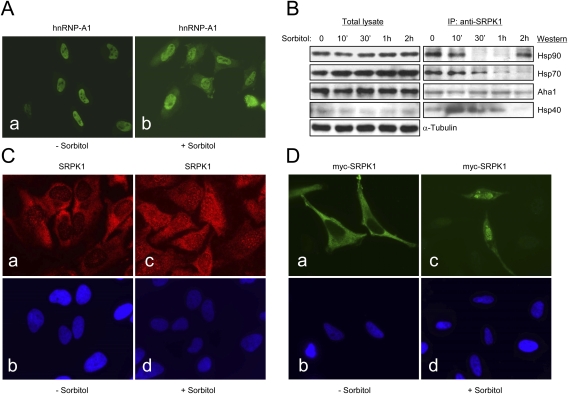
Figure 4.
Nuclear translocation of SRPK1 in response to osmotic stress. (A) Osmotic stress-induced redistribution of hnRNP A1 from the nucleus to the cytoplasm as previously reported (van der Houven van Oordt et al. 2000). (B) Stress-induced release of SRPK1 from the chaperone complexes. (Left panel) Total proteins, including α-tubulin as a loading control, in the lysate from sorbitol-treated HeLa cells at different sorbitol treatment points were determined by Western blotting. Hsp70 was slightly induced by osmotic stress (Right panel). Disassociation of SRPK1 from the Hsp70/Hsp90 chaperones in response to osmotic stress was determined by anti-SRPK1 IP from equal amounts of cell lysate at different sorbitol treatment points followed by Western blotting analysis of key chaperone components as indicated. Interestingly, Aha1 remained associated with SRPK1 during the time course. Hsp90 appeared to return to SRPK1 after 2 h, indicating dynamic interactions of SRPK1 with the Hsp70/Hsp90 machinery. (C, panels a,c) Induction of SRPK1 nuclear translocation by osmotic stress. (Panels b,d) Corresponding nuclei were stained by DAPI. (D, panels a,c) Localization of myc-tagged SRPK1 in transfected HeLa cells before and after the sorbitol treatment. (Panels b,d) Nuclei in the fields were stained by DAPI. Nuclear translocation of the exogenously expressed SRPK1 was more dramatically induced relative to endogenous SRPK1.
After 2 h of treatment with sorbitol, the level of SRPK1-associated Hsp40 was also eliminated. Interestingly, Aha1 remained attached to SRPK1, while Hsp90 became reassociated with SRPK1 (Fig. 4B). This result is highly reproducible, suggesting that Hsp90 may be recruited back to the kinase after prolonged osmotic stress. This complex role of Hsp90 has been proposed for chaperone-assisted nuclear import of p53 where the Hsp70/Hsp90-based chaperone machinery first assists p53 folding and protects it from degradation by the proteosome in the cytoplasm, and after Hsp70 is stripped, Hsp90 facilitates p53 transport to the nucleus (King et al. 2001).
To determine whether nuclear translocation of SRPK1 could be induced by the stress signal, we performed immunohistochemical analysis of SRPK1 and detected a clear increase of SRPK1 in a speckled pattern typical of SR proteins in the nucleus of HeLa cells in response to the sorbitol treatment (Fig. 4C). To further substantiate this finding, we transfected a myc-tagged SRPK1 into HeLa cells and found that the sorbitol treatment also induced nuclear translocation of the exogenously expressed SRPK1 (Fig. 4D). Interestingly, the exogenous kinase was more quantitatively shifted to the nucleus in sorbitol-treated cells. Although the reason for such response remains unclear, we suspect that the elevated responsiveness might result from a synergy between the sorbitol treatment and protein overexpression, the latter of which might also be considered as a form of stress to the cell.
Differential phosphorylation of SR proteins in sorbitol-treated cells
The induction of SRPK1 translocation to the nucleus as a function of stress signaling suggests that the status of SR protein phosphorylation might be coordinately altered. We examined this possibility by investigating the time course of SR protein phosphorylation using mAb104. This antibody was initially raised against a collection of SR proteins from amphibian oocytes and later realized for its specific recognition of common phosphoepitopes present in classic SR proteins in all higher eukaryotic cells; this antibody has thus been a very useful reagent to probe the phosphorylation state of SR proteins (Zahler et al. 1992). In response to the sorbitol treatment, we observed a steady increase in mAb104 reactivity in SRp30 (which represents a mix of SC35, ASF/SF2, 9G8, and SRp30c), while the phosphorylation of SRp75 was only modestly elevated and that of SRp40 was little affected, indicating that SR protein phosphorylation was differentially modulated by the sorbitol treatment (Fig. 5A). We also detected brighter nuclear speckles in sorbitol-treated cells relative to untreated cells with anti-SC35 (Fig. 5B), which is known to detect a phosphoepitope in this SR protein (Fu and Maniatis 1990, 1992). These data are thus consistent with hyperphosphorylation of SC35, despite that immunocytochemistry is not an quantitative assay.
Figure 5.
Hyperphosphorylation of SR proteins in sorbitol-treated cells. (A) mAb104 analysis of the phosphorylation state of typical SR proteins in response to osmotic stress. The phosphoepitoses in SRp30 were elevated by the sorbitol treatment, while SRp55 was converted to a hyperphosphorylated form. In contrast, the phosphorylation level of SRp75 was modestly elevated, and that of SRp40 was little affected. α-tubulin was probed as a loading control (note that this control was also shown in Fig. 4B because the same bunch of cells was used in both experiments). (B) Immunostaining of SC35 in nuclear speckles in mock- and sorbitol-treated cells. Although immunostaining is not a quantitative assay, the phosphoepitope detected by anti-SC35 appears brighter in sorbitol-treated cells relative to that in mock-treated cells. (C) Role of SRPKs in mediating SR protein phosphorylation before and after the sorbitol treatment. HeLa cells were transfected with control siRNA or specific siRNAs against SRPK1 and SRPK2. (Bottom panel) The levels of both kinases were determined by Western blotting. Knockdown of both kinases results in reduced SR protein phosphorylation probed by using mAb104 without the sorbitol treatment. The level of SR protein phosphorylation went further down in SRPK-depleted cells treated in the presence of sorbitol, indicating a combined effect of diminished SRPK1 and SRPK2 and activated phosphatases. We loaded a similar amount of proteins in individual lanes using α-tubulin and total SF2/ASF as loading controls, which are not included in the panel. (D) In contrast to typical SR proteins, SRp38 became partially dephosphorylated as indicated by progressive increase in its gel mobility during the course of the sorbitol treatment. (E) Sorbitol-induced partial dephosphorylation of SRp38 was confirmed by CIP treatment.
More dramatically, we found that the sorbitol treatment resulted in the appearance of a hyperphosphorylated form of SRp55, which could be converted to a hypophosphorylated form by CIP (Supplemental Fig. S2). Interestingly, SRp55 hyperphosphorylation relative to other SR proteins has been characterized previously as a unique response to treatment with the transcription inhibitor DRB, but not with other transcription inhibitors, in multiple cell types (Lai et al. 2003). Whether DRB treatment might represent a form of stress to the cell akin to sorbitol-induced SRp55 hyperphosphorylation is not clear at the moment.
To determine whether SRPKs are responsible for the observed alternation of SR protein phosphorylation induced by sorbitol, we knocked down both SRPK1 and SRPK2, which is successful as determined by Western blotting (Fig. 5C, bottom panel), and found that SR protein phosphorylation probed by the phospho-specific antibody mAb104 was reduced in the absence of sorbitol treatment and the sorbitol treatment was no longer able to elevate SR protein phosphorylation, demonstrating the role of SRPK1/2 in mediating SR protein phosphorylation both before and after the sorbitol treatment (Fig. 5C). Interestingly, we observed that the level of SR protein phosphorylation went further down in HeLa cells treated with sorbitol. This is likely contributed by diminished SR protein kinases and activation of a SR protein phosphatase(s). Consistent with this possibility, it has been reported that the phosphorylation state of SR proteins depends on the balanced activities of SRPKs and phosphatases. Thus, even though we have not yet investigated whether any specific phosphatase could be induced by sorbitol, a shift in the balance between SR proteins kinases and phosphatases likely account for the observed decrease in SR protein phosphorylation in sorbitol-treated cells.
In contrast to classic SR proteins, SRp38, an atypical SR protein responsible for splicing repression upon dephosphorylation during cell cycle or in response to heat shock (Shin and Manley 2002; Shin et al. 2004; Shi and Manley 2007), showed a modest increase in gel mobility (Fig. 5D), indicating that this particular SR protein became partially dephosphorylated in response to osmotic stress, an effect that was corroborated by CIP treatment (Fig. 5E). Collectively, these data demonstrate that stress signaling induced SRPK-mediated differential (mostly hyper-)phosphorylation of SR proteins.
Splice site selection in response to altered cellular distribution of SRPK1
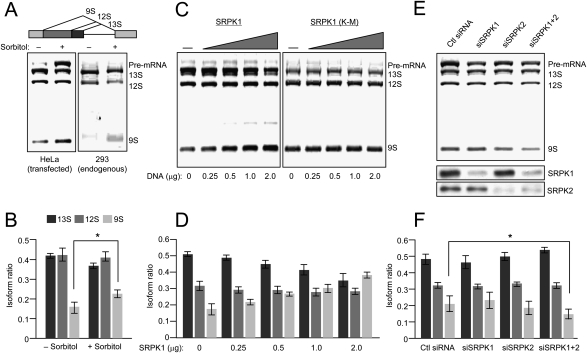
The stress signal has been shown previously to induce alternative splicing of an E1A reporter in transfected cells, which was linked to phosphorylation and cytoplasmic translocation of hnRNP A1, a well-characterized splicing repressor (van der Houven van Oordt et al. 2000). Our present data suggest that, besides cytoplasmic translocation of hnRNP A1, the stress signaling also triggered additional events related to splicing regulation, including nuclear translocation of SRPK1 and differential phosphorylation of SR proteins. We therefore used the same E1A splicing reporter to determine its response to SRPK1 nuclear translocation. We first tested E1A splicing in response to osmotic stress; but contrary to the suppression of the 9S isoform as reported previously (van der Houven van Oordt et al. 2000), we actually detected a modest activation of this isoform relative to 13S and 12S isoforms in response to sorbitol treatment in transfected HeLa cells (Fig. 6A, left, and quantified in B). This effect was more obvious when assaying the splicing pattern of the E1A endogenously expressed from an integrated adenovirus in 293T cells (Fig. 6A, right). We also noted the accumulation of the unspliced E1A pre-mRNA in transfected, sorbitol-treated HeLa cells, which might be due to splicing inhibition by hyperphosphorylated SR protein. However, for reasons that are presently unclear, this was not evident in 293T cells (Fig. 6A, right panel).
Figure 6.
The effect of sorbitol treatment on splice site selection. (A) Sorbitol induction of E1A alternative splicing in transfected HeLa (left panel) and 293T cells (right panel). The splicing pattern of the E1A reporter is illustrated above. (B) Quantification of the result from transfected HeLa cells; n = 3; (*) P < 0.05. (C) Impact of overexpressed wild-type and mutant SRPK1 on E1A alternative splicing. HeLa cells were transfected with increasing amounts of plasmids expressing wild-type (left panel) and mutant (right panel) SRPK1 along with the E1A reporter as indicated at the bottom. While wild-type SRPK1 induced the 9S isoform, the mutant SRPK1 lacked the effect, indicating the requirement for the kinase activity for the observed effect. (D) Quantification of the results from HeLa cells transfected with increasing concentrations of wild-type SRPK1 (n = 3). (E) Switch in splice site selection in response to siRNA-mediated knockdown of SRPK1 and SRPK2 in sorbitol-treated HeLa cells. The knockdown effects were verified by Western blotting as shown in the bottom two panels. (F) Quantification of the results in E; n = 3; (*) P < 0.05.
To attribute altered splice site selection to the activity of nuclear translocated SRPK1, we cotransfected the E1A reporter with increasing amounts of SRPK1. Consistently, we detected an induction of the 9S isoform in cells coexpressing wild-type SRPK1 (Fig. 6C, left, and quantified in D), the effect opposite to overexpression of a SR protein phosphatase (Cardinali et al. 1994). The kinase activity was required for the observed effect as the parallel analysis with the K-to-M mutation in the ATP-binding site of SRPK1 failed to induce the 9S isoform (Fig. 6C, right). We obtained similar results with overexpressed SRPK2 (data not shown). These results are consistent with the notion that nuclear translocated SRPK1 induced SR protein hyperphosphorylation, which, in turn, modulated alternative splice site selection in sorbitol-stressed cells.
To further demonstrate the effect of nuclear translocated SRPK1 on splice site selection in sorbitol-treated cells, we carried out a converse experiment by siRNA-mediated knockdown. We first tested the effect of SRPK1 and SRPK2 knockdown, either individually or in combination, on E1A splicing before and after the sorbitol treatment. The result indicated that, in mock-treated cells, knockdown of SRPK1 or SRPK2 had little effect, but knockdown of both kinases resulted in the accumulation of pre-mRNA. Importantly, the cells depleted of both SRPK1 and SRPK2 no longer responded to the sorbitol treatment in elevating the 9S isoform (Fig. 6E; Supplemental Fig. S3). Based on this initial observation, we performed a thorough analysis on sorbitol-treated HeLa cells to demonstrate the requirement for SRPK1 and SRPK2 in sorbitol-induced 9S isoform (Fig. 6E), and the recorded differences are statistically significant (Fig. 6F). These observations corroborate the genetic evidence that SRPKs are major kinases for SR proteins in mammalian cells as their siRNA-mediated down-regulation (Hayes et al. 2006) or inactivation by homologous recombination (P.-P. Wang and X.-D. Fu, unpubl.) both attenuated SR protein phosphorylation. Together, these results demonstrate a key role of SRPKs in controlling SR protein phosphorylation and splice site selection in response to a cellular signaling.
Discussion
Physiological importance to regulate SR protein phosphorylation in the cell
Previous studies have firmly established that the phosphorylation and dephosphorylation cycle of SR proteins is essential for pre-mRNA splicing (Cao et al. 1997). It is thus not surprising that experimental induction of both hypo- and hyperphosphorylation of SR proteins frustrates this cycle and inhibits splicing, an outcome that underscores the requirement for precise regulation of SR protein phosphorylation in the cell (Prasad et al. 1999). Despite that multiple protein kinases and phosphatases have been implicated in SR protein phosphorylation and dephosphorylation, only SRPK1 and PP1 have been characterized by RNAi-mediated knockdown (Hayes et al. 2006; Shi and Manley 2007). While a recent study demonstrated the activation of PP1 by stripping off a heat-shock-sensitive PP1 inhibitor and showed that the phosphorylation state of different SR proteins is controlled by both activated PP1 and differential accessibility of SR protein kinases (Shi and Manley 2007), little is known about how specific SR kinases might be regulated under physiological conditions. Our current study provides information on this critical gap in knowledge by documenting the regulation of SRPK1 in response to a stress signal that connects induced nuclear translocation of the kinase to SR protein phosphorylation to alternative splicing.
Molecular chaperones involved in SRPK regulation: new role for old players
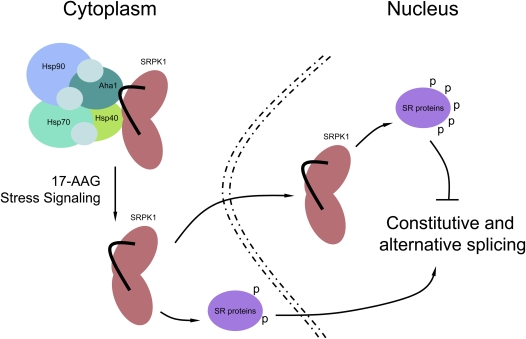
Although previous mutational analysis suggests that the unique spacer sequences in individual SRPKs control the cellular partitioning of the kinases (Ding et al. 2006), the physiological components of this control are not known. Interestingly, the spacer domain that splits the conserved kinase domains into two halves resembles the structural configuration of many receptor tyrosine kinases where the spacer domains are frequently involved in contacting other proteins to transduce signals to the cell nucleus (Pawson and Scott 1997). We now demonstrate that SRPK1 directly interacts with specific cochaperones via both the kinase and spacer domains, thereby connecting the kinase to the Hsp70/Hsp90 machinery as illustrated in the model in Figure 7.
Figure 7.
Model for the role of molecular chaperones in the regulation of SRPK1 nuclear translocation, SR protein phosphorylation, and pre-mRNA splicing.
The molecular chaperone system has been widely implicated in the regulation of transcription factors (e.g., nuclear receptors), kinases and phosphatases during cellular signaling (Pratt and Toft 2003). In general, Hsp70 and its cochaperones are the first to interact with specific clients followed by the joining of Hsp90 and its cochaperones, which together assist protein folding into active conformations and protect from protein degradation by the proteosome (Hartl and Hayer-Hartl 2002; Esser et al. 2004). In support of this function, we observed decreased kinase activity in cells depleted of the cochaperones Aha1 or Hsp40. It has been reported that, in the absence of Hsp40, Hsp70 binds to and denatures its clients (Liberek et al. 1991), and in conjunction with our earlier finding that the kinase activity of SRPK1 is essential for its translocation to the nucleus, we were unable to detect any major alteration in the cellular distribution of SRPK1 in Aha1 or Hsp40-depleted cells. We also consistently observed SRPK1 degradation after prolonged inhibition of the Hsp90 ATPase activity or treatment of the cell with sorbitol (X.-Y. Zhong and X.-D. Fu, unpubl.).
In addition to the protein folding and protective functions of the chaperone system, however, the highly dynamic interaction of the Hsp70/Hsp90 machinery with specific clients provides a regulatory mechanism in the cell. For example, Aha1 is able to stimulate the ATPase activity of Hsp90, which contributes to the proper folding of wild-type CFTR. Remarkably, attenuation of the CFTR–chaperone interactions through down-regulation of Aha1 was able to rescue misfolding of the mutant CFTR in cystic fibrosis (Wang et al. 2006). In the case of the dsRNA-dependent kinase, the chaperone complex has been shown to function both as a facilitator for protein folding and as a repressor for the kinase-mediated signaling (Donze et al. 2001). These and numerous other studies provide examples for the regulation of signaling molecules through dynamic modulation of client/chaperone interactions. We now show that this widely used strategy also functions in the regulation of SRPK1 and SR protein phosphorylation in the cell.
As depicted in Figure 7, SRPK1 forms a complex with the Hsp70/Hsp90 machinery, which may initially assist folding of the kinase into an active conformation. The complex then performs the function of restricting the kinase in the cytoplasm. Previous studies have shown that cytoplasmic SRPK1 may be responsible for initial SR protein phosphorylation critical for their subsequent import into the nucleus (Yun and Fu 2000; Lai et al. 2001). The cytoplasm-restricted SRPKs and phosphorylation-dependent nuclear import of SR proteins might be the underlying mechanism for nuclear translocation of SR proteins during zygotic activation of gene expression as reported earlier (Sanford and Bruzik 1999). Importantly, we showed here that the interaction of SRPK1 with the molecular chaperones could also be modulated by a stress signal, resulting in the release and subsequent translocation of the kinase to the nucleus. Under certain physiological conditions, this regulated release and nuclear translocation may stimulate splicing, but if the nuclear level of the kinase is too high (e.g., in stressed cells), induced hyperphosphorylation of SR proteins becomes inhibitory to splicing.
Connection of SRPK1 regulation to SR protein phosphorylation
The involvement of molecular chaperones in controlling the cellular partitioning of SRPK1 now provides mechanistic insights into several previous observations. In prior studies, heat shock was found to induce roundup of nuclear speckles, a phenotype opposite to that induced by overexpressing an SR protein kinase (Spector et al. 1991; Gui et al. 1994; Wang et al. 1998). This may reflect a cellular response to heat shock, resulting in inhibition of SRPKs. This mechanism may also explain rapid dephosphorylation of SRp38 upon heat shock. In fact, we observed diminished phosphorylation for most SR proteins, not just SRp38, in heat-shocked HeLa cells (X.-Y. Zhong and X.-D. Fu, unpubl.), which is fully consistent with a combined effect of heat-shock-induced restriction of SRPKs and activation of PP1 (Shi and Manley 2007).
Dynamic interactions of SRPK1 with heat-shock proteins may also play a critical role in facilitating the recovery of the splicing regulatory network after heat shock. In fact, this has been demonstrated in cells overexpressing Hsp27 (Marin-Vinader et al. 2006). While it is currently unclear whether this heat-shock protein is part of the chaperone complex with SRPK1 and whether Hsp27 is involved in the regulation of SR protein phosphorylation, our current findings lay a framework on further investigation of the role of the molecular chaperone system in regulated splicing via SRPKs and other families of SR protein kinases.
Signaling splicing via the regulation of SRPKs
By virtue of regulated cellular distribution, SRPK1 is a good candidate for transducing signals to control pre-mRNA processing in the nucleus. A previous study documented p38-induced translocation of the splicing repressor hnRNP A1 from the nucleus to the cytoplasm in response to osmotic stress, which was correlated with a shift in splice site selection of the E1A reporter (van der Houven van Oordt et al. 2000). It is likely that hnRNP A1 redistribution may not be the only change induced by osmotic stress that is responsible for the observed shift in splice site selection. In fact, for reasons that are currently unclear, we observed stress-induced elevation of the 9S isoform, which is opposite to the previous observation and cannot be explained by the removal of a splicing repressor. Instead, our data suggest that the stress signal had induced a complex response that also included nuclear translocation of SRPK1 and subsequent hyperphosphorylation of SR proteins.
Now, the central question lies on how specific internal or external signals may directly modulate the dynamic interactions between SRPK1 and molecular chaperones and how these interactions control splicing in the nucleus. Conceivably, this can be accomplished by modifying specific components of the client/chaperone complex, but not the kinase in the complex, as we now show by inhibiting the ATPase activity of Hsp90 with 17-AAG. Alternatively, the SR protein kinase might be a direct target for a specific post-translational modification(s) in response to signaling. Preliminary metabolic labeling experiments indicate that SRPK1 is, indeed, a phospho-protein in the cell (X.-Y. Zhong and X.-D. Fu, unpubl.), and thus, future work will be directed to the identification of specific kinases responsible for SRPK1 phosphorylation, and more importantly, to the elucidation of the mechanism for modulating the dynamic interaction of SRPK1 with the Hsp70/Hsp90 machinery. Our current studies thus establish the foundation to further understand how various signals might be transduced through SRPKs to regulate SR protein phosphorylation and alternative splicing in the nucleus.
Materials and methods
Yeast two-hybrid screening
The Matchmaker GAL4 two-hybrid system 3 (Clontech) was used for two-hybrid screen in yeast strain AH109, which harbors ADE2, HIS3, MEL1, and LacZ reporters. To generate the bait constructs, we cloned PCR-amplified full-length SRPK1 or the spacer fragment (280-491 amino acids) into the pGBKT7 vector. Yeast was transformed with pGBKT7-SRPK1 and a human HeLa cDNA library (HL4000AA, Clontech) or with pGBKT7-Spacer and a human fetal brain cDNA library (HL4029AH, Clontech). In each screen, ∼2 × 107 transformants were plated on yeast synthetic complete medium lacking tryptophan, lencine, histidine (SD/−Trp/−Leu/−His) and the transformants were also stained for the α-galactosidase activity. To confirm the specificity of interactions, we generated the pGBKT7-SRPK1ΔSpacer bait construct for transient two-hybrid assays to characterize candidates from the initial screens. Prey plasmid isolated from each positive clone was sequenced and compared against the NCBI GenBank database. The cDNA corresponding to each positive was subsequently cloned and tested with individual bait constructs to confirm their two-hybrid interactions in yeast.
Cell culture, transfection, and drug treatment
HeLa and HEK293T were cultured in DMEM media containing 10% fetal bovine serum (FBS; Hyclone) supplemented with penicillin and streptomycin. Transient transfections were performed by using the Lipofectamine 2000 transfection reagents (Invitrogen). The Hsp90 inhibitor Geldanamycin or 17-AAG (InvivoGen) was dissolved in DMSO to treat cells at the final concentration of 10 μM. Cells were stressed with 600 mM sorbitol (Sigma).
GST pull-down with recombinant proteins
Full-length cDNAs of Aha1 and DNAjc8/Hsp40 were generated by PCR and cloned into pGEX-2T vector (Amersham). Aha1 or DNAJC8 fused to GST was expressed in Escherichia coli BL21 and recombinant proteins purified on glutathione Sepharose beads (Amersham Pharmacia). Wild-type SRPK1, spacer-deleted SRPK1, and the spacer fragment from SRPK1 were each cloned into pRSET-A (Invitrogen) or pET-28a (Novagen) and expressed as His-tagged proteins and purified on Ni-resin (Invitrogen). GST or individual GST fusion proteins were immobilized on glutathione beads by mixing ∼4 μg of GST or GST fusion protein with 30 μL of glutathione Sepharose slurry (1:1) in 500 μL of PBS plus 1% Triton X-100, and 1 mM DTT for 30 min at 4°C. Beads were washed twice with PBS and once with the HEMG buffer (50 mM HEPES at pH 7.8, 50 mM KCl, 5 mM MgCl2, 0.2% Triton-X100, 1 mM DTT; 10% glycerol, 0.5 mg/mL BSA). The beads were mixed with ∼8 μg of individual His-tagged protein in 500 μL of HEMG buffer for 1 h at 4°C with agitation, washed three times with HEMG buffer, and eluted with 2× SDS loading buffer. For GST pull-down from whole-cell lysate, cultured HeLa cells were harvested, disrupted in the lysis buffer (10 mM Tris-HCl at pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 0.5% Triton-100, 1× phosphate, protease inhibitors) by sonication twice for 5 sec, and clarified by centrifugation at 15,000 rpm for 10 min.Cell lysate (300 μL [1 mg/mL]) was added to protein-bound GST beads and incubated for 1 h at 4°C. Bound proteins were either directly analyzed by SDS-PAGE or subjected to Western blotting using mouse anti-GST (1:1000; Santa Cruz Biotechnologies), mouse anti-6XHis (1:3000; H1029, Sigma) or mouse anti-SRPK1 (1:1000; BD Transduction Laboratories).
Co-IP analysis and in vitro kinase assay
Whole-cell lysate was added to 40 μL of slurry (1:1 ratio) of protein A/G Sepharose (Pharmacia) prebound with 2–3 μg of anti-SRPK1 (BD Transduction Laboratories), anti-HA antibody (H3663, Sigma), anti-Hsp70 (sc-24, Santa Cruz Biotechnologies), or anti-Hsp90 (610418, BD Biosciences). After incubation for 1 h at 4°C with rocking, antigen/antibody beads were collected and washed three times with lysis buffer. Bound proteins were eluted in 30 μL of SDS loading buffer and separated on 10% SDS-PAGE. Following transfer, the nitrocellulose membrane was blocked in buffer TBST (50 mM Tris-HCl at pH 7.8, 150 mM NaCl, 0.1% Tween-20) plus 5% nonfat milk, incubated with individual primary antibodies. After extensive rinsing with TBST, the blot was incubated with appropriate HRP-conjugated secondary antibody and analyzed using an ECL protocol. The primary antibodies used were anti-Hsp90 mouse monoclonal antibody (610418, BD Biosciences) at 1:1000 dilution, anti-Hsp70 mouse monoclonal antibody (sc-24, Santa Cruz Biotechnologies) at 1:2000, anti-α-tubulin mouse monoclonal antibody (Sigma) at 1:3000, anti-pan-Hsp40 rabbit polyclonal antibody (SPC-100C, Stress Marq, Biosciences) at 1:2000, anti-Aha1 rabbit polyclonal antibody (a gift from the laboratory of William E. Balch) at 1:4000, anti-SRPK1 (BD Transduction Laboratories) at 1:1000, anti-SRPK2 (BD Transduction Laboratories) at 1:500, the anti-SRp38 (a gift from the laboratory of James L. Manley) at 1:3000, and mAb104 at 1:5 dilution of hybridoma supernatant. To determine the SR kinase activity after IP, beads were washed once with the kinase reaction buffer (50 mM Tris-HCl at pH 7.4, 1 mM DTT, 10 mM MgCl2, 20 μM ATP). Beads were resuspended in 20 μL of the kinase reaction buffer containing 10 μCi [γ-32P] ATP and 1 μg of His-ASF/SF2 and incubated on a rotator at room temperature for 20 min. The reaction was terminated by boiling in 10 μL of 2× SDS sample buffer followed by SDS-PAGE and autoradiography.
Indirect immunofluorescence
HeLa cells grown on coverslips were either left untreated or exposed to 600 mM sorbitol for 1 h. Cells were washed once with PBS, fixed with methanol for 8 min at −20°C or 4% paraformaldehyde in PBS for 20 min at room temperature, washed once with PBS, and permeabilized with 0.2% Triton X-100 in PBS for 5 min. After blocking with 1% BSA for 30 min, coverslips were incubated for 1 h in a humid chamber with the following primary antibodies in PBS containing 1% BSA: monoclonal anti-SRPK1 (611072, BD Transduction Laboratories) at 1:1000, monoclonal anti-SC35 at 1:500 (Fu and Maniatis 1990), rabbit polyclonal anti-myc (2272, Cell Signaling Technology) at 1:500 dilution, rabbit polyclonal anti-hnRNP A1 (ARP40383, Aviva) at 1:200. After washing three times with PBS plus 0.1% BSA, the coverslips were stained with Alexa 594-conjugated donkey anti-mouse IgG (A-21203, Molecular Probes) or Alexa 488-conjugated goat anti-rabbit IgG (A-11070, Molecular Probes) in PBS and 1% BSA. After incubation for 1 h in a humid chamber, coverslips were washed three times with PBS plus 0.1% BSA, mounted in a solution containing 4,6-diamidino-2-phenylindole (DAPI) (Vectashield, Vector Laboratories), and visualized on a Zeiss Axiophot microscope.
RNAi
Control and specific siRNAs duplexes from Qiagen or Dharmacon were transfected into subconfluent HeLa cells with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. SRPKs were knocked down by a SMARTpool siRNA (Dharmacon). The sense sequences of siRNA against SRPK1 were 5′-GAACAUAACGGACCACUGGUU-3′, 5′-GAUACCAUGUGAUCCGAAAUU-3′, 5′-GCAGCUGGCUUCACAGAUUUU-3′, and 5′-ACACAUAUCUGCAUGGUAUUU-3′. The sense sequences of siRNA against SRPK2 were 5′-GCCCAGAGGUGAAACUAAAUU-3′, 5′-GAGGCAGGCUGAGUUAUUGUU-3′, 5′-GCAAAUUCUACCAAUAUUGUU-3′, and 5′-GCAGCUGACUUGUUGGUGAUU-3′. The sense sequence of siRNA against DNAJC8 was 5′-GAAUUGUGAUGGUUAGAAAdTdT-3′. The sense sequence of siRNA against AHA1 was 5′-CCUUGACCUUCAUCGACAAdTdT-3′.
E1A splicing reporter assay
The E1A minigene was cloned into the pcDNA3 vector under the control of the human CMV promoter. HeLa cells grown on six-well plates (2 × 105 cells per well) were cotransfected with 2 μg of the E1A minigene plasmid and various amounts of pCMV-myc-SRPK1 using Lipofectamine 2000 (Invitrogen). Empty vector was used to equalize the amount of total DNA transfected into the cell. Thirty hours to 40 h after transfection, total RNA was extracted with the RNeasy mini kit (Qiagen) from untreated or 600 mM sorbitol-stressed cells. Isolated RNA was treated with 100 U of RNase-free DNase I (Promega) for 30 min at 37°C in the presence of 50U of RNasin (Promega). Two micrograms of total RNA were reverse transcribed in a 20 μL reaction with SuperScript III First-Strand Synthesis System (18080-051, Invitrogen). One microliter of RT products was used for PCR amplification using the primer pair E1A-569 (5′-ATTATCTGCCACGGAGGTGT-3′) and E1A-1315 (5′-GGATAGCAGGCGCCATTTTA-3′) (Yang et al. 1994). Five microliters of each PCR reaction were loaded on 2% agarose gel and ethidium bromide-stained bands were quantified using the ImageJ software.
Acknowledgments
We are grateful to S. Taylor and colleagues for stimulating discussion during the course of this project. We thank W. Balch for anti-Aha1 and J. Manley for anti-SRp38. This work was supported by a NIH grant (GM52872) to X-D.F.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1752109.
Supplemental material is available at http://www.genesdev.org.
References
- Aubol B.E., Chakrabarti S., Ngo J., Shaffer J., Nolen B., Fu X.D., Ghosh G., Adams J.A. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc. Natl. Acad. Sci. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Jamison S.F., Garcia-Blanco M.A. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- Cardinali B., Cohen P.T., Lamond A.I. Protein phosphatase 1 can modulate alternative 5′ splice site selection in a HeLa splicing extract. FEBS Lett. 1994;352:276–280. doi: 10.1016/0014-5793(94)00973-2. [DOI] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J.L., Bell J.C., Duncan P.I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Ding J.H., Zhong X.Y., Hagopian J.C., Cruz M.M., Ghosh G., Feramisco J., Adams J.A., Fu X.D. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol. Biol. Cell. 2006;17:876–885. doi: 10.1091/mbc.E05-10-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze O., Abbas-Terki T., Picard D. The Hsp90 chaperone complex is both a facilitator and a repressor of the dsRNA-dependent kinase PKR. EMBO J. 2001;20:3771–3780. doi: 10.1093/emboj/20.14.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C., Alberti S., Hohfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Fu X.D., Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fu X.D., Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- Ghigna C., Moroni M., Porta C., Riva S., Biamonti G. Altered expression of heterogeneous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res. 1998;58:5818–5824. [PubMed] [Google Scholar]
- Goetz M.P., Toft D.O., Ames M.M., Erlichman C. The Hsp90 chaperone complex as a novel target for cancer therapy. Ann. Oncol. 2003;14:1169–1176. doi: 10.1093/annonc/mdg316. [DOI] [PubMed] [Google Scholar]
- Gui J.F., Lane W.S., Fu X.D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Hagopian J.C., Ma C.T., Meade B.R., Albuquerque C.P., Ngo J.C., Ghosh G., Jennings P.A., Fu X.D., Adams J.A. Adaptable molecular interactions guide phosphorylation of the SR protein ASF/SF2 by SRPK1. J Mol Biol. 2008;382:894–909. doi: 10.1016/j.jmb.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hayes G.M., Carrigan P.E., Beck A.M., Miller L.J. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006;66:3819–3827. doi: 10.1158/0008-5472.CAN-05-4065. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yario T.A., Steitz J.A. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. 2004;101:9666–9670. doi: 10.1073/pnas.0403533101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol. 2007;14:185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King F.W., Wawrzynow A., Hohfeld J., Zylicz M. Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J. 2001;20:6297–6305. doi: 10.1093/emboj/20.22.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.C., Tarn W.Y. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J. Biol. Chem. 2004;279:31745–31749. doi: 10.1074/jbc.C400173200. [DOI] [PubMed] [Google Scholar]
- Lai M.C., Lin R.I., Tarn W.Y. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M.C., Lin R.I., Tarn W.Y. Differential effects of hyperphosphorylation on splicing factor SRp55. Biochem. J. 2003;371:937–945. doi: 10.1042/BJ20021827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Liberek K., Skowyra D., Zylicz M., Johnson C., Georgopoulos C. The Escherichia coli DnaK chaperone, the 70-kDa heat shock protein eukaryotic equivalent, changes conformation upon ATP hydrolysis, thus triggering its dissociation from a bound target protein. J. Biol. Chem. 1991;266:14491–14496. [PubMed] [Google Scholar]
- Lin S., Fu X.D. SR proteins and related factors in alternative splicing. Adv. Exp. Med. Biol. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Lin S., Xiao R., Sun P., Xu X., Fu X.D. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol. Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Lin S., Coutinho-Mansfield G., Wang D., Pandit S., Fu X.D. The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasiewicz R., Velazquez-Dones A., Huynh N., Hagopian J., Fu X.D., Adams J., Ghosh G. Structurally unique yeast and mammalian serine-arginine protein kinases catalyze evolutionarily conserved phosphorylation reactions. J. Biol. Chem. 2007;282:23036–23043. doi: 10.1074/jbc.M611305200. [DOI] [PubMed] [Google Scholar]
- Marin-Vinader L., Shin C., Onnekink C., Manley J.L., Lubsen N.H. Hsp27 enhances recovery of splicing as well as rephosphorylation of SRp38 after heat shock. Mol. Biol. Cell. 2006;17:886–894. doi: 10.1091/mbc.E05-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Hartmuth K., Mohlmann S., Urlaub H., Ficner R., Luhrmann R. Phosphorylation of human PRP28 by SRPK2 is required for integration of the U4/U6-U5 tri-snRNP into the spliceosome. Nat. Struct. Mol. Biol. 2008;15:435–443. doi: 10.1038/nsmb.1415. [DOI] [PubMed] [Google Scholar]
- Mermoud J.E., Cohen P.T., Lamond A.I. Regulation of mammalian spliceosome assembly by a protein phosphorylation mechanism. EMBO J. 1994;13:5679–5688. doi: 10.1002/j.1460-2075.1994.tb06906.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G., Sanford J.R., Caceres J.F. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Nayler O., Schnorrer F., Stamm S., Ullrich A. The cellular localization of the murine serine/arginine-rich protein kinase CLK2 is regulated by serine 141 autophosphorylation. J. Biol. Chem. 1998;273:34341–34348. doi: 10.1074/jbc.273.51.34341. [DOI] [PubMed] [Google Scholar]
- Ngo J.C., Chakrabarti S., Ding J.H., Velazquez-Dones A., Nolen B., Aubol B.E., Adams J.A., Fu X.D., Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol. Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Ngo J.C., Giang K., Chakrabarti S., Ma C.T., Huynh N., Hagopian J.C., Dorrestein P.C., Fu X.D., Adams J.A., Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol. Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B., Yun C.Y., Wong C.F., McCammon J.A., Fu X.D., Ghosh G. The structure of Sky1p reveals a novel mechanism for constitutive activity. Nat. Struct. Biol. 2001;8:176–183. doi: 10.1038/84178. [DOI] [PubMed] [Google Scholar]
- Ohtsuka K., Hata M. Mammalian HSP40/DNAJ homologs: Cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaretou B., Siligardi G., Meyer P., Maloney A., Sullivan J.K., Singh S., Millson S.H., Clarke P.A., Naaby-Hansen S., Stein R., et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/s1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- Pawson T., Scott J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Pind M.T., Watson P.H. SR protein expression and CD44 splicing pattern in human breast tumours. Breast Cancer Res. Treat. 2003;79:75–82. doi: 10.1023/a:1023338718974. [DOI] [PubMed] [Google Scholar]
- Prasad J., Colwill K., Pawson T., Manley J.L. The protein kinase Clk/Sty directly modulates SR protein activity: Both hyper- and hypophosphorylation inhibit splicing. Mol. Cell. Biol. 1999;19:6991–7000. doi: 10.1128/mcb.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt W.B., Toft D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Roscigno R.F., Garcia-Blanco M.A. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Bruzik J.P. Developmental regulation of SR protein phosphorylation and activity. Genes & Dev. 1999;13:1513–1518. doi: 10.1101/gad.13.12.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Gray N.K., Beckmann K., Caceres J.F. A novel role for shuttling SR proteins in mRNA translation. Genes & Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Ellis J.D., Cazalla D., Caceres J.F. Reversible phosphorylation differentially affects nuclear and cytoplasmic functions of splicing factor 2/alternative splicing factor. Proc. Natl. Acad. Sci. 2005;102:15042–15047. doi: 10.1073/pnas.0507827102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Manley J.L. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol. Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- Shin C., Manley J.L. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111:407–417. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- Shin C., Feng Y., Manley J.L. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427:553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- Siebel C.W., Feng L., Guthrie C., Fu X.D. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl. Acad. Sci. 1999;96:5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L., Fu X.D., Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stickeler E., Kittrell F., Medina D., Berget S.M. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- Takeuchi M., Yanagida M. A mitotic role for a novel fission yeast protein kinase dsk1 with cell cycle stage dependent phosphorylation and localization. Mol. Biol. Cell. 1993;4:247–260. doi: 10.1091/mbc.4.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronchere H., Wang J., Fu X.D. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388:397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- van der Houven van Oordt W., Diaz-Meco M.T., Lozano J., Krainer A.R., Moscat J., Caceres J.F. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Dones A., Hagopian J.C., Ma C.T., Zhong X.Y., Zhou H., Ghosh G., Fu X.D., Adams J.A. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J. Biol. Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- Wang J., Takagaki Y., Manley J.L. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes & Dev. 1996;10:2588–2599. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]
- Wang H.Y., Lin W., Dyck J.A., Yeakley J.M., Songyang Z., Cantley L.C., Fu X.D. SRPK2: A differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Venable J., LaPointe P., Hutt D.M., Koulov A.V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Xiao S.H., Manley J.L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes & Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Xiao R., Sun Y., Ding J.H., Lin S., Rose D.W., Rosenfeld M.G., Fu X.D., Li X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell. Biol. 2007;27:5393–5402. doi: 10.1128/MCB.00288-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Bani M.R., Lu S.J., Rowan S., Ben-David Y., Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeakley J.M., Tronchere H., Olesen J., Dyck J.A., Wang H.Y., Fu X.D. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol. 1999;145:447–455. doi: 10.1083/jcb.145.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C.Y., Fu X.D. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 2000;150:707–718. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A.M., Lane W.S., Stolk J.A., Roth M.B. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zylicz M., King F.W., Wawrzynow A. Hsp70 interactions with the p53 tumour suppressor protein. EMBO J. 2001;20:4634–4638. doi: 10.1093/emboj/20.17.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]