Abstract
The human CDK8 subcomplex (CDK8, cyclin C, Med12, and Med13) negatively regulates transcription in ways not completely defined; past studies suggested CDK8 kinase activity was required for its repressive function. Using a reconstituted transcription system together with recombinant or endogenous CDK8 subcomplexes, we demonstrate that, in fact, Med12 and Med13 are critical for subcomplex-dependent repression, whereas CDK8 kinase activity is not. A hallmark of activated transcription is efficient reinitiation from promoter-bound scaffold complexes that recruit a series of pol II enzymes to the gene. Notably, the CDK8 submodule strongly represses even reinitiation events, suggesting a means to fine tune transcript levels. Structural and biochemical studies confirm the CDK8 submodule binds the Mediator leg/tail domain via the Med13 subunit, and this submodule–Mediator association precludes pol II recruitment. Collectively, these results reveal the CDK8 subcomplex functions as a simple switch that controls the Mediator–pol II interaction to help regulate transcription initiation and reinitiation events. As Mediator is generally required for expression of protein-coding genes, this may reflect a common mechanism by which activated transcription is shut down in human cells.
Keywords: CDK8 subcomplex, Med12, Med13, Mediator, transcription
The initiation of transcription is controlled in part by a macromolecular protein assembly known as the Preinitiation Complex (PIC) (Hahn 2004). This assembly, approaching 4 MDa in size, consists of the general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), RNA polymerase II (pol II), and Mediator. Upon initiation of transcription, pol II escapes the promoter and leaves behind a so-called scaffold PIC, which retains most PIC components, including Mediator, TFIIH, and TFIID (Yudkovsky et al. 2000). This scaffold complex can then recruit a second pol II enzyme (and so on) to complete multiple rounds of activated transcription. Such an efficient means of reinitiation is likely critical for rapid induction of a gene, a key feature in many diverse physiological responses. However, it is equally important that activated transcription is shut down to prevent inappropriate overexpression of activated genes. Precisely how this occurs remains unclear.
At 1.2 MDa, the Mediator complex represents a major subassembly within the PIC; accordingly, Mediator plays numerous roles in controlling PIC function and is required for expression of virtually all protein-coding genes (Conaway et al. 2005; Malik and Roeder 2005). Although the molecular mechanisms by which Mediator functions within the entire PIC are poorly defined, a number of physical and functional interactions have been identified with various PIC components. Among these, Mediator interactions with pol II and TFIIH appear to play fundamental roles in activating gene expression. Mediator interacts directly with pol II and forms a tight, binary complex with the enzyme (Davis et al. 2002); moreover, Mediator stimulates TFIIH-dependent phosphorylation of the pol II CTD, an event that correlates with pol II promoter escape (Kim et al. 1994; Sun et al. 1998). Ablation of key subunits in either Mediator, pol II, or TFIIH inhibits transcription of all protein-coding genes in yeast (Holstege et al. 1998); thus, controlling the activity of these PIC components lies at the heart of transcriptional regulation.
Notably, a “CDK8 subcomplex” (containing CDK8, cyclin C, Med12, and Med13) can directly and indirectly impact the biochemical activity of Mediator, pol II, and TFIIH. For example, CDK8 can phosphorylate the pol II CTD, which disrupts Mediator–pol II association (which requires a hypophosphorylated CTD) to negatively regulate transcription (Hengartner et al. 1998). Moreover, human CDK8 can phosphorylate cyclin H, which inhibits TFIIH and prevents transcription initiation (Akoulitchev et al. 2000). Thus, there is a link between transcriptional repression and CDK8 kinase activity. Yet Mediator–CDK8 submodule association alone appears sufficient to block Mediator interaction with pol II independent of kinase activity (Naar et al. 2002; Elmlund et al. 2006). Thus, CDK8 kinase function per se may not be required for repression. Indeed, the Med12 (240-kDa) and Med13 (250-kDa) subunits comprise a major portion of the 600-kDa CDK8 submodule, yet little is known regarding the biochemical function of these proteins.
The human Mediator complex exists in two major forms: a core Mediator complex (or simply, “Mediator”) and a CDK8–Mediator complex. The core complex displays strong coactivator function, whereas the CDK8–Mediator complex does not. Each complex is highly similar in subunit composition—25 subunits are common to each—yet only core Mediator contains the Med26 subunit, whereas CDK8–Mediator contains the additional subunits CDK8, cyclin C, Med12, and Med13 (Taatjes et al. 2004). Genetic studies in yeast suggest CDK8, cyclin C, Med12, and Med13 negatively regulate transcription (Carlson 1997); moreover, mutation of any one of these subunits results in identical or nearly identical phenotypes (Holstege et al. 1998; Loncle et al. 2007). For these reasons, it was presumed that CDK8, cyclin C, Med12, and Med13 function as a unit and may exist as a subcomplex. Indeed, a complex containing CDK8, cyclin C, Med12, and Med13 was isolated in yeast expressing a tagged version of cyclin C (Borggrefe et al. 2002).
To establish whether the CDK8 subcomplex may exist as a stable entity in human cells, we recently purified an endogenous subcomplex using conventional methods (Knuesel et al. 2009). As expected, the free CDK8 subcomplex is scarce in human cells: Most appear to be associated with Mediator, which is itself a low-abundance complex. The low abundance of the endogenous subcomplex precludes a rigorous biochemical analysis; in anticipation of this, we also generated the CDK8 subcomplex from recombinant subunits. Recombinant expression enabled purification of subcomplexes devoid of specific subunits or with point mutations that inactivate the kinase. We also established a chromatin-based in vitro transcription system with purified and recombinant human transcription factors. This system recapitulates regulated, activator-dependent transcription and provides a means to clearly define subcomplex-dependent regulatory mechanisms. Using this system, we demonstrate that the CDK8 submodule works as a simple switch to shut down activated transcription. Remarkably, this submodule-dependent switch controls the Mediator–pol II interaction despite binding ∼100 Å from the pol II–Mediator-binding domain. Moreover, the CDK8 submodule can repress transcription reinitiation from PIC scaffold complexes, invoking a “rheostat” mechanism for controlling transcript levels. These results have broad mechanistic implications, as Mediator is a genome-wide regulator of pol II-dependent transcription.
Results
Reconstitution of regulated transcription on chromatin templates
To address mechanistic questions regarding the human CDK8 subcomplex, we sought to recapitulate regulated transcription in vitro by establishing a defined transcription system consisting of highly purified and recombinant transcription factors operating on chromatin templates. The advantage of such a system is that it allows determination of how the CDK8 subcomplex might impact transcription simply by titrating the subcomplex into the reaction; moreover, the activity of various mutants could be tested alongside wild-type subcomplexes to directly compare their biochemical activities. Furthermore, a reconstituted in vitro assay allows various factors to be added at different stages of the transcription reaction (e.g., after PIC assembly) in order to address the potential importance of temporal regulation. This level of control is simply not possible with cell-based assays and enables one to address very specific mechanistic questions. Indeed, work from several laboratories has demonstrated the value of such a reconstituted system in uncovering basic regulatory functions of the transcription machinery (Naar et al. 1999; An et al. 2004; Lewis et al. 2005; Santoso and Kadonaga 2006).
To establish an in vitro transcription system, TFIIA, TFIIB, TFIIE, and TFIIF were recombinantly expressed and purified according to the protocols outlined in Supplemental Figure 1. Because TFIIH, TFIID, pol II, and Mediator are each large, multisubunit complexes, these required purification directly from human cells. Therefore, isolation of these factors required multiple chromatographic steps, including a final antibody/affinity purification step (Supplemental Fig. 1). Each factor was purified to near-homogeneity, as shown in Figure 1. Included in Figure 1 are silver-stained gels of the activators used in this study, Sp1, SREBP-1a, and gal4-p53, which were purified following recombinant expression in Escherichia coli or insect cells. Once generated, purified factors (Mediator, activators, and the general transcription factors) were then tested for activity on naked DNA templates and titrated to determine the optimum concentrations for each factor in the transcription reaction (data not shown).
Figure 1.
Purified factors used for reconstituted transcription on chromatin templates.
Next, we tested whether these purified components would recapitulate regulated transcription on physiological (chromatin) templates. Chromatin assembly was completed with supercoiled DNA templates, highly purified core histones, and a chromatin assembly fraction (S190) according to established protocols (see the Supplemental Material; Supplemental Fig. 2). Following chromatin assembly, activators were allowed to bind the template, followed by the general transcription factors and Mediator, according to the scheme outlined in Supplemental Figure 3A. After transcription, RNA was isolated and quantitated by primer extension. As shown in Supplemental Figure 3B, transcription was dependent on both activators and Mediator in this system: Reactions lacking one or both of these components were unable to generate detectable transcripts despite possessing the full complement of general transcription factors. Thus, this in vitro system mimics a cellular context insofar as transcription occurs on chromatin templates, is regulated by activators and is Mediator-dependent. Also, as expected, exclusion of TFIIH, pol II, or TFIID from the transcription system yielded no detectable transcripts (data not shown). Furthermore, we observed similar activator- and Mediator-dependent regulation at different promoters (SREBP/Sp1 and gal4-p53) (see Supplemental Fig. 3C). Given these biochemical characteristics, this reconstituted transcription system provides a suitable context in which to evaluate the role of the CDK8 subcomplex in regulating transcription initiation.
Purification and kinase activity of recombinant CDK8 subcomplexes
To generate the CDK8 subcomplex from recombinant subunits, CDK8, cyclin C, Med12, and Med13 were coexpressed in insect cells. Affinity purification yielded samples containing CDK8, cyclin C, Med12, and Med13 (Fig. 2A) that comigrated over a glycerol gradient, indicative of a 600-kDa subcomplex (Supplemental Fig. 4). Subunit identities were confirmed by Western blot and mass spectrometry (Supplemental Fig. 5) and quantitation revealed a 1:1:1:1 stoichiometry for the subunits (Knuesel et al. 2009). In order to address the potential importance of the CDK8 kinase, kinase-dead CDK8 subcomplexes were also expressed and purified. These were identical to wild-type subcomplexes except that CDK8 contained a D173A point mutation that inactivates the kinase (Akoulitchev et al. 2000). As expected, the recombinant, wild-type CDK8 subcomplex displayed potent kinase activity, whereas its kinase-dead counterpart did not (Fig. 2B).
Figure 2.
The CDK8 submodule represses transcription independently of its kinase activity. (A) Silver-stained gel showing the purified CDK8 subcomplex. (B) Kinase assays with wild-type (4wt) or kinase-dead (4kd) CDK8 subcomplexes with a known CDK8 substrate, the Rpb1 CTD. (C) Reconstituted transcription on chromatin templates. Each reaction followed the same time line (see Supplemental Fig. 3) and contained identical amounts of template, NTPs, TFIIA, IIB, IID, IIE, IIF, IIH, and pol II; Sp1, Mediator, and submodule were added as shown. (Lane 9) Peptide elution buffer used for CDK8 subcomplex purification had no impact on transcription. Representative data are shown; the graph summarizes data (mean and standard error) from multiple experiments for each experimental condition (n = 7, 7, 5, 12, 4, 4, 4, and 2 for lanes 2–9, respectively).
The CDK8 subcomplex represses activated transcription independently of its kinase activity
In order to test the biochemical activity of the CDK8 subcomplex, either the wild-type or kinase-dead subcomplex was titrated into the reconstituted transcription system. Significantly, the wild-type and kinase-dead subcomplexes behaved similarly in this assay: Each repressed transcription in a dose-dependent fashion (Fig. 2C, cf. lanes 3–5 and 6–8). To further confirm that repression of activated transcription occurred independently of CDK8 kinase activity, both wild-type and kinase-dead subcomplexes were tested at least four times for each titration point; results from these additional assays are summarized in the bar graph below the transcription data, with vertical lines representing the standard error. Experiments were also performed in which the elution buffer (1 mg/mL Glu peptide in 0.15 M HEGN) used for CDK8 subcomplex purification was added to the transcription system. These experiments showed no impact on transcript levels (Fig. 2C, lane 9). In addition, each CDK8 subcomplex was tested in alternate promoter contexts (e.g., SREBP/Sp1 or gal4-p53) with similar effects (Supplemental Fig. 6). Collectively, these results demonstrate that the free CDK8 subcomplex can repress activated transcription on chromatin templates and CDK8 kinase activity is not required for repression.
The ability of the CDK8 subcomplex to repress transcription on naked DNA templates was also tested. Each CDK8 subcomplex (wild-type or kinase-dead) was capable of repressing activated transcription in this context (see the Supplemental Material; Supplemental Fig. 7).
The CDK8 submodule inhibits Mediator coactivator function within the PIC
The chromatin-independent function observed in Supplemental Figure 7 suggested that an interaction(s) within the PIC was essential for repression by the CDK8 subcomplex. Candidate protein complexes within the PIC include TFIIH and pol II—both of which can be phosphorylated by CDK8—and the core Mediator complex, which is known to stably associate with the CDK8 submodule. To test whether any of these factors were targeted for repression by the subcomplex, reactions were supplemented with additional (i.e., beyond the levels used in a typical reaction) TFIIH, core Mediator, or pol II to determine whether any factor might overcome subcomplex-dependent repression. The results, shown in Figure 3A, indicate that additional TFIIH has no measurable effect on transcription (cf. lanes 3 and 4). In contrast, supplementation with additional core Mediator can restore activated transcription in the presence of the CDK8 submodule (Fig. 3A, cf. lanes 3 and 5). This indicates core Mediator is a primary, but perhaps not the only, target for CDK8 subcomplex repression. Indeed, supplementation of transcription reactions with additional pol II may also counteract repression by the CDK8 submodule (Fig. 3A, cf. lanes 3 and 7). However, excess pol II actually represses transcription independently of the CDK8 subcomplex (Fig. 3A, lanes 7–9; data not shown); consequently, we cannot accurately assess whether pol II can similarly overcome subcomplex-dependent repression.
Figure 3.
Additional Mediator overcomes repression by the CDK8 submodule; Med12/13 are key to submodule-dependent repression. (A) Reactions were completed as described for Figure 2C, except TFIIH or Mediator was doubled in concentration for lanes 4 or 5 and pol II was titrated to 1.5-, two- and threefold higher concentration above standard in lanes 7–9. Representative data are shown; the graph summarizes data (mean and standard error) from multiple experiments for each experimental condition (n = 5, 5, 2, 2, and 5 for lanes 2–6, respectively). (B) Experiments were completed as described for Figure 2C. CDK8 submodules added as shown. (2WT) CDK8/cyclin C binary complex; (4WT) CDK8 subcomplex; (K8/CC/12 or K8/CC/13) three-subunit complexes with Med12 or Med13, respectively; (12/13) Med12/Med13 binary complex (n = 6, 4, 5, 6, 2, 1, 2, 1, 6, 5, and 4 for lanes 3–13, respectively).
Med12 and Med13 are critical for repression
Given that the kinase activity of the CDK8 subcomplex was not required for repression of activated transcription, we considered whether other components of the CDK8 subcomplex, particularly Med12 and Med13, might play a role in repression. Although the Med12 and Med13 subunits (∼250 kDa each) comprise a majority of the 600 kDa CDK8 subcomplex, little is known about the biochemical function of these two large subunits. To explore their potential role in transcription regulation, we tested whether CDK8/cyclin C alone would repress activated transcription equally well when compared with the four-subunit CDK8 subcomplex. As shown in Figure 3B, CDK8/cyclin C were unable to repress transcription when added at similar concentrations relative to the four-subunit complex (Fig. 3B, cf. lanes 3–5). The dramatic functional difference between the four-subunit submodule and CDK8/cyclin C alone indicates that Med12/Med13 contribute significantly to the repressive function of the CDK8 subcomplex.
To further test the potential link between Med12/Med13 and transcription repression, partial subcomplexes consisting of (1) CDK8/cyclin C/Med12, (2) CDK8/cyclin C/Med13, and (3) the Med12/Med13 binary complex, were expressed and purified. Each of these subcomplexes was stable and could be isolated intact (see the Supplemental Material; Supplemental Figs. 4, 5). After isolation, each partial subcomplex was titrated into the transcription system. In contrast to CDK8/cyclin C alone, each Med12- or Med13-containing subcomplex displayed a similar ability to repress activated transcription (Fig. 3B, lanes 6–13). This striking result confirms the importance of Med12/Med13 in transcriptional repression and further highlights that the CDK8 kinase is dispensable for subcomplex-dependent repression.
Med13 links the CDK8 submodule to Mediator
Our in vitro transcription data indicated that the CDK8 subcomplex specifically repressed the coactivator function of Mediator. Thus, we hypothesized the recombinant CDK8 subcomplex was directly binding to Mediator to repress activated transcription. To test this, binding assays were performed with core Mediator and the CDK8 subcomplex. The core Mediator complex was immobilized on an anti-Med26 resin, which will specifically bind the core Mediator complex (see Supplemental Fig. 8). The immobilized core Mediator was then incubated with the purified, recombinant CDK8 subcomplex. To minimize false-positive results, a series of high-salt washes (0.5 M KCl, 0.1% NP-40; 75 column volumes) were completed following incubation; furthermore, the resin was not eluted with detergent or loading buffer. Instead, the anti-Med26 resin was eluted with peptide (1.0 mg/mL Med26 antigen in 0.15 M HEGN). Notably, the CDK8 subcomplex was retained and eluted with core Mediator (Fig. 4A, lane 5), indicating direct binding of the submodule to core Mediator. The CDK8 subcomplex did not bind a control antibody resin (Fig. 4A, lane 4).
Figure 4.
The CDK8 submodule binds directly to core Mediator via Med13. (A) Binding assays with anti-Med26 immobilized core Mediator. Western blots against indicated subunits were performed on anti-Med26 peptide eluates, except for positive control (shown in lane 3). (Lane 1) Anti-Med26 resin only. (Lane 2) Immobilized core Mediator only. (Lane 3) Positive control (CDK8 subcomplex). (Lane 4) Anti-Med26 resin + CDK8 subcomplex. (Lane 5) Immobilized core Mediator + CDK8 subcomplex. (B) 2D classes generated from EM analysis of core Mediator–CDK8 submodule assembly experiment (see the text). Panels 1 and 2 reflect the presence of free CDK8 submodules and core Mediator, respectively. Panel 3 shows a 2D class resembling CDK8-Mediator, suggesting stable interaction between the recombinant CDK8 submodule and core Mediator. Bar, 100 Å. (C) Mediator-binding assays. The complete four-subunit CDK8 subcomplex (4WT) or the partial subcomplexes (as shown) were immobilized and incubated with a partially purified fraction containing Mediator. Peptide-eluted material was analyzed by Western blot against the subunits indicated at the right.
In a different set of experiments, we mixed eluted core Mediator with the purified CDK8 submodule and examined this mixture by electron microscopy (EM). This was done to test whether intact CDK8–Mediator samples could be visualized. Because core Mediator lacks the large CDK8 subcomplex, core Mediator and CDK8–Mediator can be readily distinguished upon 2D classification from single-particle images (Taatjes et al. 2002). As expected, the sample from the core Mediator/CDK8 submodule coincubation experiment contained complexes of a size and shape consistent with the free CDK8 submodule and core Mediator (Fig. 4B, panels 1,2). In addition, however, a subset of complexes clearly resembled CDK8–Mediator (Fig. 4B, panel 3), corroborating the binding data in Figure 4A. (Representative single-particle images from this data set are shown in Supplemental Fig. 9.)
Finally, binding experiments were performed with immobilized core Mediator and the partial CDK8 subcomplexes (CDK8/cyclin C/Med12, CDK8/cyclin C/Med13, or CDK8/cyclin C). These data were inconclusive and are described in the Supplemental Material. Consequently, reciprocal experiments were completed with the partial CDK8 subcomplexes in an effort to obtain more interpretable results. The partial subcomplexes were immobilized on an antibody resin and incubated with a partially purified extract enriched in core Mediator. Following a series of high-salt washes, bound material was eluted with peptide and probed for Mediator subunits by Western blot. This assay identified Med13 as critical for core Mediator interaction: Only the partial subcomplex containing Med13 was able to retain core Mediator from the partially purified extract (Fig. 4C). Collectively, the results in Figure 4 demonstrate the recombinant CDK8 submodule can bind directly to core Mediator and this interaction is mediated by Med13.
The CDK8 subcomplex binds the Mediator leg/tail domain
Past EM studies have established clear differences between the structure of core Mediator versus CDK8-Mediator (Taatjes et al. 2002). In particular, the CDK8-Mediator complex contains an additional “foot” domain that is absent from core Mediator and is ∼500 kDa in size (derived from standard protein mass/volume estimates of 0.81 Å3/Dalton) (Taatjes et al. 2002). Based on these data, we predicted the CDK8 submodule itself would adopt a structure of size and shape similar to the foot domain in CDK8-Mediator. To test this, we completed a three-dimensional (3D) reconstruction of the purified, recombinant CDK8 submodule using EM and single-particle reconstruction techniques. Data were obtained in both tilted and untilted orientations to allow an unbiased reconstruction using random conical tilt methodology (Radermacher et al. 1987). As shown in Figure 5A, the CDK8 subcomplex does indeed adopt a size and shape consistent with the foot domain of CDK8–Mediator. Assembly of the submodule onto core Mediator is depicted in Fig. 5B; the orientation of both the CDK8 submodule (red) and core Mediator (blue) is based on docking each structure within the CDK8–Mediator EM map (Fig. 5C). Note that the length of the submodule hook domain allows an extensive interface with the leg/tail of core Mediator and the extreme end of the leg/tail domain appears to extend directly into the “socket” of the CDK8 submodule (Fig. 5B, left panel). Two views of CDK8–Mediator and the docked submodule are shown in Figure 5C, which further highlight the structural similarity between the CDK8–Mediator foot domain and the CDK8 submodule itself. For reference, the 3D structure of core Mediator is also shown in Figure 5C to identify the shared head/body and leg domains. Major differences in the head/body region of core Mediator versus CDK8–Mediator are evident upon comparing the structures (see also an overlay of the structures in Fig. 5D). In particular, the large cavity present in core Mediator is occluded in CDK8–Mediator; this region corresponds to the pol II-binding site within core Mediator (Davis et al. 2002). Additional views of the CDK8 submodule docked within CDK8–Mediator are provided in Supplemental Figure 10 and Supplemental Movie 1.
Figure 5.
The human CDK8 submodule interacts with the Mediator leg/tail domain. (A) 3D structure of the recombinant CDK8 submodule, rendered to 600 kDa. (B) Modeled interaction of CDK8 subcomplex with core Mediator. Orientations of core Mediator (semitransparent blue) and the CDK8 subcomplex (red) are based on docking each structure within the entire CDK8–Mediator EM map. The overlaid structures reveal how the hook domain might dovetail with the Mediator leg domain upon interaction. (C) Overlay of docked CDK8 submodule (semitransparent red) onto the CDK8–Mediator structure (white). The submodule overlays are oriented as in A and B. The core Mediator structure is shown for reference and specific domains are indicated (* leg is called the tail domain in the yeast Mediator). Note the CDK8–Mediator and core structures have been published and are bound to VP16 (Taatjes et al. 2002). (D) Core Mediator (blue) docked within the CDK8–Mediator (white mesh) EM map. (E) Independent 3D reconstruction of the three-subunit Med12/CDK8/cyclin C complex (green), rendered to 350 kDa. Docked structures within the entire CDK8 subcomplex (semitransparent red) are also shown. Docking for B–E was completed with Chimera (Pettersen et al. 2004).
Med13 defines the CDK8 submodule hook domain
Given the identification of Med13 as linking the CDK8 submodule to core Mediator (Fig. 4C), and the identification of a prominent hook domain within the CDK8 submodule that interfaces extensively with the leg/tail domain of core Mediator (Fig. 5B), we hypothesized that Med13 might comprise the hook domain within the submodule. To test this, a random conical tilt, 3D reconstruction of the three-subunit Med12/CDK8/cyclin C complex (i.e., Med13 is missing) was undertaken. Although the value of these data is tempered by the fact that structural shifts likely compensate for the absence of Med13 (which accounts for 42% of the CDK8 submodule mass), docking the partial subcomplex density (green) within the complete CDK8 submodule EM map (red overlay) suggests that Med13 contributes substantially to the hook domain (Fig. 5E). Med13 also appears to be important to form the socket within the CDK8 submodule, as this is absent in the Med12/CDK8/cyclin C structure. Additional views of these subcomplexes are shown in Supplemental Figure 11.
The CDK8 subcomplex inhibits reinitiation of transcription
The process of transcription initiation can be divided into a number of distinct stages, including activator binding, PIC assembly, and initiation itself. One key mechanistic question regarding the CDK8 subcomplex is the point at which it can effectively shut down activated transcription. For example, can repression occur after PIC assembly, or must the CDK8 subcomplex function at a point prior to or concomitant with PIC assembly? Addressing this question is important in order to establish whether the CDK8 subcomplex can function as a switch to shut down ongoing, activated transcription (i.e., a “rheostat” mechanism for regulating levels of transcript) or whether it simply works to prevent any initiation from occurring (i.e., an all-or-nothing switch). Although the results summarized in Figure 2C support submodule repression prior to PIC assembly, it is not established whether early incorporation into the PIC is a requirement for repression by the CDK8 subcomplex, or whether the CDK8 subcomplex can also repress transcription after a stable PIC is formed.
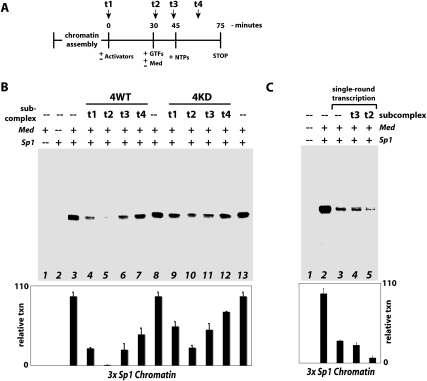
To determine whether the repressive function of the CDK8 subcomplex is sensitive to the different stages of transcription initiation, we performed a series of experiments in which CDK8 subcomplexes were introduced at different points during the transcription reaction. As outlined in Figure 6A, CDK8 subcomplexes (wild-type or kinase-dead) were added at different times: t1, prior to PIC assembly (i.e., with the activators); t2, together with core Mediator and the GTFs; t3, after PIC assembly (i.e., just prior to the addition of NTPs); or t4, after initiation of activated transcription (i.e., 15 min after addition of NTPs). As shown in Figure 6B, each CDK8 subcomplex—wild-type or kinase-dead—can repress activated transcription at each of these stages (lanes 4–7,9–12), indicating the CDK8 subcomplex can repress activated transcription during and even after PIC assembly. That repression was observed even after addition of NTPs (Fig. 6B, lanes 7,12) suggests that the CDK8 subcomplex works to inhibit pol II reinitiation events from PIC scaffold complexes. PIC scaffolds (containing Mediator, activator, TFIIA, TFIID, TFIIE, and TFIIH) remain following pol II promoter escape and work to recruit additional pol II enzymes for multiple rounds of transcription. The composition of the PIC scaffold was demonstrated most rigorously by the Hahn laboratory (Yudkovsky et al. 2000) using yeast extracts, and similar results have been observed in metazoan systems (Zawel et al. 1995; Sandaltzopoulos and Becker 1998; Yean and Gralla 1999).
Figure 6.
The CDK8 submodule can repress transcription reinitiation. (A) Time line for transcription. The different points (t1, t2, t3, t4) represent times at which the CDK8 subcomplex was added to the reaction. (B) Reconstituted transcription reactions were completed as described for Figure 2C. CDK8 subcomplexes (wild-type or kinase-dead) were added at various time points as shown. Representative data are shown; the graph summarizes data (mean and standard error) from multiple experiments for each experimental condition (n = 5, 2, 2, 2, 2, 5, 2, 2, 2, 2, and 5 for lanes 3–13). Note these data, like those in Figures 2 and 3, represent multiround transcription. (C) Single-round transcription. Experiments were completed as in B except for lanes 3–5, where Sarkosyl was added 90 sec after NTP addition to block transcription reinitiation. The CDK8 submodule was added 3 min prior to NTPs in lane 4, and together with GTFs/Mediator in lane 5 (n = 2, 2, 2, and 3 for lanes 2–5).
Because we allow transcription to proceed 30 min, a significant fraction of the total transcripts observed likely represent reinitiation from promoter-bound scaffold complexes. To further test this, single-round transcription experiments were performed in which Sarkosyl was added just after the addition of NTPs. In these experiments, the added Sarkosyl prevents reinitiation of transcription from scaffold complexes, but will not affect fully assembled PICs (Hawley and Roeder 1985, 1987; Kraus and Kadonaga 1998; Pavri et al. 2006). As shown in Figure 6C, single-round transcription accounts for ∼30% of the total transcripts in the 30-min reaction (cf. lanes 2 and 3). Thus, if fully assembled PICs are refractory to repression by the CDK8 submodule—whereas reinitiation is blocked—addition of the submodule after PIC assembly would approximate single-round transcript levels (i.e., 30% of maximum). As shown in Figure 6, this is precisely what is observed. Addition of the CDK8 subcomplex at “t3” results in transcript levels ∼30% of normal, similar to single-round levels (Fig. 6, cf. B [lanes 6,11] and C [lane 3]). Further supporting this, the CDK8 subcomplex had no significant effect on single-round transcription when added after initial PIC formation (Fig. 6C, cf. lanes 3 and 4). In fact, only when added together with pol II and the GTFs (i.e., prior to full PIC formation) does the CDK8 submodule impact single-round transcription (Fig. 6C, cf. lanes 3 and 5). Taken together, the data in Figure 6 suggest the CDK8 subcomplex can prevent reinitiation (likely from PIC scaffold assemblies), whereas fully assembled PICs are not affected.
Mutually exclusive binding of pol II or CDK8 subcomplex to Mediator
The negligible effect of the CDK8 submodule on stable, pol II-containing PICs is consistent with past biochemical studies that suggest Mediator binding to pol II or the CDK8 submodule is mutually exclusive (Naar et al. 2002; Elmlund et al. 2006). To further substantiate this, we tested whether pol II would be excluded from PICs assembled on chromatin in a CDK8 subcomplex-dependent manner. Therefore, chromatin sedimentation assays were completed in which PICs were assembled on chromatin in the presence or absence of the CDK8 submodule. Following PIC assembly, the chromatin was sedimented over a glycerol gradient; using this protocol, chromatin will migrate deeper within the gradient, whereas free (not chromatin-associated) PIC components will sediment in earlier fractions (see Supplemental Fig. 12). In the absence of the CDK8 submodule, pol II comigrates with chromatin, as expected. However, when PIC assembly occurs in the presence of the CDK8 submodule, pol II is absent from the chromatin-containing fractions and instead migrates earlier in the gradient as the free enzyme (Supplemental Fig. 12). Additional evidence for mutually exclusive binding derives from MudPIT analysis of Mediator samples purified with either an anti-CDK8 or an anti-Med1 antibody resin. Analysis of Med1-purified Mediator identifies 57 unique pol II peptides representing 10 pol II subunits, whereas CDK8-purified Mediator samples contain no detectable copurifying pol II (C. Ebmeier and D. Taatjes, unpubl.). Collectively, these results indicate that pol II association with Mediator is blocked by the CDK8 submodule, providing further validation that core Mediator interacts with pol II or the CDK8 subcomplex in a mutually exclusive fashion.
The endogenous CDK8 subcomplex mimics repression by the recombinant submodule
Despite considerable evidence suggesting the existence of free CDK8 subcomplexes in human cells, we sought to confirm this by purifying the submodule directly from HeLa nuclear extracts. A combination of conventional and affinity chromatography steps enabled isolation of an endogenous CDK8 subcomplex. As expected, the purified endogenous CDK8 subcomplex contains Med12, Med13, CDK8, and cyclin C, and is devoid of core Mediator subunits (Knuesel et al. 2009). The outcome of this purification suggests a stable population of free CDK8 subcomplexes exists in human cells, ostensibly to help regulate Mediator coactivator function. To test this, we evaluated the biochemical activity of the endogenous subcomplex in our in vitro transcription system.
As shown in Supplemental Figure 13B, the endogenous subcomplex—like the recombinant subcomplex—repressed activated transcription in a dose-dependent manner (cf. lanes 3–6). The endogenous subcomplex was also examined in order-of-addition experiments, as outlined in Supplemental Figure 13A. Again, the endogenous subcomplex was able to repress transcription when added prior to or after PIC assembly (Supplemental Fig. 13B, lanes 8–11) in a manner similar to the recombinant subcomplex. Moreover, as with the recombinant subcomplex, core Mediator—but not TFIIH—was able to restore activated transcription when added in excess (Supplemental Fig. 13B, lanes 13,14). Together, the results shown in Supplemental Figure 13 establish that the endogenous CDK8 subcomplex can repress activated transcription in a manner indistinguishable from the recombinant subcomplex.
Discussion
The isolation of an endogenous CDK8 subcomplex demonstrates its stability as an independent entity in human cells and suggests the free submodule may function autonomously to regulate gene expression. Accordingly, the free submodule can bind the core Mediator complex, and this association alone is sufficient to repress activated transcription. That repression occurs independently of CDK8 kinase activity suggests the 600-kDa CDK8 submodule may impact Mediator structure and PIC assembly. In support of this, we observe that the CDK8 submodule prevents pol II incorporation into the PIC on chromatin templates, in agreement with past studies that suggest CDK8-Mediator (in contrast to core Mediator) is unable to interact with pol II. Such mutually exclusive interactions, and their functionally divergent outcomes, provide a straightforward means to regulate Mediator activity and therefore transcription initiation itself.
The regulatory significance of the CDK8 submodule
Integrating our current findings with past studies, we hypothesize that the CDK8 submodule serves as a general checkpoint to help regulate activated transcription. Although this needs further validation with genome-wide localization studies in human cells (which to date have not been completed), this hypothesis is entirely consistent with a pair of genome-wide localization studies in yeast that indicate Mediator and CDK8 are corecruited upstream of protein-coding genes, indicative of Mediator's essential role in regulating pol II-dependent transcription (Andrau et al. 2006; Zhu et al. 2006). Each study also noted that CDK8 occupancy appeared greatest at highly expressed genes and Holstege and colleagues (Andrau et al. 2006) concluded that although overlapping, the occupancy of CDK8 was reduced relative to core Mediator, suggesting a transient association. All of these observations are consistent with the results outlined here. First, co-occupancy of CDK8 and core Mediator likely reflects a direct interaction between core Mediator and the CDK8 submodule. Indeed, CDK8–Mediator complexes can be isolated from both human and yeast cells, and direct binding of the free CDK8 subcomplex with core Mediator is demonstrated here. Second, enhanced detection of CDK8 at highly expressed genes in yeast is consistent with submodule-dependent repression following multiple rounds of activated transcription. Because the CDK8 submodule can repress reinitiation, our results predict a higher submodule population at transcriptionally active genes: Their high-level expression ultimately requires modulation by the CDK8 subcomplex. Finally, the evidence for transient CDK8–core Mediator association at yeast promoters suggests a reversible interaction between the submodule and core Mediator (Andrau et al. 2006). Past ChIP studies at select genes in human cells suggest CDK8 dissociates upon gene activation (Mo et al. 2004; Pavri et al. 2005), whereas the transcription data and binding assays shown here reveal the CDK8 submodule can also associate with core Mediator. These complementary findings highlight the reversible (both on and off) nature of the CDK8 subcomplex–Mediator interaction.
Because the CDK8 subcomplex is stable and can exist independently of Mediator, it is clear that the subcomplex itself may function autonomously in certain promoter contexts and perhaps even co-occupy promoter regions without directly inhibiting Mediator function. Signaling events may then trigger auxiliary factors to appropriately regulate the subcomplex–core Mediator association. So how might the CDK8 subcomplex–Mediator interaction be regulated? This remains unclear and will require further research. Although PARP-1 was shown to trigger CDK8 dissociation, it appears to function at only a subset of genes (Pavri et al. 2005). A more general means to regulate the submodule–Mediator interaction might involve molecular chaperones and/or post-translational modifications (Knuesel et al. 2009); alternately, the CDK8 submodule itself might be sequestered adjacent to sites of active transcription within the nucleus.
Since the CDK8 submodule can effectively block reinitiation, presumably from the PIC scaffold, this provides additional basis for a general role in regulating transcription. Indeed, the PIC scaffold likely represents a universal reinitiation intermediate (Zanton and Pugh 2006). By targeting the scaffold assembly, the CDK8 subcomplex can act subsequent to activation of transcription. Therefore, a measured output can be achieved: Submodule association after many rounds of activated transcription versus only a few rounds would provide a simple means to modulate transcript levels. Of course, the CDK8 submodule alone is not sufficient to shut down and maintain a transcriptionally silent state. Given the fundamental importance of stopping gene expression at the appropriate time, alternate mechanisms are clearly in place to supplement and perhaps substitute for repression via the CDK8 submodule (Rosenfeld et al. 2006). For example, molecular chaperones and the proteasome help control transcript levels of at least a subset of genes (Freeman and Yamamoto 2002; Ostendorff et al. 2002; Reid et al. 2003; Stavreva et al. 2004), whereas chromatin-associated factors play vital roles in shutting down and maintaining a repressed state genome-wide (Berger 2007; Li et al. 2007; Smallwood et al. 2007). Because the CDK8 submodule can effectively block activated transcription, it is likely less important at stably repressed genes.
Regulation by the CDK8 kinase
Although the results described here clearly demonstrate CDK8 kinase activity per se is not required for transcriptional repression, the kinase function undoubtedly plays key roles in regulating transcription. Recent reports have also shown a requirement for CDK8 in oncogenesis (Firestein et al. 2008; Morris et al. 2008). Repression mechanisms dependent on the kinase (e.g., phosphorylation of TFIIH) likely augment the activity of the submodule itself. For example, CDK8-dependent phosphorylation of TFIIH may play other roles in regulating transcription (e.g., modulating protein–protein interactions within the PIC or triggering TFIIH disassembly), apart from inhibiting CDK7 activity. Notably, CDK8 has been shown to phosphorylate several transcription factors in yeast, and this phosphorylation triggers nuclear export and transcription factor degradation by the proteasome (Chi et al. 2001; Nelson et al. 2003). Thus, CDK8 kinase activity may initiate disassembly and degradation of the PIC. Supporting this, CDK8-dependent phosphorylation of the Notch ICD promotes turnover of Notch and associated, promoter-bound factors in human cells (Fryer et al. 2004).
Despite its link to transcriptional repression, numerous studies have revealed CDK8 itself may also play important roles in activating gene expression (Donner et al. 2007). Interestingly, the potential dual roles of CDK8 may be dependent on distinct targets of the kinase (Akoulitchev et al. 2000), and coactivator function may derive in part from CDK8-dependent phosphorylation of histone H3 (Meyer et al. 2008). CDK8 activity may also promote cofactor exchange at the promoter, a characteristic of at least a subset of active genes (Rosenfeld et al. 2006). Because few substrates have been identified for human CDK8, additional regulatory functions may be borne out by future studies.
A structural and functional role for Med12 and Med13
Because CDK8 submodule-core Mediator association occurs with the recombinant, four-subunit assembly, no additional polypeptides are required for stable interaction. In fact, we identify Med13 as critical for linking the submodule to the core Mediator complex, thus revealing a key structural role for this protein. Accordingly, a complex consisting of Med13/CDK8/cyclin C effectively represses activated transcription; however, we also observe that Med12 can strongly affect repression. Human Med12 was shown recently to functionally interact with the methyltransferase G9a and the REST corepressor to negatively regulate neuronal gene expression (Ding et al. 2008). The ability of Med12 to repress transcription in our reconstituted system suggests the 240-kDa Med12 subunit may directly impact other components within the PIC. Future work will assess whether Med12 may play alternate roles in submodule-dependent repression. By contrast, Med12 and Med13 are evidently dispensable for repression in yeast. The expanded role for Med12/Med13 in humans defines essential functions for these large, poorly conserved proteins.
Using biochemical and biophysical approaches, we established that the CDK8 submodule represses transcription through a physical interaction with the leg/tail domain of Mediator. Yet structural studies with both human (C. Bernecky and D. Taatjes, unpubl.) and yeast Mediator (Davis et al. 2002) reveal the pol II-binding site within Mediator is >100 Å from the foot domain formed by the CDK8 submodule. How might submodule binding impact Mediator–pol II association? Based on previous structural studies with human Mediator, it is evident that the putative pol II-binding site within Mediator undergoes a conformational shift upon interaction with the CDK8 submodule (Taatjes et al. 2002). Therefore, it is likely that a submodule-dependent structural shift within the Mediator–pol II-binding domain negatively impacts pol II association. Moreover, whereas the core Mediator complex interacts strongly with the pol II CTD, this interaction is abolished upon association with the CDK8 submodule (Naar et al. 2002). Thus, a simple subcomplex-dependent allosteric switch appears to coordinate pol II-Mediator association. Further experiments will be necessary, however, to precisely define how the CDK8 submodule displaces pol II.
The localization of the CDK8 submodule to the foot domain in the human CDK8–Mediator complex is in stark contrast to the location of the CDK8 submodule in the yeast Schizosaccharomyces pombe (Elmlund et al. 2006). However, Mediator subunits are poorly conserved between yeast and humans, and S. pombe lacks subunit orthologs believed to comprise the Mediator leg/tail domain (Spahr et al. 2001; Boube et al. 2002). Consequently, the distinct architecture observed with the human CDK8-Mediator complex was not unexpected.
A model for transcriptional regulation by the CDK8 subcomplex
Collectively, our results lead us to the model proposed in Figure 7. Following PIC formation and initiation of transcription, pol II escapes the promoter to leave behind the scaffold complex (Yudkovsky et al. 2000). Based on our in vitro transcription and binding assays, the CDK8 subcomplex is capable of binding Mediator within the scaffold complex, whereas a fully assembled PIC appears impervious to submodule binding. Consequently, we hypothesize that Mediator undergoes a structural shift during the fully assembled PIC → scaffold complex transition that exposes a surface capable of interacting with the CDK8 subcomplex (Fig. 7, red oval in Mediator leg/tail domain). Yet Mediator remains competent to bind another pol II enzyme in this state, and doing so enables reinitiation from the scaffold complex (Fig. 7, bottom arrow). In the absence of CDK8 submodule binding, multiple rounds of activated transcription might occur with pol II cycling from the same PIC/scaffold assembly. However, if the CDK8 submodule is present and competent for binding Mediator, reinitiation can be blocked upon its incorporation within the scaffold complex (Fig. 7, top, dashed line). This represents a very simple means to regulate transcript levels of an activated gene; moreover, since CDK8 association with Mediator is reversible (at least at some genes), the promoter may remain in a state poised for rapid reactivation. Such built-in versatility is a hallmark of mammalian gene expression. Although the model shown in Figure 7 is almost certainly an oversimplification of events, we anticipate that further analysis will provide many more details regarding CDK8 submodule and Mediator function within the PIC.
Figure 7.
A model for transcriptional regulation by the CDK8 subcomplex. See main text for details. Scaffold PIC components (e.g., TFIID and IIH) are shown transparent with the CDK8–Mediator because their association may be destabilized in this context. The precise architecture of the PIC is not completely known, and the organization shown here is for illustrative purposes only. Individual PIC components are shown at the same relative scale.
Materials and methods
Purification of human TFIIA, IIB, IIE, and IIF
Following expression in E. coli, the protocols outlined in Supplemental Figure 1 were used for purification. Additional details can be found in the Supplemental Material.
Isolation of human pol II, TFIID, IIH, and core Mediator
Each of these factors was purified from HeLa nuclear extracts according to the protocol outlined in Supplemental Figure 1. Additional details are provided in the Supplemental Material.
In vitro transcription
Chromatin assembly and in vitro transcription was completed as described (Meyer et al. 2008). See also the Supplemental Material. For gal4-p53 transcription, the activator was added to 2 nM final concentration. In the Mediator/IIH/pol II rescue experiments, additional amounts of these factors were added 1 min after the standard amounts of GTFs/Mediator. Unless otherwise stated, experiments with the CDK8 submodule added the subcomplex together with GTFs/Mediator. For CDK8 submodule titration experiments, the subcomplex was added to ∼4, 8, and 15 nM final concentration, whereas core Mediator concentration was ∼10 nM. In the order-of-addition reactions, the CDK8 submodule was added to 15 nM at each time point. For single-round experiments, Sarkosyl was added 90 sec following the addition of NTPs, to 0.2% final concentration.
EM and image processing
EM samples were prepared and analyzed as described (Meyer et al. 2008). See the Supplemental Material for additional details.
Acknowledgments
We thank R. Tjian and C. Inouye for assistance with GTF purification, including initially providing purified pol II for in vitro transcription; and also J. Goodrich for help with TFIID purification and other useful advice. We thank E. Nogales and P. Grob for assistance with Chimera. We thank R. Roeder for the Med12 cDNA and J. Kadonaga for assistance with the single-round transcription experiments. This work was supported by the NCI (P01 CA112181) and the Ellison Medical Foundation (DJT); M.T.K., K.D.M., and C.B. were supported in part by NIH grants T32 GM07135 and T32 GM065103.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1767009.
Supplemental material is available at http://www.genesdev.org.
References
- Akoulitchev S., Chuikov S., Reinberg D. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- An W., Kim J., Roeder R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Andrau J., van de Pasch L., Lijnzaad P., Bijma T., Koerkamp M.G., van de Peppel J., Werner M., Holstege F.C.P. Genome-wide location of the coactivator Mediator: Binding without activation and transient cdk8 interaction on DNA. Mol. Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Borggrefe T., Davis R., Erdjument-Bromage H., Tempst P., Kornberg R.D. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 2002;277:44202–44207. doi: 10.1074/jbc.M207195200. [DOI] [PubMed] [Google Scholar]
- Boube M., Joulia L., Cribbs D.L., Bourbon H. Evidence for a Mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- Carlson M. Genetics of transcriptional regulation in yeast: Connections with the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- Chi Y., Huddleston M.J., Zhang X., Young R.A., Annan R.S., Carr S.A., Deshaies R.A. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes & Dev. 2001;15:1078–1092. doi: 10.1101/gad.867501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R.C., Sato S., Tomomori-Sato C., Yao T., Conaway J.W. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Davis J.A., Takagi Y., Kornberg R.D., Asturias F.A. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- Ding N., Zhou H., Esteve P., Chin H.G., Kim S., Xu X., Joseph S.M., Friez M.J., Schwartz C.E., Pradhan S., et al. Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol. Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner A.J., Szostek S., Hoover J.M., Espinosa J.M. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H., Baraznenok V., Lindahl M., Samuelsen C.O., Koeck P.J.B., Holmberg S., Hebert H., Gustafsson C.M. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc. Natl. Acad. Sci. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R., Bass A.J., Kim S.Y., Dunn I.F., Silver S.J., Guney I., Freed E., Ligon A.H., Vena N., Ogino S., et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman B.C., Yamamoto K.R. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- Fryer C.J., White J.B., Jones K.A. Mastermind recruits CycC:Cdk8 to phosphorylate the notch ICD and coordinate activation with turnover. Mol. Cell. 2004;16:509–520. doi: 10.1016/j.molcel.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Hahn S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 2004;11:394–403. doi: 10.1038/nsmb763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D.K., Roeder R.G. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J. Biol. Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- Hawley D.K., Roeder R.G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- Hengartner C.J., Myer V.E., Liao S., Wilson C.J., Koh S.S., Young R.A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- Holstege F.C., Jennings E.G., Wyrick J.J., Lee T.I., Hengartner C.J., Green M.R., Golub T.R., Lander E.S., Young R.A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- Kim Y., Bjorklund S., Li Y., Sayre M.H., Kornberg R.D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Knuesel M.T., Meyer K.D., Donner A.J., Espinosa J.M., Taatjes D.J. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of Mediator. Mol. Cell. Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W.L., Kadonaga J.T. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes & Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.A., Sims R.J., Lane W.S., Reinberg D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell. 2005;18:471–481. doi: 10.1016/j.molcel.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Loncle N., Boube M., Joulia L., Boschiero C., Werner M., Cribbs D.L., Bourbon H. Distinct roles for Mediator cdk8 module subunits in Drosophila development. EMBO J. 2007;26:1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S., Roeder R.G. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Meyer K.D., Donner A.J., Knuesel M., York A.G., Espinosa J.M., Taatjes D.J. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 2008;27:1447–1457. doi: 10.1038/emboj.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X., Kowenz-Leutz E., Xu H., Leutz A. Ras induces mediator complex exchange on C/EBPb. Mol. Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- Morris E.J., Ji J., Yang F., Di Stefano L., Herr A., Moon N., Kwon E., Haigis K.M., Naar A.M., Dyson N.J. E2F1 represses b-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar A.M., Beaurang P.A., Zhou S., Abraham S., Solomon W., Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- Naar A.M., Taatjes D.J., Zhai W., Nogales E., Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes & Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C., Goto S., Lund K., Hung W., Sadowski I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 2003;421:187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- Ostendorff H.P., Peirano R.I., Peters M.A., Schluter A., Bossenz M., Scheffner M., Bach I. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- Pavri R., Lewis B., Kim T.K., Dilworth F.J., Erdjument-Bromage H., Tempst P., de Murcia G., Evans R., Chambon P., Reinberg D. PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol. Cell. 2005;18:83–96. doi: 10.1016/j.molcel.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Radermacher M., Wagenknecht T., Verschoor A., Frank J. Three-dimensional reconstruction from a single-exposure random conical tilt series applied to the 50s ribosomal subunit of Escherichia coli. J. Microsc. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Reid G., Hubner M.R., Metevier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M.G., Lunyak V.V., Glass C.K. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Sandaltzopoulos R., Becker P.B. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol. Cell. Biol. 1998;18:361–367. doi: 10.1128/mcb.18.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso B., Kadonaga J.T. Reconstitution of chromatin transcription with purified components reveals a chromatin-specific repressive activity of p300. Nat. Struct. Mol. Biol. 2006;13:131–139. doi: 10.1038/nsmb1048. [DOI] [PubMed] [Google Scholar]
- Smallwood A., Esteve P., Pradhan S., Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes & Dev. 2007;21:1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahr H., Samuelsen C.O., Baraznenok V., Ernest I., Huylebroeck D., Remacle J.E., Samuelsson T., Kieselbach T., Holmberg S., Gustafsson C.M. Analysis of Schizosaccharomyces pombe mediator reveals a set of essential subunits conserved between yeast and metazoan cells. Proc. Natl. Acad. Sci. 2001;98:11985–11990. doi: 10.1073/pnas.211253898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva D.A., Muller W.G., Hager G.L., Smith C.L., McNally J.G. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. Mol. Cell. Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhang Y., Cho H., Rickert P., Lees E., Lane W., Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- Taatjes D.J., Naar A.M., Andel F., Nogales E., Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- Taatjes D.J., Marr M.T., Tjian R. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- Yean D., Gralla J.D. Transcription reinitiation rate: A potential role for TATA box stabilization of the TFIID:TFIIA:DNA complex. Nucleic Acids Res. 1999;27:831–838. doi: 10.1093/nar/27.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N., Ranish J.A., Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- Zanton S.J., Pugh B.F. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes & Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L., Kumar K.P., Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes & Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- Zhu X., Wiren M., Sinha I., Rasmussen N.N., Linder T., Holmberg S., Ekwall K., Gustafsson C.M. Genome-wide occupancy profile of Mediator and the srb8-11 submodule reveals interactions with coding regions. Mol. Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]