Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that direct post-transcriptional repression of protein-coding genes. In vertebrates, each highly conserved miRNA typically regulates hundreds of target mRNAs. However, the precise relationship between expression of the miRNAs and that of their targets has remained unclear, in part because of the scarcity of quantitative expression data at cellular resolution. Here we report quantitative analyses of mRNA levels in miRNA-expressing cells of the zebrafish embryo, capturing entire miRNA expression domains, purified to cellular resolution using fluorescent-activated cell sorting (FACS). Focus was on regulation by miR-206 and miR-133 in the developing somites and miR-124 in the developing central nervous system. Comparison of wild-type embryos and those lacking miRNAs revealed predicted targets that responded to the miRNAs and distinguished miRNA-mediated mRNA destabilization from other regulatory effects. For all three miRNAs examined, expression of the miRNAs and that of their predicted targets usually overlapped. A few targets were expressed at higher levels in miRNA-expressing cells than in the rest of the embryo, demonstrating that miRNA-mediated repression can act in opposition to other regulatory processes. However, for most targets expression was lower in miRNA-expressing cells than in the rest of the embryo, indicating that miRNAs usually operate in concert with the other regulatory machinery of the cell.
Keywords: Dicer, gene regulatory networks, miRNA targets, microRNA, zebrafish development
MicroRNAs (miRNAs) are ∼23-nucleotide (nt) endogenous RNAs, which pair to sites in the messages of protein-coding genes to direct the translational repression or destabilization of these mRNAs (Bartel 2004). miRNAs are one of the most abundant classes of gene regulators in animals and have been demonstrated to play important regulatory roles, such as the control of cell death, developmental timing, and neuronal patterning in flies and nematodes (Ambros 2004). In vertebrates, individual miRNAs have been found to play important roles in many processes, including hematopoietic lineage differentiation and function, as well as brain, heart, and mesoderm development (Chen et al. 2004; Giraldez et al. 2005, 2006; Choi et al. 2007; Rodriguez et al. 2007; Thai et al. 2007; van Rooij et al. 2007; Xiao et al. 2007; Zhao et al. 2007; Johnnidis et al. 2008; Ventura et al. 2008).
The first genetically identified miRNAs displayed strong mutant phenotypes that could be explained by the misregulation of single target genes, suggesting that each miRNA was responsible for switching off the expression of just a few target genes (Lee et al. 1993; Wightman et al. 1993; Reinhart et al. 2000). This initial paradigm appeared to hold as the plant miRNAs and their targets were discovered (Llave et al. 2002; Reinhart et al. 2002; Rhoades et al. 2002). However, it was overturned in animals when bioinformatic studies indicated that many messages have been under selective pressure to maintain pairing to each of the highly conserved miRNAs (Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005) and experiments showed that hundreds of direct targets respond to the introduction or removal of a highly expressed miRNA (Lim et al. 2005; Giraldez et al. 2006; Baek et al. 2008; Selbach et al. 2008). As the widespread scope of metazoan miRNA targeting began to come into focus, so, too, did the possibility that most interactions might not be binary developmental switches of the type observed initially for the genetically identified interactions (Bartel and Chen 2004). Indeed, despite the striking organ- and tissue-specific expression of many miRNAs in zebrafish (Wienholds et al. 2005), a major role in determining organogenesis was excluded in experiments with mutant zebrafish that express no detectable miRNAs yet appear to have all the major organs and correctly differentiated cell types (Giraldez et al. 2005).
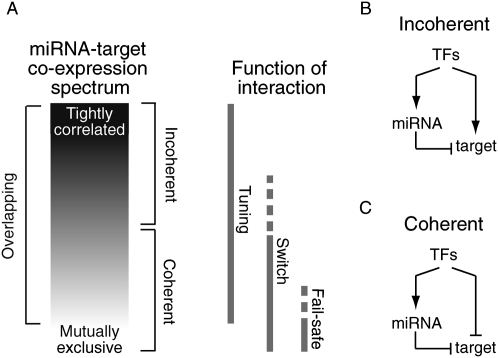
When considering the roles of miRNAs in animals, it is useful to know the relationship between the expression of miRNAs and that of their target mRNAs, for which we define “expression” as “presence” (i.e., not necessarily as active ongoing transcription). Theoretically, the expression of miRNAs and their targets could display a degree of coexpression anywhere between the two extremes of tight coexpression (perfect correlation or coregulation) and nonoverlapping expression (Fig. 1A; Bartel and Chen 2004; Hornstein and Shomron 2006). A few miRNAs expressed from excised introns are predicted to target their host genes; such miRNA–target pairs are presumed examples of tight coexpression. An experimentally validated example of tight coexpression is found with the miR168 targeting of Arabidopsis ARGONAUTE1 (AGO1). In this example, the miRNA is expressed everywhere in the plant that its target is required, as evidenced by the ability to rescue the ago1-null phenotype with an AGO1 transgene expressed from the MIR168 promoter (Vaucheret et al. 2004, 2006). Nonoverlapping expression of miRNAs and their targets has been reported in a genome-wide survey of in situ expression patterns in Drosophila (Stark et al. 2005) and for several miRNAs during development (Ronshaugen et al. 2005; Stark et al. 2005, 2008; Li et al. 2006). Between these extremes, the expression of a target and the miRNA can overlap to various degrees. The target can be preferentially expressed with the miRNA, in which case the targeting interaction is considered “incoherent” because miRNA-directed target repression is opposing the overall outcome of all the other regulatory processes responsible for inducing both the target and miRNA (Fig. 1B; Tsang et al. 2007). Alternatively, the target can be expressed in cells that express the miRNA but at higher levels in those cells that do not express the miRNA, in which case the targeting interaction is considered overlapping but “coherent,” with the miRNA working in concert with other regulatory processes to yield lower target expression in miRNA-expressing cells (Fig. 1C; Farh et al. 2005; Sood et al. 2006); both scenarios discussed in Hornstein and Shomron (2006). Here we also speak of “coherent expression” and “incoherent expression,” which we define as expression patterns implying coherent and incoherent regulation, respectively.
Figure 1.
Overview of miRNA–target expression and miRNA function. (A) miRNA–target expression can fall anywhere between the two extremes of tightly correlated and nonoverlapping, where preferential coexpression and anti-correlation indicate incoherent and coherent regulation, respectively. Different regions of the coexpression spectrum are expected to correspond to different functional roles for the miRNA-mediated repression (diagrammed to the right). (B) Schematic representing the regulatory circuitry for incoherent regulation, in which miRNA-mediated repression opposes the overall action of transcription factors and other regulatory processes that affect mRNA levels (abbreviated in aggregate as TFs). (C) Schematic representing the regulatory circuitry for coherent regulation, in which miRNA-mediated repression reinforces the overall action of transcription factors and other regulatory processes that affect mRNA levels (abbreviated in aggregate as TFs).
To the extent that overlapping expression implies miRNA-mediated repression, knowing whether expression is predominantly overlapping or nonoverlapping, or if it implies coherent or incoherent regulation, can shed light on the underlying function of miRNA-dependent target regulation (Fig. 1A). For example, target expression restricted only to miRNA-expressing cells suggests a tuning function for that interaction, because the alternative of a binary off-switch would snuff out consequential protein production in all cells that the target is expressed, rendering the target gene functionless. Tuning interactions are those for which the miRNA acts as a rheostat to dampen protein output to a more optimal level but one that is still functional in the cell, thereby enabling more customized output in different cell types as well as more uniform output within each cell type (Bartel and Chen 2004; Karres et al. 2007). Tuning is also employed in negative feedback loops to adjust the relative concentrations of miRNA and target (Xie et al. 2003; Vaucheret et al. 2004, 2006). Falling at the opposite end of the coexpression spectrum is nonoverlapping expression. Nonoverlapping expression implies either a previous function in helping to dampen output of mRNAs no longer expressed, or a fail-safe function, in which the miRNA ensures that spurious mRNAs arising from leaky transcription do not produce consequential amounts of protein (Bartel and Chen 2004; Hornstein et al. 2005; Stark et al. 2005). In this and other switch-like interactions, the miRNA participates in reducing target output to inconsequential levels. Between the extremes, function is less clearly implied, with incoherent expression perhaps suggestive of tuning interactions and coherent expression suggestive of either tuning or switch interactions, depending on the targets and their thresholds for optimal and consequential expression. Moreover, the different functions do not exclude each other, with, for example, a given targeting interaction providing tuning function in one cell type and fail-safe in another, or changing over time, as can occur with developmental switches, when a miRNA can be induced to dampen and then accelerate the clearance of pre-existing messages (Giraldez et al. 2006; Bushati et al. 2008).
Although examples for each of these scenarios for miRNA/target expression are known, their relative importance and prevalence has remained controversial, with successive mammalian studies indicating a prevalence of coherent but overlapping expression (Farh et al. 2005; Sood et al. 2006), and a Drosophila study indicating a prevalence of “mutually exclusive” expression, which is defined as nonoverlapping expression, at least as detected by in situ hybridization, while allowing for some low-level, undetectable expression of targets, or the presence of target messages inherited from transcription at an earlier developmental stage (Stark et al. 2005). More recently, a prevalence, at least for some miRNAs, for incoherent expression also has been reported (Tsang et al. 2007). Some of these discrepancies can be attributed to differing computational approaches, which scored either enrichment and depletion (Farh et al. 2005; Stark et al. 2005; Sood et al. 2006) or conservation rate (Tsang et al. 2007). But the more fundamental differences might stem from limitations in the data sets, which are either quantitative but primarily at organ resolution (Farh et al. 2005; Sood et al. 2006) or have cellular resolution but are not quantitative (Stark et al. 2005). A more satisfactory answer requires a systematic analysis of transcript levels in the miRNA-expressing tissues, at cellular resolution.
Here we report a genome-wide quantitative analysis of mRNA levels in miRNA-expressing cells of the developing zebrafish. We captured the entire miRNA expression domain in the embryo, purified to cellular resolution using fluorescent-activated cell sorting (FACS). For all three miRNAs examined, muscle-specific miR-206 and miR-133, and neuron-specific miR-124, the miRNAs and their predicted targets exhibited primarily coherent but overlapping expression patterns, with a few incoherent examples also found. The physiological relevance of these predicted interactions was supported by evolutionary conservation and specific up-regulation in dicer mutants, thereby bolstering our conclusion that coherent but overlapping expression encompasses the majority of miRNA–target interactions during vertebrate development.
Results
High-resolution isolation of miRNA-expressing cells
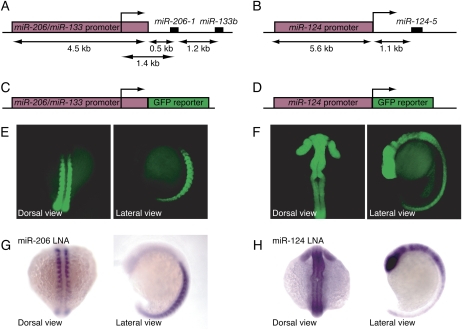
To identify miRNA-expressing cells we generated transgenic fish with GFP reporter genes driven by miRNA promoters. We chose the promoters of two zebrafish miRNA primary transcripts with very different expression domains that are conserved among diverse vertebrates: muscle-specific, polycistronic mir-206-1/mir-133b, and brain/CNS-specific mir-124-5. Using available expressed sequence tags (ESTs) and our data from rapid amplification of cDNA 5′ ends (5′-RACE), transcriptional starts were identified, and genomic fragments 4.5 kb and 5.6 kb upstream of the transcriptional starts were cloned as the presumptive promoter regions for mir-206-1/mir-133b and mir-124-5, respectively (Fig. 2A,B). Transient injection assays confirmed that the promoter-GFP fusions (Fig. 2C,D) expressed GFP in the same domains as the respective miRNAs; i.e., brain, retina, and spinal cord for miR-124 and somites for miR-206/133 (Wienholds et al. 2005; data not shown). The same pattern was observed for stable transgenic lines carrying the promoter fusions (Fig. 2E,F), with the GFP expression from the fusions accurately recapitulating endogenous miRNA expression (Fig. 2G,H). The miR-124 reporter was detectable from 9 h post-fertilization (hpf) in neuroepithelial cells, whereas the miR-206/miR-133 reporter was visible from 10 hpf in the unsegmented paraxial mesoderm and in somites, consistent with primary transcript in situ hybridization (data not shown). Thus, our reporter constructs allowed us to specifically label miRNA-expressing cells with GFP in vivo.
Figure 2.
Generation of miRNA reporter transgenic zebrafish lines. (A,B) Overview of the genomic loci of mir-206-1/mir-133b and mir-124, highlighting the promoter fragments upstream of the pre-miRNA hairpins (red boxes). (C,D) miRNA promoter reporter fusions. (E) The mir-206/mir-133 promoter drives expression of the GFP reporter gene in somites. (F) The miR-124 promoter drives expression of GFP in the spinal cord, brain, and retina. (G,H) Expression of miR-206 and miR-124, visualized by in situ hybridization, corresponded to that of GFP in the respective reporter lines.
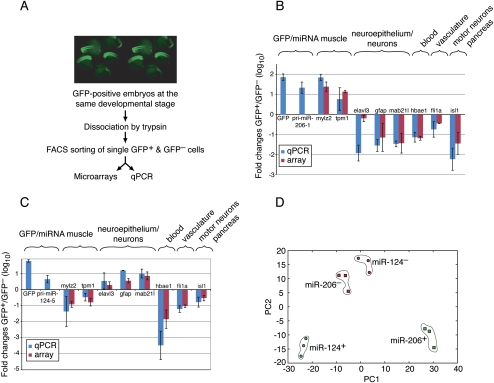
To isolate single cells expressing miRNA reporters, we dissociated several hundred embryos in trypsin and sorted the GFP+ from the GFP− cells by FACS (Fig. 3A; Supplemental Fig. 1). Embryos were dissociated at the 16- to 18-hpf stage, which corresponded to the onset of neuronal differentiation and the middle of somitogenesis, and was a stage in which both mature miR-124 and mature miR-206 were detectable (Fig. 2G,H). Using embryos at the same developmental stage for both the miR-124 and miR-206/miR-133 sorting enabled a comparison of miRNA target transcripts in different tissues. Two sequential rounds of FACS sorting were performed, yielding populations of GFP+ and GFP− cells with purities exceeding 99% (Supplemental Fig. 1).
Figure 3.
FACS sorting of single cells expressing miR-124 and miR-206/miR-133. (A) Schematic representation of the experimental setup. (B) Expression of marker genes in GFP+ and GFP− cells from embryos expressing miR-206/miR-133 reporter. Shown are fold differences between GFP+ and GFP− cells obtained by qRT–PCR (blue bars) and by microarray analyses (red bars) for markers characteristic of the indicated tissues. (C) Expression of marker gene in GFP+ and GFP− cells from embryos expressing the miR-124 reporter, details as in B. Error bars show standard deviations between three biological samples. (D) Principle component analysis (PCA) of microarray data. The intensity vectors for miRNA-expressing cells (green) and non-miRNA-expressing cells (red) are projected onto the first two principal components, revealing dense clustering of biological replicates of each condition and substantial separation between conditions.
GFP+ and GFP− cells for both reporter constructs were examined by quantitative RT–PCR (qRT–PCR) for expression of cell-type markers of muscle or neuronal cells. As expected, GFP+ cells from the fish with GFP driven from the mir-206/133 promoter were highly enriched in GFP, mir-206/133 primary transcript, and genes expressed in fish body muscle, such as myosin (mylz2) and tropomyosin1 (tpm1) (Fig. 3B). Moreover, these miR-206 GFP+ cells were depleted of markers characteristic of other tissues and cells, including markers from cell types that are tightly associated with muscle, such as neurons and neuroepithelial cells (elavl3, gfap, mab21l1), blood (hbae1), endothelial cells making blood vessels (fli1a), and motor neurons (isl1) (Fig. 3B). Similarly, miR-124 GFP+ cells were enriched in GFP, mir-124 primary transcript, and neuronal and neuroepithelial markers (elavl3, mab21l1, and gfap), whereas they were depleted of muscle (mylz2 and tpm1), blood (hbae1), and endothelial (fli1a) markers (Fig. 3C). Together, these results showed that FACS sorting enabled a clean and precise separation of cells from closely associated tissues, which is difficult to achieve with manual dissection of tissues and organs.
Predicted miRNA targets are expressed at lower levels in miRNA-expressing cells
To analyze expression profiles of miRNA-expressing cells, we hybridized Affymetrix microarrays with RNA isolated from miR-124 GFP+ and GFP− cells and from miR-206/miR-133 GFP+ and GFP− cells, analyzing three biological replicates for each. Each biological sample was collected from independent FACS experiments and tested for purity by the flow cytometer and qRT–PCR. Biological replicates for each condition showed consistent results, with correlation coefficients of the probe signal intensities ∼0.99 and average error rates (average coefficient of variation) between replicates below 15% (Supplemental Fig. 2). Array results were also validated using qRT–PCR for selected cell-type markers, which showed high consistency in all cases tested (Fig. 3B,C). Moreover, principle component analysis (PCA) showed that the variation between biological replicates was small compared with the differences between cell types, indicating that differences between cell types could be meaningfully assessed given the resolution of the data. PCA is a statistical method to reduce the dimensionality of data while retaining most of the variation (i.e., information) of the data set (Ringner 2008). We applied it here to display in two dimensions the global similarities and differences between GFP+ and GFP− cells measured across thousands of transcripts in our microarray experiments. For example, plotting the data according to the first two principal components showed that biological replicates for each condition fell in tight clusters, indicating their highly similar expression profiles (Fig. 3D). These results also showed that the difference between miR-124 GFP+ cells and miR-206/miR-133 GFP+ cells was greater than the difference between the respective GFP− cells (Fig. 3D), consistent with the expectation that the difference between muscle and nerve cells (GFP+ cells) would be much greater than the difference between the heterogeneous mixtures of cells comprising the rest of the embryo (GFP− cells). Taken together, the high quality and reproducibility for different miRNAs expressed in different tissues confirmed the general applicability of our approach and that our experimental setup enabled a comparison between gene expression programs of miRNA-expressing cells and those of other cells.
Messages that respond to miRNAs were predicted based on the presence of short conserved or nonconserved sites complementary to the seed region of the miRNA (Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005). Our analysis focused on 7mer sites that fall in 3′ untranslated regions (UTRs) and have perfect Watson-Crick pairing to the miRNA seed (nucleotides 2–7) supplemented by either a match to miRNA nucleotide 8 (7mer-m8 sites) or an A across from miRNA nucleotide 1 (7mer-A1 sites). A search for these 7mer sites also captures 8mer sites, which have both the match to nucleotide 8 and an A across from nucleotide 1. We chose these 7–8mer sites because, of the site types with abundance sufficient for our purposes, these are most effective in mediating down-regulation (Brennecke et al. 2005; Lim et al. 2005; Grimson et al. 2007; Nielsen et al. 2007; Baek et al. 2008). Our analysis considered only those sites falling in 3′UTRs, because these tend to be more effective than those found within 5′UTRs or ORFs (Grimson et al. 2007; Baek et al. 2008).
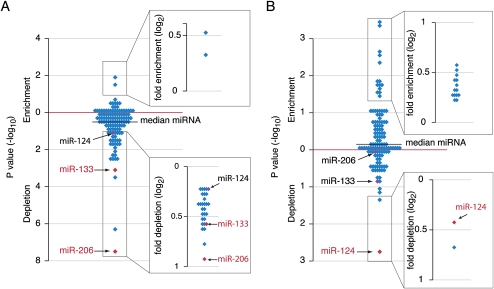
We first examined whether predicted targets of miR-206 and miR-133 were preferentially expressed or depleted in miR-206/miR-133 GFP+ cells, which express these two miRNAs. Although miR-133 and miR-206 are transcribed from the same promoter (as one polycistronic primary transcript), they have different seed regions crucial for target recognition, and as a consequence, have distinct sets of predicted targets. Only 25% of the miR-206 predicted targets were expressed at higher levels in miR-206 GFP+ cells than in miR-206 GFP− cells, whereas ∼75% were expressed at higher levels in GFP− cells (identical expression is rarely seen). Because overall about half the transcripts were expressed at higher levels in GFP+ cells, this represented a twofold depletion among messages preferentially expressed with the miRNA. Similarly, the miR-133 predicted targets were 1.6-fold depleted among these messages. Indeed, compared with predicted targets of all annotated zebrafish miRNAs, those of miR-206 and miR-133 were among the most depleted, with miR-206 targets displaying the most significant depletion (P < 10−7 and P < 10−3 for miR-206 and miR-133, respectively, hypergeometric P values) (Fig. 4A). As further evidence for the specificity of this depletion, we observed the reciprocal results in miR-124 GFP+ cells, with miR-124 predicted targets, not miR-206 or miR-133 predicted targets, depleted among transcripts expressed preferentially miR-124 GFP+ cells (1.4-fold depletion, P = 0.0014) (Fig. 4B). When examining the depletion signal in miR-124 GFP+ cells for predicted targets all zebrafish miRNAs, the confidence in the depletion of miR-124 predicted targets was the most significant (Fig. 4B), even though some miRNAs have overlapping expression domains in neuronal cells.
Figure 4.
Analyses of predicted target expression in sorted cells. (A) Enrichment and depletion of predicted miRNA targets among genes that are preferentially expressed in miR-206/miR-133 GFP+ cells (≥1.15-fold). Plotted in the main chart are hypergeometric P values for targets of each of the 122 zebrafish miRNA families examined. Plotted in the inset are the depletion values for those miRNAs with predicted targets significantly depleted in GFP+ cells (P ≤ 0.05). (B) Enrichment and depletion of predicted miRNA targets among genes that are preferentially expressed in miR-124 GFP+ cells (≥1.15-fold), plotted as in A.
In the analyses of Figure 4, all detected transcripts, even those with slight differences between GFP+ and GFP− cells (≥1.15-fold), were scored as preferentially expressed in either GFP+ or GFP− cells. We obtained analogous results when restricting analyses to messages significantly up-regulated or down-regulated in miRNA-expressing cells (i.e., those messages with at least 1.5-fold differences between GFP+ and GFP− cells, each with confidence of differential regulation exceeding 95%). At these more stringent cutoffs, only 4% of miR-206 targets were among the messages preferentially coexpressed with the miRNA, whereas 24% were among the messages preferentially expressed outside the miRNA-expressing cells. This corresponded to a sixfold difference between targets that were preferentially expressed in GFP+ versus GFP− cells and a threefold depletion among messages preferentially expressed with the miRNA. Analogous results were observed for miR-133 (5% vs. 21%; 1.4-fold depletion) and miR-124 (8% vs. 21%, 1.8-fold depletion). Quantification by qRT–PCR, sampling miR-206 and miR-124 predicted targets over a range of depletion or enrichment, generally agreed with the microarray data, although the magnitude of the fold change measured by qRT–PCR was often larger than that measured by expression arrays (Supplemental Fig. 3A,B).
In rapidly dividing cells mRNAs tend to have shortened 3′UTRs with fewer miRNA-binding sites due to alternative cleavage and polyadenylation (Sandberg et al. 2008). To test whether such variations of 3′UTR lengths could affect the trends reported in Figure 4, we repeated the enrichment/depletion analyses considering separately sites in the first third, middle third, and last third of the 3′UTRs, reasoning that sites in the last and middle thirds would be more likely to be missing in truncated UTRs. Predicted targets for all three miRNAs followed the same trends that we observed with full-length 3′UTRs, irrespective of which UTR region we considered (Supplemental Fig. 4A,B), which indicated that alternative cleavage and polyadenylation did not confound the observed depletion.
Predicted targets have diverse expression levels in miRNA-expressing cells
Our results showed that transcripts containing miRNA target sites tended to be expressed at higher levels outside of the miRNA-expressing cells, as expected for predominantly coherent regulation. However, two considerations complicate this interpretation. First, we do not know which of the predicted targets are actual targets of the miRNAs; not all messages with 7- or 8mer sites respond to the miRNA (Grimson et al. 2007; Baek et al. 2008). The second consideration is the phenomenon of “anti-targeting,” an evolutionary process whereby messages highly expressed in the cells that express a miRNA selectively avoid sites to that miRNA (Bartel and Chen 2004; Farh et al. 2005; Stark et al. 2005). This selective avoidance does not occur for the remainder of the messages, leading to a surplus of inconsequential sites in messages that are not expressed or do not produce a useful product in the miRNA-expressing cells. Selective avoidance can lead to depletion of half the sites in preferentially coexpressed messages (Farh et al. 2005), and thus in principle, could explain all of our expression trends observed in GFP+ and GFP− cells, obscuring the trends for the actual targets. To address these considerations, we examined whether the messages with sites were expressed in the GFP+ cells and also focused on the messages more likely to be functional targets, as evaluated by site conservation and tissue-specific up-regulation in fish lacking miRNAs.
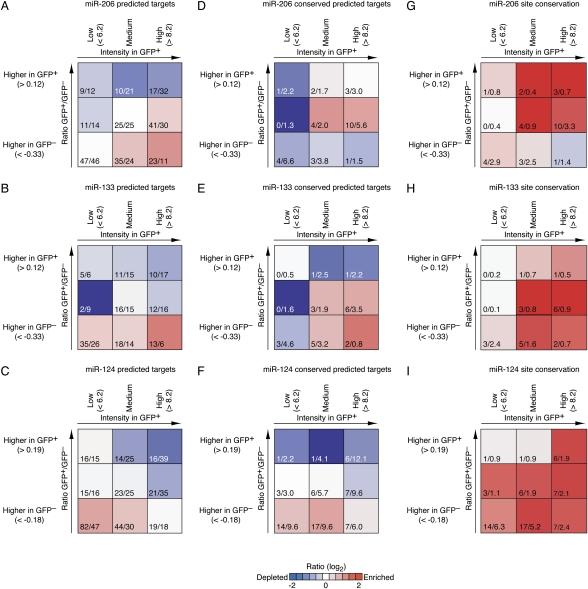
When examining whether the predicted targets were expressed at high or low levels in the miRNA-expressing GFP+ cells, we considered only the messages with sufficient signal above background in either GFP− or GFP+ cells to be declared “present” by the array-analysis package (3395 unique messages for the miR-206/133 experiment, and 3405 for the miR-124 experiment). We also restricted the analyses to 7mer sites perfectly complementary to nucleotides 2–8. Because 8mer sites also satisfy the criterion used to identify these 7mer sites, this restriction focused on the most effective sites, the 7mer-m8 and 8mer sites, while avoiding the complications associated with including overlapping sites in the analyses of signal and background. Reasoning that results might differ for the set of messages preferentially expressed with the miRNA compared with that preferentially expressed elsewhere, we considered these sets separately, assigning the 3395 messages for the miR-206/133 experiment and 3405 messages for the miR-124 experiment into three equally populated bins based on expression ratios between GFP+ and GFP− cells. Binning these genes separately based on observed intensities in GFP+ cells created a matrix of nine cells (Fig. 5A–C). The three columns of this matrix separated the transcripts with respect to expression intensity (Supplemental Fig. 5), binning the third with the lowest expression in GFP+ cells (including those that were not detected in GFP+ cells; left column), the third with the highest expression (right column) and the third with no strong trend (including those at the mode of normalized log intensities; middle column), and the three rows separated these transcripts with respect to the ratios (GFP+ vs. GFP−).
Figure 5.
Predicted target density and site conservation at different expression levels. All unique messages with detectable expression in either GFP+ or GFP− cells were binned into three equally populated bins, each according to its GFP+/GFP− array signal ratio (rows) and into three equally populated bins according to signal intensity in GFP+ cells (columns), yielding a gene density map with matrix of nine cells. (A–C) Gene density maps of predicted conserved and nonconserved targets for the indicated miRNAs. Fractions indicate the number of observed target sites (numerator) and the number of target sites expected based on the background distribution (denominator). Enrichment or depletion when dividing observed by expected is indicated by colors (red indicates enrichment, blue depletion). Significance of individual ratios was also assessed by hypergeometric P values (Supplemental Fig. 6) and using Z scores (Supplemental Fig. 7). (D–F) Gene density maps considering only those predicted targets with sites conserved in other fish; otherwise as in A–C. (G–I) Preferential conservation of sites, evaluated in each cell of the gene density map. For each cell, the numerator is the number of conserved target sites, as in D–F, whereas the denominator is the background expectation, calculated as the number total target sites (numerator in A–C) multiplied by the frequency that control motifs for the miRNA are conserved. Colors indicate miRNA preferential conservation when compared with the conservation of these control sequences. Note that this enables detection of preferential conservation in each cell, independent of target abundance in the cell; i.e., independent of relative enrichment or depletion (as shown in A–F). Matrix cells with very low counts (cells for which both the observed and expected counts of <1) are not colored.
For each cell of the matrix, we determined the relative enrichment or depletion of miRNA predicted targets by a ratio, compared with the mean abundance of control sites (Fig. 5), and assessed the significance of each ratio by a hypergeometric P value (Supplemental Fig. 6) and by Z scores based on the variability of abundances observed for the control sequences (Supplemental Fig. 7). For all three miRNAs, target genes were enriched in the bottom row and depleted in the top row (Fig. 5A–C), confirming our finding that predicted targets are expressed at lower levels in cells that express the cognate miRNA (Fig. 4A,B). miR-206 predicted targets were most enriched in the lower right corner (Fig. 4A,B), indicating that although these predicted targets were preferentially expressed where the miRNA is not expressed, they nonetheless tended to be present at high levels in cells that express the miRNA. In contrast, miR-124 predicted targets were most enriched in the lower left corner, which corresponded to genes with both preferential expression outside of neurons and low expression in neurons (Fig. 5C). The pattern for miR-133 fell between the other two, with enrichment across the bottom row. We obtained the same picture when controlling for differences in 3′UTR GC content (Supplemental Fig. 8), and when excluding all transcripts with annotated alternative splice forms in zebrafish or human, indicating that neither correlation of array intensities with transcript GC content nor differential miRNA targeting of alternative splice forms affected our results for these three miRNAs.
Conserved targets tend to be expressed in miRNA-expressing cells
The enrichment in the lower left corner of the matrices in Figure 5, observed for miR-124 and to some extent for miR-133, could be explained either by an abundance of fail-safe interactions or by the preferential accumulation of inconsequential sites in messages of this matrix cell, accentuated by the selective depletion elsewhere. Because fail-safe interactions provide beneficial function, they would tend to be preferentially conserved in other species, whereas inconsequential sites accumulating in messages that either are not expressed in miRNA-expressing cells or do not produce a beneficial or harmful product in these cells would be conserved no more than expected by chance. To examine if the expression of messages with conserved sites differed from that of all predicted targets, we repeated the enrichment/depletion analyses, focusing only on those messages with conserved sites, defining a conserved site as one that is present at a similar position (within a window of 100 nt) in at least one of the aligned fish sequences (Fig. 5D–F; Supplemental Fig. 7D–F).
When compared with enrichment of all predicted targets (Fig. 5A–C), enrichment of conserved predicted targets shifted upward and/or rightward (Fig. 5D–F). This was the result expected if messages with inconsequential sites preferentially resided in the bottom left portion of the graphs and the conservation filter helped to exclude them from consideration. Despite the upward shift, a tendency toward coherent expression, as indicated by enrichment in the bottom row compared with the top row, was retained for both miR-133 and miR-124 (Fig. 5E,F).
Fail-safe targets and targeted transcripts remaining from previous developmental stages were expected to be included among the conserved targets in the lower left corner. For miR-206 and miR-133, enrichment for conserved targets was not observed in this corner, whereas for miR-124 some enrichment was observed but it did not appear greater than the lower middle matrix cell, which was comprised of messages expressed at medium levels in GFP+ cells yet higher levels in GFP− cells. Therefore, expression of targets of biological importance, as inferred through conserved sites, had detectable overlap with expression of the miRNAs. Indeed, because our expression cutoffs assigned some target–miRNA pairs with overlapping expression to the lower left corner, the fraction of biologically important targets with perfectly nonoverlapping expression could be quite small.
Messages with excess site conservation have a variety of expression patterns
We next sought to determine whether the sites matching the miRNAs were conserved more frequently than expected by chance. An excess beyond chance conservation was observed in most of the matrix cells (Fig. 5G–I)—even in some matrix cells that were depleted in conserved predicted targets (Fig. 5D–F; Supplemental Fig. 7D–F). An informative example was the upper right corner of the miR-124 matrices. Although this corner had less than half the predicted targets expected based on control sequences (16 observed, compared with 39 expected) (Fig. 5C) and only half the expected conserved predictions (six observed, compared with 12 expected) (Fig. 5F), of the 16 predicted targets populating this corner, six were conserved, which was about four more than expected by chance (Fig. 5I). A similar pattern of relatively high conservation rates for the few sites present in the upper right area of the matrices was observed for miR-206 and miR-133, indicating that each of the miRNAs probably has a few targets for which it acts incoherently with other regulatory processes to dampen the expression of preferentially coexpressed messages. This being said, the overall picture gleaned from tallying the sites above background for the aggregate preferential conservation data was one of coherent expression (Figs. 5G–I).
Analyses of conserved sites do not predict all the biologically important targeting. For example, some biologically important targets are missed because they have emerged recently, or because of imperfections in UTR annotations or alignments. These factors that lessen sensitivity are more pronounced in zebrafish than in mammals or flies, because zebrafish has less thorough UTR annotations and fewer sequenced species at optimally informative evolutionary distances. However, we have no reason to suspect that the conserved targeting we detected might populate the expression sectors of Figure 5 differently than the biologically important targeting that we missed. With this in mind, we conclude that biologically important targeting tends to be coherent but overlapping, as proposed from analyses of array data from tissues, organs, and cells that were purified less extensively (Farh et al. 2005; Sood et al. 2006).
The high enrichment for predicted targeting sometimes observed in messages expressed at low levels with the miRNA (Fig. 5B,C, bottom left corner) at least partially diminished when considering site conservation (Fig. 5E,F), suggesting that one contribution of this enrichment was the indirect effect of anti-targeting: an accumulation of sites in messages in which these sites are inconsequential. Nonetheless, we cannot rule out nonoverlapping expression for a detectable minority of conserved targets (Fig. 5F,I, bottom left corner). Also in contrast to the general trend, some incoherent targeting was detected (Fig. 5G,I, top right corner), despite the strong overall depletion of predicted targets with this expression profile (Fig. 5A,C, top right corner). We suggest that for some miRNAs, selective avoidance of fortuitous sites could eliminate a sufficient number of nonconserved sites in messages preferentially expressed with the miRNA, such that the few remaining sites are conserved at a higher proportion than sites in other messages. Because of this possibility, estimating target abundance by calculating the proportion of conserved sites (signal divided by background) (Tsang et al. 2007), might be misleading. Using the appropriate metric of excess conservation (signal minus background), we found that incoherent expression of miRNAs and their targets occurred, but not as frequently as coherent expression.
Coherent expression is diminished but retained in the absence of miRNAs
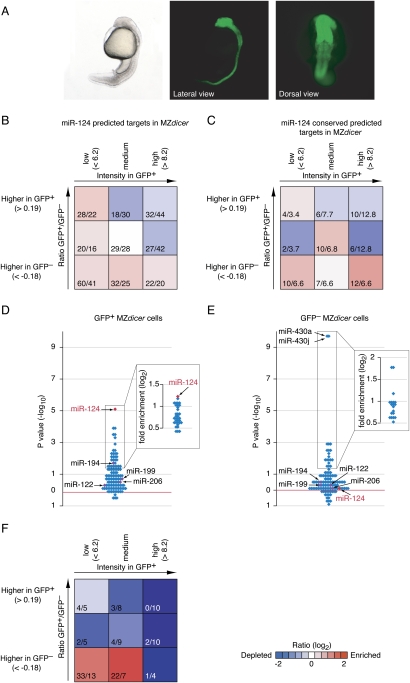
An unresolved issue arising from Figures 4 and 5, as well as previous reports of coherent expression of miRNAs and their targets in tissues from wild-type animals (Farh et al. 2005; Stark et al. 2005; Sood et al. 2006), is the degree to which the lower expression of targets in miRNA-expressing cells was a result of miRNA-mediated destabilization of target messages. Indeed, in an extreme scenario, miRNA-mediated target destabilization could transform expression that is predominantly incoherent into the pattern observed, which is predominantly coherent. To test this possibility, and to learn which messages respond to the loss of the miRNAs, we repeated the miR-124 experiment using maternal–zygotic dicer (MZdicer) embryos, which do not produce miR-124 or any other mature miRNAs because they produce defective Dicer and also lack the functional Dicer normally contributed to the egg by the mother.
MZdicer mutants have been an invaluable tool for examining miRNA function and targeting during vertebrate development (Giraldez et al. 2005, 2006). Despite defects in development, MZdicer mutants form all major cell types and organs (Giraldez et al. 2005), allowing us to study the impact of miRNA loss on predicted target expression during early embryonic development. To focus on the impact of losing miR-124, we crossed the Dicer mutant (Wienholds et al. 2003) with our line containing the miR-124 promoter fusion and used the resulting line to generate MZdicer mutants that express GFP from the mir-124-5 promoter.
In MZdicer mutants, the miR-124 reporter fusion was expressed in the same domain as in wild-type embryos, including the brain, developing retina, and spinal cord (Fig. 6A). These results demonstrated miRNA-independent regulation of the mir-124 promoter, further illustrating that tissue-specific expression programs can be correctly regulated in absence of all mature miRNAs, as can tissue specification and formation. GFP+ and GFP− cells were sorted from embryos at the developmental stage of 14–18 somites, the same stage used for wild-type embryos. Analytical flow cytometery and qRT–PCR confirmed the purity of the collected GFP+ and GFP− cells (Supplemental Fig. 9; data not shown). Array results from three biological replicates from GFP+ mutant cells formed a tight cluster in PCA analyses, as did those from three biological replicates from GFP− mutant cells, indicating the highly similar expression profiles between replicates and a clear distinction between different cell types, consistent with retained cell type identity and expression programs in dicer mutants (Supplemental Fig. 10; Giraldez et al. 2005).
Figure 6.
The response of predicted targets to the loss of miRNAs. (A) The GFP reporter gene driven by the mir-124 promoter in MZdicer fish at 30 hpf is expressed in the brain, retina, and neural tube, as observed in wild-type zebrafish. (B) Gene density map displaying enrichment and depletion of conserved and nonconserved predicted targets in the absence of miRNAs; otherwise as in Figure 5C. (C) Gene density map displaying enrichment and depletion of conserved predicted targets in the absence of miRNAs, otherwise as in Figure 5F. (D) Enrichment of predicted targets among genes that are preferentially up-regulated in MZdicer GFP+ cells compared with wild-type GFP+ cells (greater than or equal to twofold). Plotted are hypergeometric P values, as in Figure 3. For miRNAs with significantly enriched predicted targets (P ≤ 0.05), fold enrichment is shown in the insets. No miRNAs had predicted targets that were significantly depleted. (E) Enrichment of predicted targets among genes that are preferentially up-regulated in MZdicer GFP− cells compared with wild-type GFP− cells (greater than or equal to twofold), plotted as in D. No miRNAs had predicted targets that were significantly depleted. (F) Gene density map displaying enrichment and depletion of miR-124-regulated transcripts compared with all transcripts, evaluated in wild-type fish (matrix cells defined as in Fig. 5). miR-124-regulated transcripts were defined as transcripts that contain miR-124 sites and are up-regulated in MZdicer GFP+ cells compared with wild-type GFP+ cells (greater than or equal to twofold).
Analyzing expression of the predicted targets revealed the overall tendency of coherent expression observed previously in wild-type embryos, but this tendency was muted somewhat in the MZdicer cells, presumably reflecting the absence of mRNA destabilization directed by miR-124 and other miRNAs (Fig. 6B; Supplemental Fig. 11A). A similar picture of a muted but retained tendency for coherent expression was observed also for the predicted targets with conserved sites (Fig. 6C; Supplemental Fig. 11B).
Many miR-124 predicted targets are tissue-specifically up-regulated in the absence of the miRNA
When compared with the array data from sorted wild-type cells, the array data from sorted mutant cells provided the opportunity to identify predicted targets that are derepressed in the absence of miR-124, thereby providing experimental support for the authenticity of these predicted interactions. Among the 436 transcripts significantly up-regulated in MZdicer GFP+ cells compared with wild-type GFP+ cells (greater than or equal to twofold, P ≤ 0.05), 32 had 7mer-m8 (or 8mer) UTR sites matching miR-124, which was a significant enrichment over the 14 expected by chance (P < 10−5) (Fig. 6D). In contrast, predicted targets of miR-124 were not enriched in transcripts significantly up-regulated in MZdicer GFP− cells (P = 0.69) (Fig. 6E), as expected in these cells that do not express miR-124.
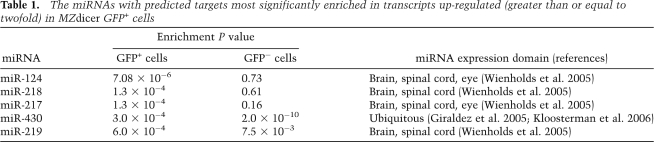
We also checked whether transcripts up-regulated in GFP+ MZdicer cells were enriched for targets of other miRNAs. As expected for miR-124-positive neuroepithelial cells, up-regulated mRNAs were enriched for predicted targets of brain-specific miRNAs (miR-217, miR-218, and miR-219) and ubiquitously expressed miRNAs (miR-430) (Table 1), whereas they were not as enriched for predicted targets of miRNAs expressed elsewhere, including muscle (miR-206), liver (miR-122), gut/gall bladder (miR-194), or epithelium (miR-199) (Fig. 6D). As observed for predicted targets of miR-124, this enrichment for predicted targets of miR-217 and miR-218 was specific in that it was not observed in transcripts up-regulated in GFP− mutant cells compared with GFP− wild-type cells, although as reported for the entire embryo at earlier developmental stages (Giraldez et al. 2006), these GFP− mutant cells were still enriched for targets of ubiquitously expressed miR-430 (Fig. 6E; Table 1). Interestingly, we did not detect enrichment of targets of other tissue-specific miRNAs (miR-122, miR-206, miR-194, and miR-199) in transcripts up-regulated in GFP− mutant cells, even though these miRNAs normally are expressed in a subset of these cells. These results highlighted the importance of cell sorting to achieve the tissue and cell specificity required to detect derepression of targets of tissue-specific miRNAs in Dicer mutants.
Table 1.
The miRNAs with predicted targets most significantly enriched in transcripts up-regulated (greater than or equal to twofold) in MZdicer GFP+ cells
Coherent expression of most of the predicted targets substantially derepressed in the Dicer mutant
We next considered the relative and absolute expression of the 71 predicted targets that are derepressed in MZdicer mutants (≥1.5-fold up-regulation; P ≤ 5%), binning the transcripts based on their expression in wild-type embryos, as in Figure 5C. A striking enrichment in the bottom left corner and bottom center matrix cell was observed, indicating propensity for coherent expression of these responsive messages (Fig. 6F). Several types of messages reside in this bottom left corner, including those with overlapping expression and those with nonoverlapping expression. The detection of highly responsive messages in the bottom left corner implies overlapping expression between miR-124 and these responsive messages, which might be switch targets, tuning targets, or neutral targets; i.e., messages with sites that are functional but inconsequential nonetheless because the repression serves no beneficial or harmful purpose.
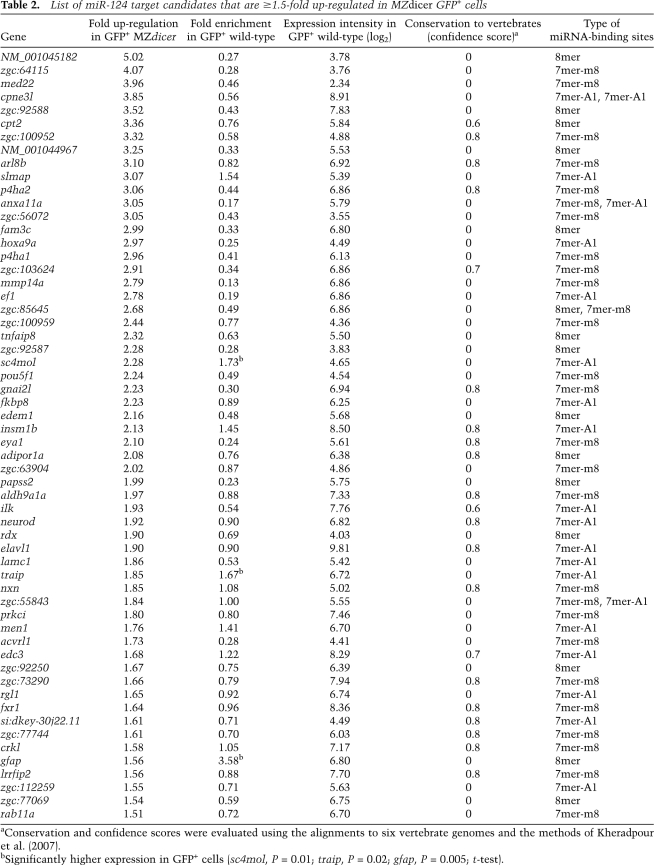
Our data comparing the expression of messages in wild-type and Dicer-deficient cells provided information valuable for considering the biological functions of miR-124 during zebrafish development. With this in mind, we compiled a list of predicted targets confidently up-regulated in Dicer-deficient cells that would normally express miR-124 (Table 2). To be more inclusive, the list included genes with 7mer-A1 UTR sites, in addition to the predicted targets with 7mer-m8 and 8mer UTR sites considered in our other analyses (Figs. 4–6). Confidence of site conservation was evaluated using control sequences and a branch length score (BLS) (Kheradpour et al. 2007).
Table 2.
List of miR-124 target candidates that are ≥1.5-fold up-regulated in MZdicer GFP+ cells
Discussion
A general method for identifying miRNA target candidates
Our approach for studying miRNA–target coexpression, which combined miRNA-deficient zebrafish, GFP reporter fusions indicating miRNA expression, and FACS sorting, also enabled the identification of candidate messages that are targeted by the miRNA under physiologically relevant conditions. These candidate targets were identified by their tissue-specific derepression that was obscured when analyzing RNA from the whole embryo but revealed after cell sorting. Although not every message with a target site that is up-regulated constitutes a direct target of the miRNA, up-regulated messages with sites were much more abundant than expected by chance, which indicated that most of the candidates were authentic direct targets. When considering the cell type specificity that most miRNAs display (Wienholds et al. 2005), we anticipate that our approach will be a valuable tool for investigating miRNA function in animals. In addition to specific stages during normal development, it can be applied under selected conditions such as disease, stress, or after drug treatment, when miRNA targeting and/or function might be expected to change.
Examples of rare incoherent regulation—a transcription factor feedback loop and a switch target
As expected based on the analyses of messages with sites (Figs. 4–6), most responsive messages with sites were expressed at lower levels in miR-124-expressing cells than in other cells (Table 2). However, we identified seven messages that were confidently derepressed in MZdicer mutants yet showed a tendency to be preferentially coexpressed with miR-124 in wild-type fish (Table 2). Some of these are known to be crucially involved in neurogenesis, such as a zinc-finger transcription factor insulinoma-associated 1b (insm1b), which is a transcriptional repressor of a proneural bHLH gene neurogenic differentiation (neurod) (Liu et al. 2006). Both genes neurod (positive regulator of neurogenesis) and insm1b (negative regulator of neurod) have conserved miR-124 target sites in their 3′UTRs and are derepressed in MZdicer mutants (1.9-fold for neurod and 2.1-fold for insm1b) (Table 2), strongly suggesting that both genes are direct targets of miR-124. In addition, both genes are highly expressed in miRNA-expressing cells. A similar target/miRNA relationship was described for miR-430 in the Nodal pathway, where it regulates both the pathway agonist squint and the antagonist lefty (Choi et al. 2007). Since coexpression of miR-124 and its targets insm1b and neurod is maintained in differentiated neurons at later stages of development, it is likely that these miRNA–target interactions represent a tuning mode of miRNA action, in which miR-124 helps establish optimal concentrations of Insm1b and Neurod proteins, both in absolute and relative levels (Fig. 1A). In addition to providing another means of gene control, tuning interactions in which high transcription is coupled with miRNA-mediated translational repression can help reduce stochastic fluctuations in gene expression (Bartel and Chen 2004; Raser and O'Shea 2005).
Another example of apparently incoherent regulation is that of glial fibrillary acidic protein (gfap), a class III intermediate filament gene expressed throughout the entire neuroepithelium at earlier stages of development and in astrocytes and radial glial cells of the CNS at later stages (Sprague et al. 2006). gfap is preferentially expressed in miR-124-expressing cells and 1.6-fold up-regulated in GFP+ MZdicer mutant cells (Table 2). gfap could represent a classical switch target (Fig. 1A) that is coexpressed with miR-124 at earlier stages and then reduced to inconsequential levels in cells expressing the miRNA, or miR-124 could clear gfap mRNA after the gene is transcriptionally silenced. In this case, miR-124 might help to establish lineage specification by destabilizing transcripts that should be down-regulated in neurons but maintained in glial cells, a role similar to that for Drosophila miR-124, which has been proposed to help discriminate between neuronal and epithelial cells (Stark et al. 2005).
miRNAs might counteract prevalent ubiquitous low-level transcription
We show that coherently regulated predicted targets that are either undetectable or detected at low levels in miRNA-expressing cells (among the lower third of messages analyzed) can still be physiologically relevant and detectably regulated, as indicated by site conservation and derepression in Dicer mutants. Some of these predicted targets might be relevant at an earlier developmental stage and not efficiently or rapidly cleared in the absence of the miRNA, consistent with the preferential targeting of epidermal genes by miR-124 in developing neurons in flies (Stark et al. 2005). Another scenario, which we suspect applies to more targets, is that prevalent low-level transcription requires that the protein levels of many genes be dampened post-transcriptionally. Indeed, of the 5824 RefSeq transcripts that were detected according to a consistent “present call” of the Affymetrix analysis package in miR-206-positive muscle cells or miR-124-positive neuronal cells, only 121 (2%) were uniquely present in muscle and only 280 (5%) were unique to neurons, indicating that many genes are present at low levels in most cells. This is consistent with findings that a large fraction of the genome is detectably transcribed in a wide range of species, as reported by tiling-array (Manak et al. 2006; Birney et al. 2007; Kapranov et al. 2007) and high-throughput sequencing of RNA fragments (Nagalakshmi et al. 2008; Wilhelm et al. 2008).
Transcripts acquire or reject miRNA target sites over the course of evolution
We show that single miRNAs operate in different modes and provide a framework to describe miRNA–target coexpression and associated miRNA functions. The proposed modes and functions depend on the individual target genes and describe the regulatory relationship of miRNA–target pairs, rather than classify individual miRNAs. This is consistent with an evolutionary scenario, in which different transcripts retain or discard binding sites for an expressed miRNA dependent on whether the functional consequences are beneficial or detrimental. This applies to coexpressed transcripts for which the protein levels need to be fine-tuned or stochastic fluctuations need to be avoided (either incoherent or coherent regulation), to transcripts that need to be cleared or safely turned off during development (coherent regulation), and to those that need to be coexpressed with the miRNA at high levels (anti-targets). Once beneficial miRNA–target interactions appear, they are maintained by means of their selective advantage and can be observed through conserved miRNA-binding sites prevalent in both tuning and switch targets.
Materials and methods
Generation of miRNA reporter transgenic lines
Seven mir-124 genes are annotated in zebrafish. We chose mir-124-5 because of the presence of several annotated EST clones upstream of the mir-124-5 hairpin (EH445664, DT872908, EH579759, CN507738). 5′-RACE (GenRacer kit, Invitrogen) identified a transcriptional start of the mir-124-5 primary transcript, which is located 1.1 kb from the mir-124-5 hairpin. A 5.6-kb fragment upstream of the mir-124-5 transcriptional start was amplified from genomic DNA using primers 1 and 2 (Supplemental Table 1).
For polycistronic mir-206-1/mir-133b, we were not able to identify the transcriptional start precisely by 5′-RACE due to the repetitive sequence in this region. Analyses of EST clones (BM259633, CF999541, CF348604, DR729161) mapped the approximate start of the mir-206-1/mir-133b primary transcript to 1.4 kb from the mir-206 hairpin. A 4.5-kb upstream fragment that includes ∼0.9 kb of the primary transcript was amplified from genomic DNA using primers 3 and 4 (Supplemental Table 1). Both promoter fragments were fused to GFP reporter genes. The miRNA promoter fusions were flanked by I-SceI meganuclease recognition sites to increase the efficiency of transgenensis (Thermes et al. 2002).
GFP reporter constructs were injected into one-cell embryos. Approximately 1.0 nL of 50 ng/μL of circular DNA and 0.2 units/μL of ISce-I meganuclease (New England BioLabs) were injected into the oocyte. Embryos with strong transient GFP expression were raised to the sexual maturity. To identify stable transgenic lines, F0 fish were mated to wild-type fish. Embryos from these crosses were screened for GFP expression and raised to maturity thereby establishing stable transgenic lines. Multiple independent carriers with germline transmissions for each miRNA showed identical expression of GFP reporter constructs. Two independent founder fish were used for miR-206/miR-133 to set up the F1 generation used to produce the embryos for our analyses, and five independent carriers were used for miR-124.
To generate MZdicer fish expressing a GFP reporter under control of the mir-124 promoter, heterozygous dicerhu715/+ fish (Wienholds et al. 2003) were crossed to the Tg(mir-124:GFP) fish. F2 embryos from this cross were used as donor embryos for replacing the germline of wild-type recipient embryos (Giraldez et al. 2005). Fertile adults with a dicer mutant germline and GFP expression driven by the mir-124 promoter were intercrossed to produce MZdicer Tg(mir-124:GFP) embryos. Attempts to generate the analogous line expressing GFP from the mir-206/mir-133 promoter were unsuccessful, which could be explained by the promoter fusion of the line used having integrated into Chromosome 17, the same chromosome that contains the Dicer locus.
Cell collection and flow cytometry
Several hundred GFP+ embryos were decorionated by Pronase (Roche) treatment and washed five times with E3 solution (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 10−5 % Methylene Blue). Embryos anesthetized in 0.02% tricaine were dissociated in Trypsin-EDTA solution (Sigma) for 5–10 min. Fetal calf serum (to 5%) was added to inactivate the trypsin, and cells were centrifuged (7 min at 1300 rpm), washed once with PBS, and resuspended in PBS before filtering through a cell strainer (70 μL Nylon; Falcon). Propidium iodide (PI; 1:200; Invitrogen) was added as a marker to identify dead cells. FACS was performed based on forward scatter, side scatter, GFP fluorescence, and PI exclusion, using a MoFlo flow cytometer. To achieve ∼99% cell purity (Supplemental Fig. 1), both GFP+ and GFP− cells were subjected to a second round of sorting.
RNA analyses
Colormetric in situ hybridization, using miR-206 and miR-124 DIG-labeled LNA probes, was performed as described (Wienholds et al. 2005).
Cells isolated by FACS sorting were centrifuged (10 min at 1300 rpm) and total RNA was isolated using Trizol Reagent (Ambion). Isolated RNA was treated with DNase I (Ambion) and then purified with RNeasy columns (Qiagen). For qRT–PCR, cDNA was synthesized (∼100 ng of total RNA input, oligo-dT pimer, Thermoscript RT kit, Invitrogen), and qPCR was performed using the SYBR green mix on a PRISM 7000 sequence detection system (Applied Biosystems). β-Actin and β-catenin were used for normalization. Primers for qPCR are listed (Supplemental Table 1).
Total RNA, DNase treated and purified on RNeasy columns, was used in a labeling reaction using the small-sample protocol (50 ng total RNA; Affymetrix). Labled RNA was hybridized to Affymetrix GeneChip Zebrafish Genome Arrays. Probes were mapped to Refseq transcripts according to Netaffix. Before labeling the quality of RNA was confirmed by analyzing 20–50 ng of RNA with a Bioanalyzer 2100 (Agilent).
miRNA target prediction and analyses
We predicted miRNA targets as transcripts with 7mer-A1, 7mer-m8 3′UTR sites (Lewis et al. 2005). We used whole-genome alignments of zebrafish (danRer4) to six vertebrate genomes (human [hg18], mouse [mm8], opossum [monDom4], Xenopus [xenTro2], Tetraodon [tetNig1], Fugu [fr1]), obtained as a multiple-sequence alignment from UCSC (Supplemental Table 2; Kent et al. 2002). We also obtained the corresponding mapping of RefSeq transcripts and 3′UTRs to the zebrafish genome from the UCSC genome browser. Enrichment and conservation of each seed-matched site was evaluated with respect to 10 control motifs of similar characteristics (Lewis et al. 2003; Kheradpour et al. 2007), which controlled for potential biases in overall 3′UTR conservation, nucleotide composition, or length. For these analyses using control motifs we considered only matches complementary to nucleotides 2–8 to avoid dependencies between partially overlapping motifs.
Our results were robust with respect to alignment or species choice, as indicated by performing the analyses with different settings: We predicted miRNA targets using a whole-genome alignment of zebrafish (danRer4) and four fish species (Tetraodon [tetNig1], Fugu [fr3], Stickleback [gasAcu1], and Medaka [aryLat1]), obtained as pairwise alignments from the UCSC genome browser (Supplemental Table 3; Kent et al. 2002). We also repeated the enrichment analyses on zebrafish RefSeq transcripts independent of any alignment, defining the 3′UTR of Refseq transcripts as the sequence downstream from the longest ORF (Giraldez et al. 2006).
We compared the relative enrichment or depletion of predicted targets for each of the zebrafish miRNAs among the preferentially coexpressed transcripts (ratio GFP+/GFP− ≥ 1.15). Note that we did not require the transcripts to be significantly enriched (e.g., P ≤ 0.05), because this would have biased the classification (preferentially coexpressed vs. preferentially not coexpressed) to more strongly enriched transcripts, which would have favorably distorted our analysis (i.e., lead to stronger trends). To reduce biases due to different 3′UTR lengths, conservation, or nucleotide composition, we restricted the transcripts to those that are targeted by at least one miRNA, essentially testing for preferential miRNA targeting of specific miRNAs versus all miRNAs. For each zebrafish miRNA, we calculated the fold enrichment or depletion among the preferentially coexpressed transcripts and assessed the significance by hypergeometric P values (Fig. 4).
We binned all transcripts into three equally populated bins according to their absolute expression (i.e., intensity) in GFP+ cells and into three equally populated bins according to their preferential expression in GFP+ cells (ratio GFP+/GFP−). We then intersected these bins to create a matrix of nine cells that separated the transcripts by both absolute and relative expression levels in GFP+ cells. For each bin, we calculated the following miRNA target statistics: enrichment of target genes, enrichment of target sites, enrichment of conserved target sites, and target site conservation, each assessed by a ratio, a hypergeometric P value, and by the actual number of targets or sites above the background expectation. For miR-124, we also calculated the enrichment of transcripts that respond to the miRNA, as determined by their specific up-regulation in MZdicer mutant miR-124-expressing cells. In addition, we calculated the significance of enrichment/depletion and site-conservation in each matrix cell with respect to the set of shuffled controls using Z scores (i.e., taking the differences between individual control 7mers into account). For this, we determined the fraction of predicted target genes in each matrix cell (enrichment analyses) or the fraction of conserved sites in each matrix cell (conservation analyses) for the miRNA targets and each of the corresponding controls separately. We then calculated the mean and standard deviation of the fractions and calculated the Z score as (F − FC)/stdevC (F is fraction for miRNA targets; FC is average fraction for controls; stdevC is standard deviation for controls).
To test and exclude influences due to alternative splicing, we repeated the analysis (1) only with transcripts for which all alternative transcripts (Refseq transcripts with overlapping genomic coordinates) in zebrafish had the same number of miRNA target sites, (2) only with transcripts without any alternative forms in zebrafish, and (3) only with transcripts without any alternative forms in either zebrafish or human. To test for influences of transcript GC content on the array intensity values, we repeated the analysis with equal GC contents across bins. To achieve this, we first binned all transcripts into ten bins according to their GC content and then populated the three intensity bins of the gene density map by transcripts with intensity values in the bottom, middle, and top one-third from each GC content bin.
We determined transcripts that are significantly up-regulated in MZdicer mutants, comparing GFP+ cells or GFP− cells between wild-type and MZdicer mutant zebrafish (ratio MZdicer/wild type ≥ 2; P value ≤0.05, t-test). We then assessed the specific enrichment (fold enrichment and hypergeometric P value) of predicted targets for each zebrafish miRNA among the up-regulated transcripts that are predicted targets for at least one miRNA to reduce biases due to 3′UTR length, conservation, or nucleotide composition.
We assessed target gene expression outside the miRNA expression domain and at stages not surveyed by our approach using the collected in situ hybridization data of the zfin database (http://www.zfin.org; Sprague et al. 2006).
Acknowledgments
We thank the Kellis laboratory, especially P. Kheradpour and M. Kellis, for their support, Calvin Jan and Amanda Dickinson for providing helpful comments on the manuscript, and Antonio Giraldez for providing dicerhu715/+ fish. A. Shkumatava and A. Stark were supported by Human Frontiers Science Program long-term post-doctoral fellowships. This work was supported by a grant from the NIH to D.B. D.B. is an HHMI Investigator.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1745709.
Supplemental material is available at http://www.genesdev.org.
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel D.P., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Birney E., Stamatoyannopoulos J.A., Dutta A., Guigó R., Gingeras T.R., Marguiles E.J., Weng Z., Snyder M., Dermitzakis E.G., Thurman R.E., et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA–target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., Cohen S.M. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081. [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Choi W.Y., Giraldez A.J., Schier A.F. Target protectors reveal dampening and balancing of nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson M.J., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Mansfield J.H., Yekta S., Hu J.K., Harfe B.D., McManus M.T., Baskerville S., Bartel D.P., Tabin C.J. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., Brummelkamp T.R., Fleming M.D., Camargo F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Karres J.S., Hilgers V., Carrera I., Treisman J., Cohen S.M. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradpour P., Stark A., Roy S., Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Steiner F.A., Berezikov E., de Bruijn E., van de Belt J., Verheul M., Cuppen E., Plasterk R.H. Cloning and expression of new microRNAs from zebrafish. Nucleic Acids Res. 2006;34:2558–2569. doi: 10.1093/nar/gkl278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang F., Lee J.A., Gao F.B. MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes & Dev. 2006;20:2793–2805. doi: 10.1101/gad.1466306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu W.D., Wang H.W., Muguira M., Breslin M.B., Lan M.S. INSM1 functions as a transcriptional repressor of the neuroD/β2 gene through the recruitment of cyclin D1 and histone deacetylases. Biochem. J. 2006;397:169–177. doi: 10.1042/BJ20051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie Z., Kasschau K.D., Carrington J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser J.M., O'Shea E.K. Noise in gene expression: Origins, consequences, and control. Science. 2005;309:2010–2013. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Ringner M. What is principal component analysis? Nat. Biotechnol. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronshaugen M., Biemar F., Piel J., Levine M., Lai E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes & Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sood P., Krek A., Zavolan M., Macino G., Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J., Bayraktaroglu L., Clements D., Conlin T., Fashena D., Frazer K., Haendel M., Howe D.G., Mani P., Ramachandran S., et al. The zebrafish information network: The zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–D585. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A., Bushati N., Jan C.H., Kheradpour P., Hodges E., Brennecke J., Bartel D.P., Cohen S.M., Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes & Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai T.H., Calado D.P., Casola S., Ansel K.M., Xiao C., Xue Y., Murphy A., Frendewey D., Valenzuela D., Kutok J.L., et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Thermes V., Grabher C., Ristoratore F., Bourrat F., Choulika A., Wittbrodt J., Joly J.S. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech. Dev. 2002;118:91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- Tsang J., Zhu J., van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crete P., Bartel D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Mallory A.C., Bartel D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Koudijs M.J., van Eeden F.J., Cuppen E., Plasterk R.H. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wilhelm B.T., Marguerat S., Watt S., Schubert F., Wood V., Goodhead I., Penkett C.J., Rogers J., Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Xiao C., Calado D.P., Galler G., Thai T.H., Patterson H.C., Wang J., Rajewsky N., Bender T.P., Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau K.D., Carrington J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]