Abstract
Th17 cells are involved in the pathogenesis of many autoimmune diseases, but it is not clear whether they play a pathogenic role in type 1 diabetes. Here we investigated whether mouse Th17 cells with specificity for an islet antigen can induce diabetes upon transfer into NOD/SCID recipient mice. Induction of diabetes in NOD/SCID mice via adoptive transfer of Th1 cells from BDC2.5 transgenic mice was prevented by treatment of the recipient mice with a neutralizing IFN-γ–specific antibody. This result suggested a major role of Th1 cells in the induction of disease in this model of type 1 diabetes. Nevertheless, transfer of highly purified Th17 cells from BDC2.5 transgenic mice caused diabetes in NOD/SCID recipients with similar rates of onset as in transfer of Th1 cells. However, treatment with neutralizing IL-17–specific antibodies did not prevent disease. Instead, the transferred Th17 cells, completely devoid of IFN-γ at the time of transfer, rapidly converted to secrete IFN-γ in the NOD/SCID recipients. Purified Th17 cells also upregulated Tbet and secreted IFN-γ upon exposure to IL-12 in vitro and in vivo in NOD/SCID recipients. These results indicate substantial plasticity of Th17 commitment toward a Th1-like profile.
Introduction
Th17 cells have achieved prominence for their causative role in many autoimmune diseases such as EAE, rheumatoid arthritis, and myocarditis. Nevertheless, Th17 are clearly not responsible for all autoimmune syndromes. Mice with T cell–specific unresponsiveness to TGF-β are unable to generate Th17 cells (1), yet they die of autoimmunity at the age of 2 weeks, following massive infiltration of lung, liver, stomach, pancreatic islets, and thyroid glands (2).
There are data suggesting that Th1 cells play a major role in diabetes, driving the development of disease via IFN-γ (3). This includes the observations that blockade of IFN-γ (4) or absence of STAT4 (5, 6) prevent disease, whereas IL-12 promotes accelerated diabetes (7). However, the exact role of IFN-γ in disease pathogenesis has been difficult to resolve, as discrepant observations have been found using NOD mice expressing targeted mutations in either IFN-γ or its receptor (3, 8, 9). Differences between observations using antibody blockade versus targeted mutation could be ascribed to redundancy, while the differing findings in receptor targeted mutants have been attributed to linkage disequilibrium of the receptor α chain with an insulin dependent diabetes resistance allele (9, 10). Diabetes develops slowly in female NOD mice with an onset at around 12 weeks of age, but the kinetics are accelerated in an adoptive transfer system, in which CD4 T cells from BDC2.5 mice, expressing transgenic TCR with specificity for an islet antigen (11), are injected into NOD/SCID recipients (12). It is known that TGF-β (13, 14) as well as IL-10 (13) are regulatory countermeasures that delay the onset of diabetes. In fact, current immunotherapeutic approaches using anti-CD3 application are thought to confer protection through mechanisms involving TGF-β production, possibly by regulatory T cells (14). Given the close link of TGF-β with Th17 T cells that differentiate in the presence of TGF-β and IL-6, cytokines that are frequently found in inflammatory conditions, the question arises as to whether Th17 cells would be pathogenic in the NOD environment or whether they might in fact be protective. A recent publication (15) has suggested that Th17 cells play a pathological role in the development of type 1 diabetes. In this study, it was shown that transfer of BDC2.5 T cells polarized to Th17 appeared to transfer diabetes, but as these cells contained some IFN-γ–secreting cells, the possibility that diabetes was in fact due to contaminating Th1 cells expanding within a lymphopenic environment could not be excluded. In order to definitively address this question of the role of Th17 cells in diabetes development, we made use of the well-established adoptive transfer system of T cells from BDC2.5 mice into NOD/SCID recipients. Our results indicate that Th17 cells, even when 99% pure and devoid of any IFN-γ–secreting cells, upregulate T-box expressed in T cells (Tbet) and IFN-γ under the influence of IL-12 in vitro and likewise upregulate Tbet and convert to secrete IFN-γ in the NOD environment, causing diabetes with only a minor delay compared with Th1 BDC2.5 cells. Thus, although Th17 cells appear to be pathogenic in causing diabetes, our study clearly shows that it is the conversion to a Th1-like profile that underlies disease development, not the Th17 profile in itself. Our data indicate that there is substantial plasticity in the Th17 profile, which can be influenced by the local cytokine milieu in which inflammatory immune responses are taking place.
Results
Th17 BDC2.5 cells transfer diabetes with a similar kinetic to Th1 cells.
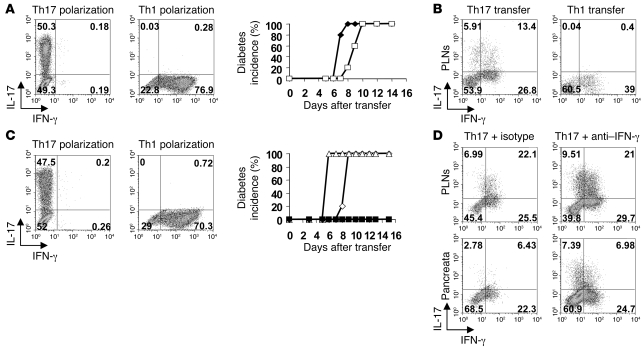
In order to test whether Th17 cells are pathogenic in the context of diabetes, naive T cells from BDC2.5 mice were polarized under Th1 or Th17 conditions in vitro (Figure 1A, left panels) and then adoptively transferred into NOD/SCID hosts. Surprisingly, mice that received polarized BDC2.5 Th17 cells developed diabetes similarly to those transferred with Th1 cells, albeit with a minor but consistent delay (Figure 1A, middle panel). Although, under Th17 culture conditions, the majority of BDC2.5 cells expressed IL-17 (see Figure 1A, left panel), analysis of their cytokine profile by intracellular staining, when recovered 8 days later from pancreatic lymph nodes (PLNs), showed a substantial proportion of IFN-γ–secreting cells (Figure 1B, left panel). In contrast, transfer of Th1 cells did not result in the emergence of Th17 cells (Figure 1B, right panel).
Figure 1. Th17 cells cause diabetes in NOD/SCID mice.
(A) Intracellular staining for IL-17 and IFN-γ of polarized Th17 or Th1 BDC2.5 cells was performed on day 4 of culture. Cells were transferred into NOD/SCID recipients and incidence of diabetes for Th1 (n = 5) (filled symbols) and Th17 (n = 5) (open symbols) transfer is shown. (B) Intracellular staining of CD4+Vβ4+ T cells in the PLNs on day 8 after transfer is shown. (C) Intracellular staining was performed as in A, followed by transfer into antibody-treated NOD/SCID recipients. Th17 were transferred into isotype control–treated (n = 4) (open diamonds) or anti–IFN-γ–treated (n = 5) (filled squares) hosts. Th1 were transferred into isotype-treated (n = 4) (open triangles) or anti–IFN-γ–treated (n = 5) (filled diamonds) hosts. (D) Representative FACS plots of CD4+Vβ4+ T cells in PLNs and pancreata of isotype–treated (left panels) or anti–IFN-γ–treated (right panels) hosts 10 days after transfer. Numbers indicate the percentage of cells in each quadrant.
Diabetes transfer is prevented by anti–IFN-γ but not by anti–IL-17.
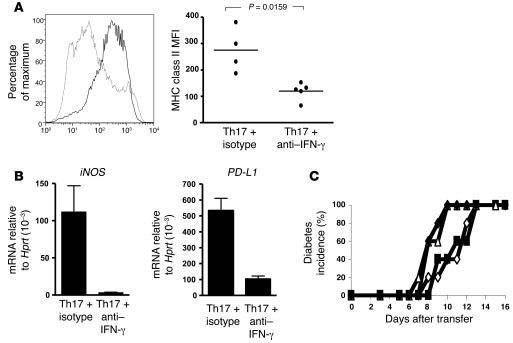
As some studies have suggested that IFN-γ may play a role in β cell destruction, we injected the adoptive hosts at the time of transfer with neutralizing antibody to IFN-γ (Figure 1C). This treatment prevented the onset of diabetes, whether the hosts had received Th1- or Th17-polarized cells (Figure 1C, middle panel). Cytokine analysis of PLNs and pancreata showed that while in the presence of anti–IFN-γ a somewhat higher proportion of IL-17 production was maintained in hosts of the Th17 cells, there was nevertheless a sizeable fraction of these cells with intracellular staining for IFN-γ (Figure 1D). Thus, neutralizing IFN-γ, while preventing the onset of disease, did not prevent the induction of IFN-γ in the Th17-polarized cells. Blockade of IFN-γ, however, had a striking effect on the profile of host CD11b-expressing antigen presenting cells, which failed to upregulate MHC class II (Figure 2A) and did not express iNOS or programmed death ligand 1 (PD-L1) (Figure 2B). IFN-γ–mediated activation of macrophages and their production of reactive oxygen intermediates is thought to be causally involved in the destruction of β islets (16). In contrast to the effect of neutralizing IFN-γ on the transfer of diabetes by Th17 cells, anti–IL-17 treatment did not inhibit their ability to transfer disease (Figure 2C); this puts into question whether Th17 cells themselves can be pathogenic in this model.
Figure 2. Anti–IFN-γ treatment inhibits activation of CD11b+ cells but not the effect on diabetes by anti–IL-17A administration.
(A) Histogram of MHC class II expression (left panel) on CD11b-expressing cells isolated from the pancreata of isotype- (thick line) or anti–IFN-γ–treated (thin line) NOD/SCID recipients, and MFI of MHC class II on individual samples (right panel). Horizontal bars represent mean MHC class II MFI, and individual points represent MHC class II MFI of CD11b-infiltrating cells from individual mice. (B) mRNA expression of iNOS and programmed death ligand 1 (PD-L1) is shown. Mean ± SEM. (C) Th17 and Th1 BDC2.5 T cells were adoptively transferred into antibody-treated NOD/SCID recipients. Th1 was transferred into isotype control–treated (open triangles) or anti–IL-17A–treated (filled diamonds) hosts. Th17 was transferred into isotype-treated (open diamonds) or anti–IL-17A–treated (filled squares) hosts. Mice were injected with 2 mg of isotype (OX-1) or anti–IL-17A (MM17-F3) on days 0, 2, 4, and 6. n = 5.
Diabetes transfer by Th17 is not due to contaminating Th1.
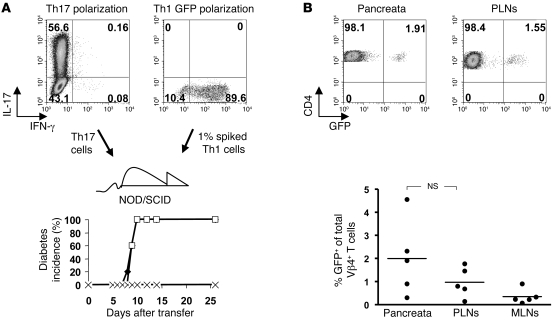
Despite the fact that the transferred Th17 cells were highly polarized (on average 40%–60%), there was still a sizeable fraction of cells that had not committed to IL-17 production. Furthermore, a small number of the IL-17 producers coproduced IFN-γ. It was therefore conceivable that either the double producers or the uncommitted fraction of the transferred cells could have selectively expanded and caused the IFN-γ–mediated effects. In order to assess whether a small proportion of IFN-γ producers could selectively expand in the NOD environment, we “spiked” a Th17 inoculum with a 1% Th1 contaminant of Th1-polarized BDC2.5 cells. These cells could be identified in vivo, because they were derived from a BDC2.5NOD strain, expressing GFP on its T cells (17). Nevertheless, although we could detect the GFP Th1 cells in PLNs and pancreata of these mice, they had not expanded significantly beyond the ratio in which they had been transferred and were unable to induce diabetes if transferred in these low numbers on their own (Figure 3).
Figure 3. Spiked Th1 cells do not outgrow Th17.
(A) Day 4 intracellular staining of Th17 (upper-left panel) or GFP+ Th1 (upper-right panel) cells. Cells were transferred as Th17 (n = 5) (filled diamonds), Th17 plus 1% GFP+ Th1 (n = 5) (open squares), or 1% GFP+ Th1 alone (n = 5) (× symbols), and diabetes development is shown (bottom panels). (B) Percentage of GFP+ Vβ4+ T cells in tissues on day 9 after transfer. Top panels show representative FACS plots of CD4 versus GFP gated on all Vβ4 T cells recovered from pancreas (left panel) and PLNs (right panel) from a Th17 plus 1% GFP Th1 recipient mouse. Horizontal bars represent mean percentage of GFP+ of total Vβ4+ T cells from 5 samples, and individual points represent percentage of GFP+ of total Vβ4+ T cells of individual mice. Numbers indicate the percentage of cells in each quadrant.
Highly purified Th17 cells transfer diabetes and convert to IFN-γ production.
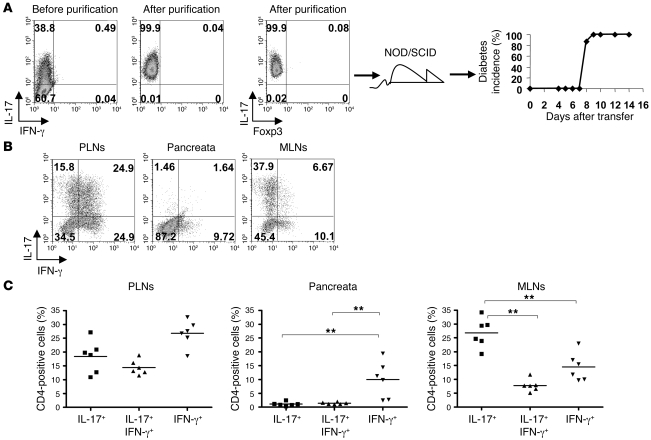
Although the emergence of IFN-γ producers in the Th17 inoculum is unlikely to be due to selective expansion of contaminants, the possibility remained that the sizeable fraction of uncommitted cells contaminating the transfer inoculum could have differentiated into Th1 cells in vivo. Thus, in order to ascertain whether Th17 cells are capable of converting to a Th1 profile, we proceeded to further purify the Th17 population prior to transfer, taking advantage of a recently described IL-17 capture assay (18). At the same time as positively selecting IL-17 producers, we negatively selected any IFN-γ producers by incubating the cells with capture constructs for IL-17 and IFN-γ and sorted the pure IL-17–producing populations by FACS. The purities obtained were routinely more than 98%, and a representative experiment in Figure 4 shows the complete absence of IFN-γ producers or cells expressing forkhead box P3 (Foxp3) following purification. Despite the high purity of the transferred Th17 cells (Figure 4A, left panel), the adoptive hosts developed diabetes with the same kinetics as seen previously (Figure 4A, far-right panel), and again, cytokine analysis of PLNs and pancreata showed a substantial proportion of the cells had converted to secrete IFN-γ, whereas in the mesenteric lymph nodes (MLNs), the IL-17 profile were better preserved (Figure 4, B and C). Thus, it seems that Th17 cells are indeed able to change their effector profile in the NOD environment.
Figure 4. Purified Th17 T cells convert in vivo.
(A) FACS plots of tetramer staining for IL-17 versus IFN-γ before (left dot plot) or after (middle dot plot) purification. IL-17 versus Foxp3 profile of purified cells (right dot plot). Diabetes onset in NOD/SCID recipients of highly purified Th17 cells (filled diamonds) (right panel). Numbers indicate the percentage of cells in each quadrant. (B) Representative FACS plots gated on CD4+Vβ4+ T cells from PLNs, pancreata, and mesenteric lymph nodes (MLNs) 8 days after transfer of highly purified Th17 cells, showing IL-17 versus IFN-γ intracellular staining. Numbers indicate the percentage of cells in each quadrant (A and B). (C) Summary graphs showing the percentage of cells producing IL-17, IL-17 plus IFN-γ, or IFN-γ alone. Horizontal bars represent mean percentage of CD4+ cells positive for stated cytokines from 6 samples, and individual symbols represent percentage of CD4+ cells positive for stated cytokines for individual mice. **P < 0.01, Mann-Whitney U test (n = 6). These data are representative of 3 independent experiments.
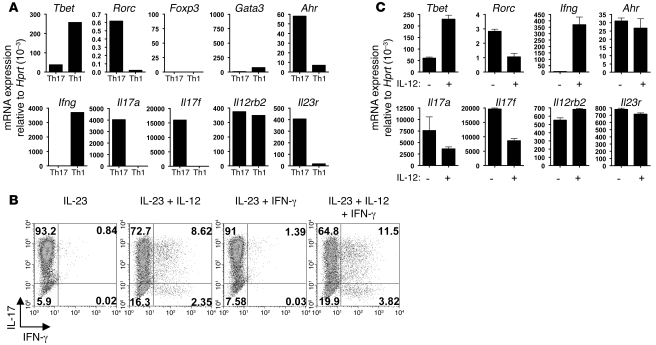
Th17 cells produce IFN-γ following culture with IL-12.
We next determined the expression of transcription factors, receptors, and cytokines typical for either Th17 or Th1 cells by quantitative PCR, starting with cells that were highly purified by the capture assay. Pure Th17 cells expressed RAR-related orphan receptor γ t (RORγt also known as Rorc) and aryl hydrocarbon receptor (AhR) but not GATA binding protein 3 (GATA-3) or Foxp3. Their expression of Tbet was minimal compared with pure Th1 cells, but it was not completely absent. There was no trace of IFN-γ in pure Th17 cells, and they expressed IL-17A and IL-17F as well as the IL-23 receptor as expected for this population. Interestingly, these Th17 cells also highly expressed the IL-12Rβ2 chain, which is part of the IL-12 receptor, whereas both IL-23 and IL-12 receptors shared the IL-12Rβ1 chain (Figure 5A). This suggests that Th17 cells could respond to IL-12 in the microenvironment, and this could be responsible for their conversion to secrete IFN-γ. In order to directly assess this, purified Th17 cells were further cultured in vitro in the presence of IL-23 to sustain their survival and with the addition of either IL-12 or IFN-γ or both factors. Analysis after 3 days of culture under these conditions clearly showed the emergence of IFN-γ double and single producers derived from the Th17 inoculum after culture in the presence of IL-12 (Figure 5B), and quantitative PCR for transcription factors showed that Th17 cells cultured with IL-12 substantially upregulated Tbet and Ifng and downregulated Rorc, Il17a, and Il17f, whereas expression of Ahr, Il23r, and Il12rb2 was preserved (Figure 5C). Such conversion was not restricted to Th17 cells from NOD mice, as comparable results were obtained using highly purified cells from C57BL/6 mice (see Supplemental Figure 1; available online with this article; doi: 10.1172/JCI37865DS1).
Figure 5. In vitro conversion of highly purified Th17 cells.
(A) mRNA expression relative to Hprt for stated transcription factors, cytokines, and receptors, comparing highly purified Th17 cells and Th1 cells. (B) Representative FACS plot showing intracellular staining of highly purified Th17 cells, cultured in the presence of IL-23 and the cytokines indicated, on day 3 following purification. Numbers indicate the percentage of cells in each quadrant. (C) mRNA expression relative to Hprt, comparing highly purified Th17 cells cultured with IL-23, with (+) or without (–) IL-12, for stated transcription factors, cytokines, and receptors. Mean ± SEM.
Adoptively transferred Th17 cells acquire transcriptional markers for Th1 program in vivo.
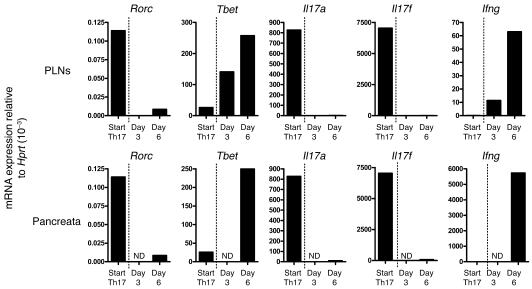
Having shown that highly pure Th17 cells can convert to IFN-γ production in vitro and that this is accompanied by upregulation of Tbet and downregulation of Rorc, Il17a, and Il17f, we examined whether such changes could be observed over time in vivo when purified Th17 cells are transferred into NOD/SCID recipients. As can be seen from Figure 6, the transferred highly purified Th17 population of cells expressed Rorc and a minimal amount of Tbet. Analysis of the PLNs on day 3 after transfer revealed a downregulation of Rorc and an upregulated expression of Tbet. There was also a loss of Il17a and Il17f expression and upregulation of Ifng at this time point. Analysis of the PLNs at day 6 showed an even greater increase in expression of Ifng and Tbet. The transferred cells were not present in the pancreata in sufficient numbers at day 3 to permit analysis but by day 6 changes seen in the PLNs were mirrored in the pancreata.
Figure 6. Kinetic analysis of transcriptional markers for Th1 program, following adoptive transfer of purified Th17 cells.
Purified Th17 cells were transferred into NOD/SCID recipients, and 5 mice were sacrificed on both day 3 and 6 after transfer. PLNs and pancreata were harvested, and CD4+Vβ4+ T cells were pooled and then isolated by MoFlo cell sorting followed by RNA extraction. Whole transcriptome amplification (QIAGEN) was then performed to create a cDNA template for real-time PCR. Depicted are mRNA expression levels relative to Hprt for stated transcription factors, cytokines, and receptors, comparing starting Th17 population (Start Th17) and (upper row) PLNs residing T cells and (bottom row) pancreata infiltrating T cells on day 3 and 6 after transfer. ND, none detected.
Discussion
Th17 cells have been implicated in a range of autoimmune conditions, including EAE and rheumatoid arthritis (19–21). The development of type 1 diabetes has usually been ascribed to a Th1 response with disease transfer in animal models being mediated by Th1 clones or lines (22, 23). TGF-β is an essential cofactor in the differentiation of Th17 and has been shown to ameliorate diabetes pathology. TGF-β and IFN-γ appear to counterregulate Th1 and Th17 development, respectively, at least under in vitro conditions (24, 25). While IFN-γ promotes diabetes pathology, TGF-β plays a role in limiting it (26, 27).
We have used the BDC2.5TCR transgenic mouse to investigate the role of Th17 cells in type 1 diabetes and found, using highly purified populations of cells, that although Th17 cells appear to transfer disease to NOD/SCID recipients, this is in fact due to a conversion of these cells to Th1. Analysis of the pancreata and PLNs of recipients showed a rapid conversion of the transferred Th17 cells to IFN-γ production, whereas transferred Th1 cells did not convert to Th17 cells. Indeed the ability of Th17-polarized T cells to transfer disease was inhibited by anti–IFN-γ treatment of recipients. Blockade of IFN-γ had a major effect on the functional maturation of antigen presenting cells expressing CD11b, preventing upregulation of MHC class II and iNOS as well as the expression of programmed death ligand 1.
Since the early days of Th17 T cell discovery, IL-17 secretion was suggested to be unstable and shown to default to a Th1 cytokine response following prolonged culture (28), although in many cases, technical difficulties in generating and maintaining these cells may have contributed to this notion. Nevertheless, the reported coexpression of IL-17 and IFN-γ under inflammatory conditions in vivo (29, 30) was remarkable, considering the strong inhibitory effect IFN-γ has on the initial differentiation of Th17 cells (25, 31). Double producers of IL-17 and IFN-γ were also found in the gut of patients with Crohn disease and coexpressed transcriptional markers for Th17 cells, such as RORγt, as well as Th1 traits, such as Tbet and IL-12Rβ2 (32).
However, definitive proof for conversion from Th17 to Th1 profile remained elusive due to the problems of purifying Th17 T cells on the basis of their cytokine profile. These limitations were overcome with the development of cytokine reporter mice, which should allow rigorous purification based on the expression of a cytokine reporter construct, although such mice are not available on a NOD background and the use of a single reporter construct does not obviate the problem of cells producing both IL-17 and IFN-γ. We therefore made use of a tetramer cytokine capture assay developed by Streeck et al. (18) to carefully isolate Th17 cells that were devoid of any IFN-γ signal assessed both by intracellular staining and RT-PCR analysis. Similar cytokine capture assays were used previously to demonstrate that cytokine-secreting effector T cells can differentiate to memory cells (33). The data we obtained indeed suggest that even highly pure Th17 cells easily convert to a Th1 profile, expressing increased amounts of Tbet and IFN-γ and reduced RORγt and IL-17 upon transfer into NOD/SCID mice. We could rule out that the emergence of a Th1 response in the PLNs and the pancreata is due to expansion of a contaminating Th1 population, as a 99% pure population of Th17 cells converts in vivo to a Th1 cytokine profile, and, furthermore, deliberate spiking of our Th17 populations with GFP-expressing Th1 cells did not reveal their outgrowth. Our data, therefore, suggest that these polarized Th17 cells are surprisingly plastic. Interestingly, however, there was no conversion of pure Th17 cells into a regulatory phenotype (data not shown), although inducible Tregs (iTregs) and Th17 cells develop in reciprocal fashion in vitro (34), depending on the cytokine regulated balance of RORγt and Foxp3 (35, 36). This suggests either that the cytokine microenvironment was not conducive or that the pathways leading to Th17 or iTreg effector differentiation are not malleable anymore once the cells have differentiated. Lexburg and colleagues, who obtained a comparable purity of Th17 cells, also demonstrated such conversion in vitro (37) but suggested that Th17 cells that develop in vivo are not converting to the same degree. It is currently unclear up to what stage of differentiation this apparent malleability of Th17 cells is applicable. Thus, it could be argued that in vitro generated Th17 cells resemble effector cells, whereas Th17 cells isolated from nonimmunized mice are more likely to be memory cells. Nevertheless, recent discoveries relating to plasticity of effector T cell fates indicate that some subsets may be more malleable than others, e.g., Th2 cells that will not divert to a Th1 fate anymore can nevertheless switch off their Th2 program upon exposure to TGF-β and activate a distinct program characterized by production of IL-9 (38).
As our highly purified polarized Th17 cells could be shown to express the IL-12Rβ2 chain in addition to the IL-23R and had small but detectable Tbet but no IFN-γ expression, we explored the possibility that IL-12 could provide a means for deviating the cells from Th17 to Th1. Our in vitro studies showed that in the presence of IL-23 and absence of IL-12, the Th17 cells retained Th17 polarization, whereas inclusion of IL-12 permitted the emergence of cells expressing both IL-17 and IFN-γ or IFN-γ alone. Kinetic analysis of cytokine and transcription factor expression in the PLNs and pancreata following Th17 transfer revealed a comparable conversion in vivo. This plasticity of Th17 cells in vivo suggests that caution should be used in the interpretation of data based on polarized T cell transfer and provides an alternative explanation for the recent publication, suggesting that Th17 cells may play a role in the development of type 1 diabetes (15). The data also highlight the need for rigorous purification in order to draw conclusions about precursor-product relationships.
NOD mice and other autoimmune prone mice such as New Zealand Black (NZB) and New Zealand White (NZW) have a polymorphism in their IL-12p40 chain (39), which has been associated with IL-12 overproduction and attributed to the insulin dependent diabetes susceptibility locus, Idd4 (40). Since the IL-12p40 chain is shared with IL-23 (41), it is conceivable that this polymorphism affects the expression of IL-23 as well, but potential changes in IL-23 expression in NOD mice have not been analyzed to date. Our data suggest that Th17 cells have the potential to convert toward a Th1 profile and, as seen by the differential maintenance of IL-17 versus IFN-γ expression in the pancreata and mesenteric lymph nodes, that the cytokine microenvironment in vivo may play a role in determining the outcome of such T cell differentiation. It is therefore possible that effector cell differentiation and lineage commitment in vivo are more malleable for some effector T cell subsets than originally thought on the basis of the more stringent commitment of Th1 and Th2 cells (42) and irreversible commitment may be less common during exposure of cells to different environmental stimuli in vivo. This could be of substantial relevance for the development of immune therapies targeted to individual cytokines.
Methods
Mice.
NOD/SCID, BDC2.5NOD, and C57BL/6 mice and GFP.BDC2.5NOD (17) mice were maintained under barrier conditions in the Biological Services facility of the Department of Pathology at the University of Cambridge. All animal experiments were done with the approval of the University of Cambridge and according to institutional guidelines and United Kingdom Home Office regulations.
Antibodies and flow cytometry.
Anti–IFN-γ antibody (XMG1.2), anti-CD3 (145.2C11), anti–IL-4 (11B11), and rat IgG1 isotype control (MAC221) were grown from hybridomas in our own laboratory. Biotinylated anti–IL-17A (MM-17F3) (19), for use in tetramers, was obtained from Jacques Van Snick (Ludwig Institute for Cancer Research). The endotoxin levels were less than 1 EU/mg protein, and the preparations were stored at –20°C until use. Commercially available antibodies used were anti-CD28 (no azide, low endotoxin) (clone 37.51); APC- or PerCP-conjugated anti-CD4 (RM4-5); PE-conjugated anti-CD25 (PC61); FITC-conjugated anti-CD44 (IM7); FITC- or PE-conjugated anti-Vβ4 (KT4); PerCP-Cy5.5–conjugated CD11b (M1/70); PE-, PE-Cy7–, or FITC-conjugated anti–IFN-γ (XMG1.2); biotinylated anti–IFN-γ (R46A2); PE-conjugated anti–IL-17A (TC11-18H10); and PE-, FITC-, or PE-Cy7–conjugated IgG1 isotype controls and all were obtained from BD Pharmingen. Anti–IL-17A FITC (eBIO17B7), anti-Foxp3 FITC (FJK-16s), and anti-CD45.2 (clone 104) were obtained from eBioscience. FITC-conjugated anti–MHC Class II (OX-6) was obtained from Serotec. Intracellular staining was done as described previously (43).
In vitro T cell differentiation.
Naive CD4 T cells, sorted by flow cytometry (CD4+CD25–CD44lo), were cultured in Iscove’s modified Dulbecco medium (IMDM) supplemented with 10% heat-inactivated FCS, 2 × 10–3 M l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 5 × 10–5 M mercaptoethanol (all Sigma-Aldrich). For generation of Th17- and Th1-polarized cells, naive BDC2.5NOD T cells were cultured on anti-CD3 (2 μg/ml) and anti-CD28 (10 μg/ml) coated 24-well plates in the presence of either 25 ng/ml IL-6 (Peprotech), 10 ng/ml IL-1β (Invitrogen), 2 ng/ml human TGF-β (R&D Systems), 50 μg/ml anti–IFN-γ (XMG1.2), and 10 μg/ml anti–IL-4 (1B11) blocking antibodies for the Th17 condition or 10 ng/ml IL-12 (Peprotech) for the Th1 condition.
Cells were cultured for 4 days and then transferred to fresh plates without stimulus and rested for 4 days. Thereafter, cells were harvested and restimulated on anti-CD3 and anti-CD28 coated plates in the presence of 20 ng/ml IL-23 (R&D Systems) for 2 days before purification by the cytokine capture assay.
For in vitro conversion of highly purified Th17 cells, the cells were placed on anti-CD3 and anti-CD28 coated 96-well plates in the presence of 20 ng/ml IL-23 (R&D Systems), with or without 10 ng/ml IL-12 (Peprotech) and 100 U/ml IFN-γ (R&D Systems) for 3 days before harvesting for FACS or RT-PCR.
Isolation of pure T cell subsets by cytokine capture assay.
The method for tetramer formation and IL-17 capture was adapted from Streeck et al. (18). To make IL-17 and IFN-γ tetramers, either biotinylated anti–IL-17A (MM-17) or biotinylated anti–IFN-γ (R46A2; BD Pharmingen) antibodies were mixed with biotinylated anti-CD45.2 in a 3:1 molar ratio. Avidin-neutravidin (5 μg) (Invitrogen) was added to the biotinylated antibodies and then vortexed and incubated at 4°C for 10 minutes. A further 5 μg of avidin-neutravidin was added for a further 10 minutes, and the aliquots were stored at 4°C until use. To obtain purified Th17 cells, naive T cells were differentiated, rested, and restimulated as previously described. Cells were harvested and washed 3 times in 2% FCS PBS, and 4 × 106 cells were incubated on ice for 2 minutes with 1 aliquot of IL-17 tetramer, before further incubation for 15 minutes with 1 aliquot of IFN-γ tetramer. Ten milliliters of culture medium was added to each tube and the tubes then slowly rotated for 1 hour and 30 minutes at 37°C to allow cytokine capture, followed by washing and staining with PE-conjugated anti–IL-17A and FITC-conjugated anti–IFN-γ (both BD Pharmingen) for 15 minutes at 4°C. PE-positive, FITC-negative cells were sorted using a MoFlo Cell Sorter (Beckman Coulter) to achieve a purity of 95%. Further purification to more than 99.5% was achieved by performing a second sort for PE-positive, FITC-negative cells.
Adoptive transfer experiments and in vivo antibody treatment.
Polarized or highly purified cells were harvested and washed twice before resuspension in sterile PBS, and 5 × 104 cells were adoptively transferred i.p. into NOD/SCID recipients.
For GFP spiking experiments, 2 × 105 Th17 cells were seeded with 2 × 103 Th1 GFP cells and transferred in sterile PBS i.p. into NOD/SCID recipients. In vivo treatment with neutralizing antibody was done with either 2 mg/mouse of anti–IFN-γ (XMG1.2) or control IgG1 isotype (MAC221) or 2 mg/mouse of anti–IL-17A (MM17-F3) or control mouse IgG1 (OX-1) administered i.p. on days 0, 2, 4, and 6, following transfer of Th17 cells. All recipients were tested daily, as early as day 5 onward, for glucose in urine using Diastix (Bayer Diagnostics).
Purification of mRNA and quantitative PCR analysis.
RNA was extracted using RNeasy Mini Kit (QIAGEN) following standard manufacturer’s protocol. Using a QuantiTect Reverse Transcription Kit with real-time SYBR green PCR (QIAGEN), cDNA was used as a template for the amplification of genes of interest and the housekeeping gene (Hprt). Where indicated, Whole Transcriptome Amplification (QIAGEN) of small RNA samples was performed according to manufacturer’s instructions to yield cDNA for real-time PCR. The following primers (all from QIAGEN) for transcription factors, cytokines, and receptors were tested: Tbet, Rorc, Foxp3, Gata3, Ahr, Il17a, Il17f, Ifng, Il12rb2, and Il23r. Amplification plots and dissociation stages were analyzed using a 7500 Fast Real-Time PCR Detector System with 7500 Fast Software System version 1.4.6 (both from Applied Biosystems). Gene expression was calculated using the comparative method for relative quantification upon normalization to Hprt gene expression.
Statistics.
Statistical analysis was performed with GraphPad Prism software, using a Kaplan-Meier survival curve and log-rank test. For analysis of nonparametric data sets, a Mann-Whitney U test was performed. Results were deemed significant if P values were less than 0.05.
Supplementary Material
Acknowledgments
We thank Nigel Miller, Chris Bland, Oliver Burton, and Stephen Newland for valuable contributions to this project. We also thank Nick Holmes for critical reading of this manuscript. This work was funded by the Wellcome Trust. David Bending is funded by the Biotechnology and Biological Sciences Research Council (BBSRC).
Footnotes
Authorship note: David Bending and Hugo De La Peña contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: AhR, aryl hydrocarbon receptor; Foxp3, forkhead box P3; PLN, pancreatic lymph node; RORγt, RAR-related orphan receptor γ t; Tbet, T-box expressed in T cells.
Citation for this article: J. Clin. Invest. 119:565–572 (2009). doi:10.1172/JCI37865
References
- 1.Veldhoen M., Hocking R.J., Flavell R.A., Stockinger B. Signals mediated by transforming growth factor-beta initiate autoimmune encephalomyelitis, but chronic inflammation is needed to sustain disease. Nat. Immunol. 2006;7:1151–1156. doi: 10.1038/ni1391. [DOI] [PubMed] [Google Scholar]
- 2.Li M.O., Sanjabi S., Flavell R.A. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Wang B., et al. Interferon-gamma impacts at multiple points during the progression of autoimmune diabetes. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13844–13849. doi: 10.1073/pnas.94.25.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicoletti F., et al. The effects of a nonimmunogenic form of murine soluble interferon-gamma receptor on the development of autoimmune diabetes in the NOD mouse. Endocrinology. 1996;137:5567–5575. doi: 10.1210/en.137.12.5567. [DOI] [PubMed] [Google Scholar]
- 5.Boyton R.J., et al. Stat4-null non-obese diabetic mice: protection from diabetes and experimental allergic encephalomyelitis, but with concomitant epitope spread. Int. Immunol. 2005;17:1157–1165. doi: 10.1093/intimm/dxh293. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z., et al. Autoimmune diabetes is blocked in Stat4-deficient mice. J. Autoimmun. 2004;22:191–200. doi: 10.1016/j.jaut.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 7.Trembleau S., et al. Interleukin 12 administration induces T helper type 1 cells and accelerates autoimmune diabetes in NOD mice. J. Exp. Med. 1995;181:817–821. doi: 10.1084/jem.181.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hultgren B., Huang X., Dybdal N., Stewart T.A. Genetic absence of gamma-interferon delays but does not prevent diabetes in NOD mice. Diabetes. 1996;45:812–817. doi: 10.2337/diabetes.45.6.812. [DOI] [PubMed] [Google Scholar]
- 9.Serreze D.V., et al. Interferon-gamma receptor signaling is dispensable in the development of autoimmune type 1 diabetes in NOD mice. Diabetes. 2000;49:2007–2011. doi: 10.2337/diabetes.49.12.2007. [DOI] [PubMed] [Google Scholar]
- 10.Kanagawa O., Xu G., Tevaarwerk A., Vaupel B.A. Protection of nonobese diabetic mice from diabetes by gene(s) closely linked to IFN-gamma receptor loci. J. Immunol. 2000;164:3919–3923. doi: 10.4049/jimmunol.164.7.3919. [DOI] [PubMed] [Google Scholar]
- 11.Katz J.D., Wang B., Haskins K., Benoist C., Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-E. [DOI] [PubMed] [Google Scholar]
- 12.Mueller R., Bradley L.M., Krahl T., Sarvetnick N. Mechanism underlying counterregulation of autoimmune diabetes by IL-4. Immunity. 1997;7:411–418. doi: 10.1016/S1074-7613(00)80362-3. [DOI] [PubMed] [Google Scholar]
- 13.Phillips J.M., Parish N.M., Drage M., Cooke A. Cutting edge: interactions through the IL-10 receptor regulate autoimmune diabetes. J. Immunol. 2001;167:6087–6091. doi: 10.4049/jimmunol.167.11.6087. [DOI] [PubMed] [Google Scholar]
- 14.You S., et al. Adaptive TGF-beta-dependent regulatory T cells control autoimmune diabetes and are a privileged target of anti-CD3 antibody treatment. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6335–6340. doi: 10.1073/pnas.0701171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain R., et al. Innocuous IFNgamma induced by adjuvant-free antigen restores normoglycemia in NOD mice through inhibition of IL-17 production. J. Exp. Med. 2008;205:207–218. doi: 10.1084/jem.20071878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchings P., et al. Transfer of diabetes in mice prevented by blockade of adhesion-promoting receptor on macrophages. Nature. 1990;348:639–642. doi: 10.1038/348639a0. [DOI] [PubMed] [Google Scholar]
- 17.Raine T., Zaccone P., Mastroeni P., Cooke A. Salmonella typhimurium infection in nonobese diabetic mice generates immunomodulatory dendritic cells able to prevent type 1 diabetes. J. Immunol. 2006;177:2224–2233. doi: 10.4049/jimmunol.177.4.2224. [DOI] [PubMed] [Google Scholar]
- 18.Streeck H., et al. Rapid ex vivo isolation and long-term culture of human Th17 cells. J. Immunol. Methods. 2008;333:115–125. doi: 10.1016/j.jim.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 19.Uyttenhove C., Van Snick J. Development of an anti-IL-17A auto-vaccine that prevents experimental auto-immune encephalomyelitis. Eur. J. Immunol. 2006;36:2868–2874. doi: 10.1002/eji.200636662. [DOI] [PubMed] [Google Scholar]
- 20.Sonderegger I., et al. Neutralization of IL-17 by active vaccination inhibits IL-23-dependent autoimmune myocarditis. Eur. J. Immunol. 2006;36:2849–2856. doi: 10.1002/eji.200636484. [DOI] [PubMed] [Google Scholar]
- 21.Rohn T.A., et al. Vaccination against IL-17 suppresses autoimmune arthritis and encephalomyelitis. Eur. J. Immunol. 2006;36:2857–2867. doi: 10.1002/eji.200636658. [DOI] [PubMed] [Google Scholar]
- 22.Healey D., et al. In vivo activity and in vitro specificity of CD4+ Th1 and Th2 cells derived from the spleens of diabetic NOD mice. J. Clin. Invest. 1995;95:2979–2985. doi: 10.1172/JCI118006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz J.D., Benoist C., Mathis D. T helper cell subsets in insulin-dependent diabetes. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- 24.Park H., et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mangan P.R., et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 26.Moritani M., et al. Abrogation of autoimmune diabetes in nonobese diabetic mice and protection against effector lymphocytes by transgenic paracrine TGF-beta1. J. Clin. Invest. 1998;102:499–506. doi: 10.1172/JCI2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grewal I.S., et al. Expression of transgene encoded TGF-beta in islets prevents autoimmune diabetes in NOD mice by a local mechanism. J. Autoimmun. 2002;19:9–22. doi: 10.1006/jaut.2002.0599. [DOI] [PubMed] [Google Scholar]
- 28.Mathur A.N., et al. T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood. 2006;108:1595–1601. doi: 10.1182/blood-2006-04-015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanov I.I., et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 30.Luger D., et al. Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J. Exp. Med. 2008;205:799–810. doi: 10.1084/jem.20071258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrington L.E., et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat. Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 32.Annunziato F., et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohning M., et al. Long-lived virus-reactive memory T cells generated from purified cytokine-secreting T helper type 1 and type 2 effectors. J. Exp. Med. 2008;205:53–61. doi: 10.1084/jem.20071855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bettelli E., et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 35.Zhou L., et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang X.O., et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lexberg M.H., et al. Th memory for interleukin-17 expression is stable in vivo. Eur. J. Immunol. 2008;38:2654–2664. doi: 10.1002/eji.200838541. [DOI] [PubMed] [Google Scholar]
- 38.Veldhoen M., et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat. Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 39.Ymer S.I., et al. Polymorphisms in the Il12b gene affect structure and expression of IL-12 in NOD and other autoimmune-prone mouse strains. Genes Immun. 2002;3:151–157. doi: 10.1038/sj.gene.6363849. [DOI] [PubMed] [Google Scholar]
- 40.Simpson P.B., et al. Cuttine edge: diabetes-associated quantitative trait locus, Idd4, is responsible for the IL-12p40 overexpression defect in nonobese diabetic (NOD) mice. J. Immunol. 2003;171:3333–3337. doi: 10.4049/jimmunol.171.7.3333. [DOI] [PubMed] [Google Scholar]
- 41.Oppmann B., et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–725. doi: 10.1016/S1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 42.Murphy E., et al. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J. Exp. Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veldhoen M., Hocking R.J., Atkins C.J., Locksley R.M., Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.