Abstract
The Argonaute superfamily is a large family of RNA-binding proteins involved in gene regulation mediated by small noncoding RNA and characterized by the presence of PAZ and PIWI domains. The family consists of two branches, the Ago and the Piwi clade. Piwi proteins bind to 21–30-nucleotide-long Piwi-interacting RNAs (piRNAs), which map primarily to transposons and repeated sequence elements. Piwi/piRNAs are important regulators of gametogenesis and have been proposed to play roles in transposon silencing, DNA methylation, transcriptional silencing, and/or post-transcriptional control of translation and RNA stability. Most reports to date have concentrated on the Piwi family members in the male germline. We have identified four Piwi proteins in Xenopus and demonstrate that two, namely, Xiwi1b and Xili, are expressed in the oocyte and early embryo. Xiwi1 and Xili are predominantly found in small, separate complexes, and we do not detect significant interaction of Piwi proteins with the cap-binding complex. Putative nuclear localization and export signals were identified in Xiwi1 and Xili, supporting our observation that Xiwi1, but not Xili, is a nucleo-cytoplasmic protein. Furthermore, by immunoprecipitation of small RNAs, we establish Xiwi1 as a bona fide Piwi protein. These results suggest that the Piwi/piRNA pathway is active in translationally repressed oocytes. This is a significant finding as the Xenopus model provides an excellent tool to study post-transcriptional mechanisms.
Keywords: Argonaute, piRNA, gene silencing, oocyte
INTRODUCTION
The Argonaute superfamily is a large family of proteins involved in gene regulation mediated by small non-coding RNA and characterized by the presence of PAZ and PIWI domains (Cerutti et al. 2000). The family consists of two branches based on sequence similarity—the Ago proteins, which are related to the Arabidopsis protein Argonaute, and the Piwi clade—characterized by homology with the Drosophila Piwi protein (Farazi et al. 2008; Hock and Meister 2008; Hutvagner and Simard 2008). Much is already known about the Ago clade and the important role its members play in siRNA and miRNA pathways (for review, see Peters and Meister 2007).
However, Piwi proteins are less well understood, although considerable progress has been made recently to elucidate their function and mechanism of action. Expression of Piwi proteins in Drosophila is seen both in the germline as well as in somatic cells (Cox et al. 1998; Nishida et al. 2007), while in vertebrates, significant expression has only been documented in the germline (Kuramochi-Miyagawa et al. 2001; Qiao et al. 2002; Tan et al. 2002; Lau et al. 2006; Carmell et al. 2007; Houwing et al. 2007; Sugimoto et al. 2007).
Piwi proteins were initially identified as regulators of germline stem cell division in Drosophila (Lin and Spradling 1997; Cox et al. 1998). In Drosophila, Piwi protein mutants are defective in gametogenesis (Klattenhoff and Theurkauf 2008). In the mouse, knockout males are sterile, and females remain fertile even though expression of at least one Piwi protein has been documented in the ovary (Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007; Watanabe et al. 2008), while in zebrafish, loss of Ziwi triggers germ cell apoptosis in premeiotic cells (Houwing et al. 2007). Similarly to Ago proteins, Piwi proteins have been recently shown to bind to small RNAs, named Piwi-interacting RNAs (piRNAs) (Aravin et al. 2006; Girard et al. 2006; Lau et al. 2006; Kuramochi-Miyagawa et al. 2008). These 21–30 nucleotide (nt) RNAs map primarily to transposons and repeated sequence elements (Brennecke et al. 2007; Gunawardane et al. 2007; Batista et al. 2008; Das et al. 2008). Although piRNA biogenesis is still poorly understood, it has been suggested that at least in Drosophila they are generated from primary transcripts of piRNA clusters (Brennecke et al. 2007), and subsequently amplified in a feedback mechanism involving Piwi proteins, called the ping-pong amplification loop (Aravin et al. 2007a; Klattenhoff and Theurkauf 2008), features of which are conserved in zebrafish and mouse (Aravin et al. 2007b; Houwing et al. 2007).
Piwi proteins have been shown to be responsible for transposon silencing, as mutations in genes encoding these proteins lead to an increase in transposon transcription and a decrease in the production of piRNA sequences directed against them (Sarot et al. 2004; Kalmykova et al. 2005; Saito et al. 2006; Aravin et al. 2007b; Houwing et al. 2007). Mutations in Mili and Miwi2 result in the early loss of transposon element DNA methylation and in a sterility phenotype in testis, suggesting that piRNAs guide de novo methylation and transcriptional repression (Carmell et al. 2007; Aravin and Bourc'his 2008; Kuramochi-Miyagawa et al. 2008). Furthermore, in Drosophila, Piwi can exert opposite effects on different genomic regions, possibly reflecting the influence of local chromatin context (Yin and Lin 2007). Several reports have also implied Piwis in post-transcriptional control, at the level of translation and/or RNA stability (Lin 2007).
While Piwi proteins in the male germline are well characterized, comparatively little is known about which Piwi proteins are expressed in the vertebrate female germline, their intracellular location, or which proteins, if any, they interact with; all features that would inform on function (see Discussion).
This is the first report to demonstrate the expression of two Piwi proteins, Xiwi1 and Xili, in the Xenopus oocyte and in early embryogenesis. Xiwi1 and Xili are predominantly found in small, largely distinct, complexes, and we do not detect interaction of Piwi proteins with the cap-binding complex. Putative nuclear localization and export signals were identified in Xiwi1 and Xili proteins, supporting our observation that Xiwi1, but not Xili, is a nucleo-cytoplasmic protein. Furthermore, by immunoprecipitation of small RNAs, we establish Xiwi1 as a bona fide Piwi protein. These results suggest that the piRNA pathway is active in translationally repressed oocytes.
RESULTS
Identification of Piwi family proteins in Xenopus
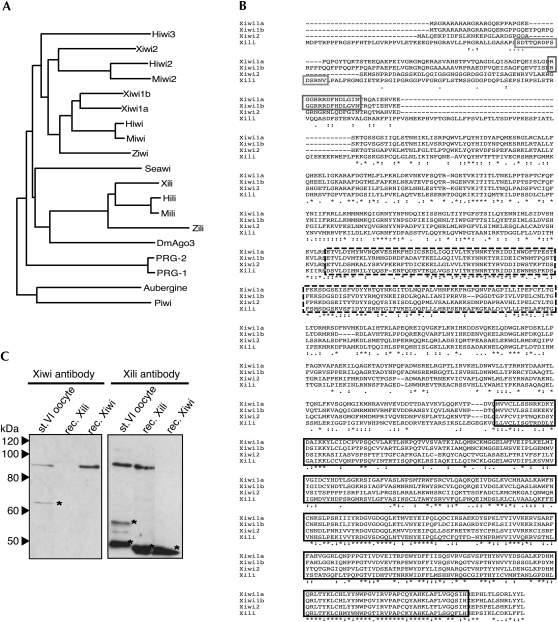
Analysis of the Gurdon Institute Xenopus tropicalis full-length database (Gilchrist et al. 2004; http://informatics.gurdon.cam.ac.uk/online/xt-fl-db.html) yielded sequences of four putative Piwi family members. Phylogenetic analysis allowed us to name them: Xiwi1a/Xiwi1b by sequence similarity to Miwi; Xili by sequence similarity to Mili; and Xiwi2 by sequence similarity to Miwi2 (Fig. 1A,B). Xiwi1a and Xiwi1b are paralogs, not splicing variants, and are both most closely related to mammalian Piwi orthologs. They share 73% overall sequence identity, while overall sequence identity between Xiwi1b and Xili is only 42%. The sequence identity between PIWI domains is 81% and 53%, respectively, and for the PAZ domain – 58% and 45%, respectively. Xiwi1a and Xiwi1b are more similar to each other than to any other proteins of the Piwi clade. The predicted molecular weights of Xiwi1a, Xiwi1b, Xili, and Xiwi2 are 97 kDa, 99 kDa, 106 kDa, and 96.4 kDa, respectively. RT-PCR and real-time RT-PCR analysis revealed that of the four Xenopus Piwi mRNAs, only Xiwi1b and Xili mRNAs were robustly expressed in the oocyte (J. Armisen, M. Gilchrist, A. Wilczynska, N. Standart, and E.A. Miska, in prep.). Therefore, peptide antibodies were generated against Xiwi1 and Xili proteins (Fig. 1A).
FIGURE 1.
Identification of Xenopus Piwi proteins. (A) Phylogenetic tree of Piwi family proteins. Unrooted phylogenetic tree based on ClustalW alignment of Piwi domains as predicted by the Prosite database (Hulo et al. 2008). The sequences used are PRG-1, C. elegans NM_059720; PRG-2, C. elegans NM_068593; Piwi, D. melanogaster NM_057527; Aubergine, D. melanogaster NM_057386; DmAgo3, D. melanogaster EF211827; Seawi, S. purpuratus NM_214600; Ziwi, D. rerio NM_183338; Zili, D. rerio EF186090; Miwi, M. musculus NM_021311; Miwi2, M. musculus NM_177905; Mili, M. musculus NM_021308; Hiwi, H. sapiens NM_004764; Hiwi2, H. sapiens NM_152431; Hili, H. sapiens NM_018068; Hiwi3, H. sapiens NM_001008496. (B) ClustalW alignment of four putative Xenopus tropicalis Xiwi1a, Xiwi1b, Xili, and Xiwi2 protein sequences. The Xiwi1a sequence was derived from IMAGE clone 7689388. The Xiwi1b sequence was assembled from scaffolds Xt7.1-CAAN9738.3.5 and Xt7.1-XZG3113.5 of the Gurdon Institute X. tropicalis full-length database. Xili was derived from IMAGE clone 7676520, and Xiwi2 was assembled from scaffolds Xt7.1-CAAN6696.5, Xt7.1-CAAN4060.3, and Ensembl Gene prediction ENSXETP00000014018. (Highly related, albeit partial, X. laevis EST sequences were also retrieved from databases.) Sequences highlighted with the solid gray box denote peptides used for generating specific antibodies against Xiwi1 and Xili, respectively. (Dashed black box) PAZ and (solid black box) PIWI domains. (C) Antibodies raised against Xiwi1 and Xili are specific to the proteins and do not cross react. Polyclonal antibodies were tested on a Western blot containing one oocyte equivalent of stage VI Xenopus oocyte lysate, 4.5 ng of recombinant His-tagged Xili protein, and 1.5 ng of recombinant His-tagged Xiwi1. (*) Nonspecific bands detected by the antibodies. Proteins were resolved on a 15% polyacrylamide SDS gel.
Xiwi1 and Xili antibodies each identify a single major protein, of the predicted size, in stage VI Xenopus oocyte lysate, and specifically recognize the corresponding recombinant form of the protein, as shown by Western blotting (Fig. 1C).
Xiwi1 and Xili are expressed throughout oogenesis and in early embryos
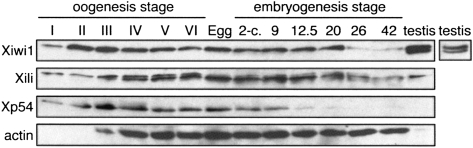
We next assessed the expression of Xiwi1 and Xili throughout oogenesis and early embryogenesis in Xenopus. Xiwi1 in the female germline is detectable by Western blotting as a single band, as compared to two in testis (Fig. 2). Given the size distribution of the two forms in testis and the RT-PCR data (discussed above), we propose that Xiwi1b but not Xiwi1a is present in the female germline and early embryo. Xiwi1b protein levels initially increase as the oocyte grows, decrease following stage III, and are maintained until stage 20 of embryogenesis (the neural tube stage). In contrast, Xili protein levels increase steadily up to stage VI of oogenesis and are maintained longer into embryogenesis (Fig. 2). Intriguingly, we note that in oogenesis, the ratio of levels of the two Piwi proteins is continually changing and is different from that in adult testis. Comparison of expression levels of Xiwi1 and Xili with fixed titers of Escherichia coli–expressed recombinant protein by Western blot revealed that in stage VI oocytes, the total amount of Xiwi1 protein was ∼10 ng per oocyte, and Xili 2.5 ng per oocyte (data not shown). This result places Piwi proteins in a group of low-abundance RNA-binding proteins in the oocyte, such as Staufen (Allison et al. 2004), and of much lower abundance than major translational regulators such as CPEB or Xp54 (Minshall et al. 2007). Our analysis suggests that in comparison with Xenopus testis, Xili protein is expressed at relatively high levels in late oogenesis and early embryogenesis, while Xiwi1 levels are significantly lower in the female than in the male germline. For these experiments, Xp54 and actin, which span oogenesis and embryogenesis, were used as loading controls (Ladomery et al. 1997; Allison et al. 2004; Minshall et al. 2007).
FIGURE 2.
Expression of Xiwi1 and Xili throughout Xenopus oogenesis and embryogenesis. Two oocyte equivalents for each stage of oogenesis, two egg equivalents, two embryo equivalents for each stage of embryogenesis, and 20 μg of testis lysate protein (equivalent of one stage VI oocyte) were resolved on 10% SDS-PAGE. The stages of embryogenesis correspond to stage 9, midblastula; stage 12.5, gastrula; stage 20, neural fold closure; stage 26, tail bud stage; stage 42, tadpole-like stage. Stage I may be underrepresented here because of the technical difficulties of accurately estimating cell number with these small cells. An alternative image of Xiwi1 protein in testis is presented for better visualization of the protein doublet. Note that Xili appears as a doublet in oocytes, for unknown reasons, due to better resolution of high-molecular-weight proteins compared to Figure 1C.
Xiwi1 and Xili are predominantly present in small but distinct protein complexes
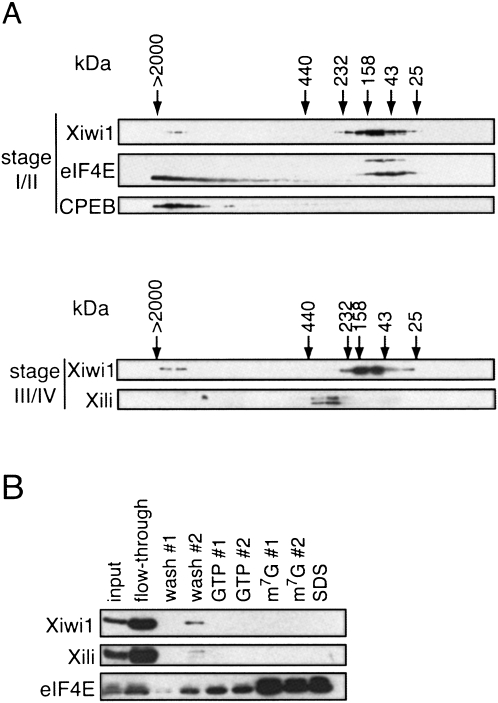
We next addressed the question of whether Xiwi1 and Xili are complexed with other proteins in the Xenopus oocyte. To this end, we first analyzed fractions from Superose 6 HR 10/30 FPLC gel filtration of stage I–II oocyte lysates by Western blot. The majority of Xiwi1 protein cofractionated with complexes of a small size (∼100–150 kDa), suggesting that for the most part, the protein is not associated with many protein partners (Fig. 3A, upper panels). It is noteworthy, though, that the cap-binding protein eIF4E1a (both isoforms, which differ by an 18-amino-acid-long repeat) (Minshall et al. 2007) partially comigrates with Xiwi1, which may indicate a potential interaction between the two proteins. Also, a small proportion is detected in large complexes (>2 MDa) that copurify with the previously described large CPEB complex, as indicated by the presence of both CPEB and the noncanonical 24-kDa eIF4E1b (Minshall et al. 2007). The presence of Xiwi1 in the large complex was sensitive to treatment of the lysate with RNase prior to gel filtration (data not shown), suggesting that the interaction of a minor portion of the cellular pool of Xiwi1 with large mRNP complexes occurs not through direct protein–protein interactions but, rather, with RNA. Since Xili protein is expressed very weakly in stage I/II oocytes, we analyzed gel filtrations fractions from stage III/IV lysates for both Xiwi1 and Xili (Fig. 3A, lower panels). Interestingly, compared to Xiwi1, Xili cofractionates with somewhat larger complexes (∼300 kDa). Neither Xiwi1 nor Xili small complexes were sensitive to prior RNase treatment (data not shown). Their presence in different sized complexes is significant as the two proteins themselves are of very similar size (99 kDa/106 kDa for Xiwi1/Xili, respectively) and suggests that Xili interacts with proteins different from or additional to Xiwi1.
FIGURE 3.
(A) Xiwi1 and Xili are predominantly present in small complexes. Lysate prepared from (upper panel) stage I/II and (lower panel) stage III/IV oocytes was gel-filtered as described previously, on separate columns (Minshall et al. 2007). Alternate fractions were resolved by 15% SDS-PAGE and analyzed by Western blot with the indicated antibodies. For stage I/II fractions, Xiwi1 was detected using the anti-Miwi antibody (Deng and Lin 2002). For stage III/IV fractions, the Western blot was probed with the anti-Xiwi1 and anti-Xili antibodies described above. The molecular weight standards used were chymotrypsin A (25 kDa), ovalbumin (43 kDa), aldolase (158 kDa), catalase (232 kDa), and ferritin (440 kDa). (B) Xiwi1 and Xili do not interact with the cap-binding complex. m7GTP-Sepharose chromatography was performed using stage III/IV oocyte lysate. Following binding, the beads were washed and then eluted with GTP- and m7GpppG-containing buffer and finally with SDS buffer. Input represents ∼0.25% of the input lysate; the flow-through and wash fractions represent 1% of each fraction; the GTP and m7G fractions represent 12% of each fraction; and SDS represents 50% of the SDS fraction.
Xiwi1 and Xili do not interact with the cap-binding complex
Miwi has been previously described to interact with the cap-binding complex in mouse testis (Grivna et al. 2006). To establish whether the same association can be seen for the Xenopus Piwi family members, m7GTP-Sepharose affinity chromatography was performed using lysates prepared from stage III/IV oocytes (Fig. 3B). Following the binding of the lysate proteins to m7GTP-Sepharose, the beads were washed, then consecutively eluted with GTP- and cap analog-containing buffer, and any proteins remaining bound to the beads were removed with SDS-containing buffer. The samples were separated by SDS-PAGE and analyzed by Western blot. Since both Xiwi1 and Xili proteins remained in the unbound flow-through and wash fractions, as compared to eIF4E protein, which bound preferentially to m7GpppG, as reported previously (Minshall et al. 2007), we conclude that Piwi family proteins in the Xenopus female germline do not associate with the cap-binding complex.
Subcellular localization of Xiwi1 and Xili
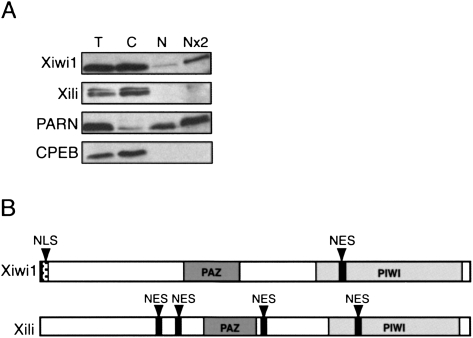
Piwi family proteins have been reported to function in both nuclear and cytoplasmic processes. In order to determine the subcellular localization of the Xenopus Piwi proteins, we analyzed nuclear and cytoplasmic stage VI oocyte fractions by Western blot. We conclude that Xiwi1 is distributed between the nucleus and the cytoplasm at a ratio of about 1:3, whereas Xili protein is exclusively cytoplasmic (Fig. 4A). PARN was used here as a nucleus-specific marker, and CPEB as a cytoplasm-specific maker (Copeland and Wormington 2001; Minshall et al. 2007). In support of this observation, amino acid sequence analysis of Xiwi1b and Xili revealed that Xiwi1b contains a putative nuclear localization signal (NLS) based on a search in NLSdb (Nair et al. 2003) in the N-terminal region and a putative nuclear export signal (NES) based on the consensus sequence (L/M)-X2-3-(L/F)- X2-3-(L/I/V)-X-(L/I) (Kutay and Guttinger 2005) within the Piwi domain. In contrast, Xili contains four putative NES sequences distributed throughout the protein and no predicted NLS (Fig. 4B). If both the Xiwi1b NLS and NES were functional, this would provide a possible mechanism for the differences in the localization patterns.
FIGURE 4.
Xiwi1 localization is partially nuclear, while Xili is exclusively present in the cytoplasm. (A) Equivalents of (T) one total stage VI oocyte, (C) one cytoplasm, (N) one nucleus, and (Nx2) two nuclei were resolved on 10% SDS-PAGE and analyzed by Western blot. PARN was used to assess the content of the nuclear fraction, and CPEB was used as a marker of the cytoplasm. Xili appears as a doublet due to better resolution of high-molecular-weight proteins compared to Figure 1C. PARN was used to assess the content of the nuclear fraction, and CPEB was used as a marker of the cytoplasm. (B) Predicted (NES) nuclear export signals were identified in the coding sequence of both Xiwi1 (616MGGELWSVEI625) and Xili (258LPVKLAQTVEL268, 304MRRVFKILDLKL313, 490MKDLTQQIHL499, 701LGGELWGVDI710), according to the consensus (L/M)-X2-3-(L/F)- X2-3-(L/I/V)-X-(L/I) (Kutay and Guttinger 2005). One putative nuclear localization signal (NLS) was identified in the coding sequence of Xiwi1 (4RARARARGRARG15) using NLSdb (Nair et al. 2003).
Xiwi1 interacts specifically with a subpopulation of small RNAs
Piwi family proteins have been recently shown to bind a specific subset of the small noncoding RNA population called piRNAs. While there appears to be no sequence conservation among piRNAs, they are 21–30 nt long, and their one defining feature is their affinity for Piwi proteins (Aravin et al. 2006; Girard et al. 2006; Lau et al. 2006; Kuramochi-Miyagawa et al. 2008). A number of distinct groups of small RNAs can be readily detected in Xenopus oocytes, including several discrete bands around 30 nt and 40 nt in length (Fig. 5, total small RNA lane). (The 30-nt fraction in total RNA corresponds to tens of thousands of discrete sequences mapping to unique and repetitive regions of the X. tropicalis genome [J. Armisen, M. Gilchrist, A. Wilczynska, N. Standart, and E.A. Miska, in prep.].) Therefore, we examined whether Xenopus Piwi proteins bind a particular subpopulation of these small RNAs in the oocyte. We extracted RNA coimmunoprecipitated together with Xiwi1 and submitted it to 5′-end labeling and resolved it on a denaturing polyacrylamide gel. Indeed, the RNA fraction immunoprecipitated with the Xiwi1 antibody was enriched in a population of RNA ∼30 nt long compared to RNA extracted from a control immunoprecipitation using preimmune serum. This result confirms that Xiwi1 protein is a Piwi family member.
FIGURE 5.
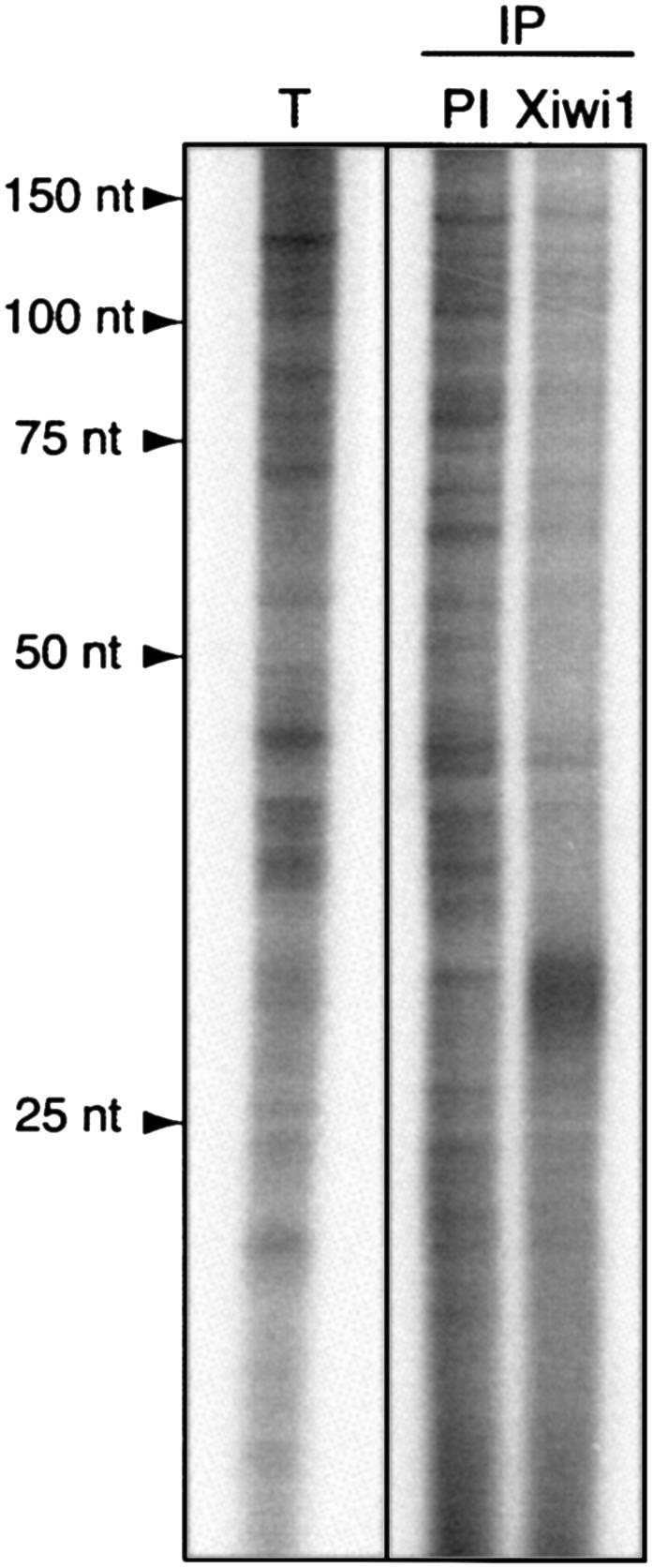
Xiwi1 interacts specifically with RNAs ∼30 nt in length. RNAs immunoprecipitated with an antibody directed against Xiwi1 and the corresponding preimmune serum and total small RNA were extracted and 32P-end-labeled. A specific fraction migrating at ∼30 nt is enriched in anti-Xiwi immunoprecipitates. (IP) Immunoprecipitated samples; (PI) immunoprecipitation with preimmune serum corresponding to the anti-Xiwi serum. Total small RNA was isolated using the mirVana kit (Ambion).
DISCUSSION
Most reports to date have focused on the expression of Piwi family members in the male germline. However, several publications have demonstrated the presence of these proteins in the female germline in Drosophila as well as in vertebrates (Cox et al. 2000; Houwing et al. 2007, 2008; Ro et al. 2007; Aravin and Bourc'his 2008; Tam et al. 2008; Watanabe et al. 2008). Here we report for the first time the expression of two members of the Piwi family in the Xenopus female germline. We have identified four Piwi family members altogether in Xenopus and demonstrated that two, namely, Xiwi1b and Xili, are expressed in the oocyte and early embryo. We show that Xiwi1 is a bona fide Piwi family member by demonstrating that the protein specifically binds ∼30-nt-long RNAs, in keeping with observations in other vertebrates (Aravin et al. 2006; Girard et al. 2006; Kuramochi-Miyagawa et al. 2008).
Our findings contrast with reports of Piwi protein expression in the mouse female germline, where expression of only the Xili ortholog Mili was observed in oocytes, while Miwi, Miwi2, and Mili were all detected in mouse testis (Aravin et al. 2008; Watanabe et al. 2008). On the other hand, our data are in agreement with those of Houwing and colleagues in zebrafish. Ziwi, an ortholog of Miwi and Xiwi1, is expressed in both oocytes and testis (Houwing et al. 2007), as well as Zili, the only other zebrafish Piwi protein, reported recently (Houwing et al. 2008). Both Ziwi and Zili are required for germ cell maintenance and differentiation, bind piRNAs, and participate in a conserved amplification ping-pong loop, first described in Drosophila.
Few protein partners have been described for Piwi proteins. Indeed, Ziwi protein is found in complexes not larger than 300 kDa in oocytes (Houwing et al. 2007), while in testis, the rat ortholog of Xiwi—Riwi—is also present in small complexes (50–160 kDa) and cofractionates with rRecQ protein, although no evidence was given of direct interaction between the two proteins (Lau et al. 2006). Additionally, both Miwi and Mili coimmunoprecipitate with MHV, another protein essential for spermatogenesis (Kuramochi-Miyagawa et al. 2004). In Drosophila, Piwi binds the heterochromatin 1a protein (Brower-Toland et al. 2007). Finally, it was recently reported that Zili and Ziwi proteins interact in zebrafish oocytes, as demonstrated by coimmunoprecipitation (Houwing et al. 2008).
Our data partly support these findings. We detect the majority of Xiwi1 protein in small complexes up to 200 kDa in size that comigrate with the initiation factor eIF4E1a. Direct association of testis Miwi with the cap-binding complex on m7GTP-Sepharose has been reported (Grivna et al. 2006). However, in a similar assay, we do not find evidence of any significant interaction between Xenopus Xiwi1 or Xili and the cap-binding complex in the oocyte. We do note that Grivna et al. (2006) only saw a proportion of total Miwi bind to the cap-binding complex. The difference may also reflect the unique mode of translational regulation in oocytes, in which many maternal mRNAs are stored in a deadenylated and translationally silent state (Seydoux 1996). While translation is also tightly regulated in spermatocytes, these cell-type-specific differences may account for the discrepancy between our observations and those made in mouse testis.
Importantly, the two Xenopus Piwi proteins fractionate with different sized complexes in gel filtration chromatography, indicating different or additional protein partners for Xili compared to Xiwi1. While it cannot be excluded that they interact, it is clear that the majority of Xili and Xiwi are in distinct complexes. In zebrafish, only a small proportion of Zili and Ziwi coimmunoprecipitate (Houwing et al. 2008), in line with our observation.
We detect a minor fraction of Xiwi1 in gel-filtration fractions of much larger molecular weight (∼2 MDa), suggesting that the protein is at least transiently associated with large ribonucleoprotein complexes. Strikingly, in these large fractions, Xiwi1 comigrates with the previously characterized CPEB RNP complex (Minshall et al. 2007), and this association occurs through RNA. Piwi proteins have been previously shown to associate with mRNP complexes and polysomes in an RNA-dependent manner in mouse testis (Grivna et al. 2006). It remains to be established whether Xiwi1 detected in these large complexes in oocytes is associated with translationally repressed mRNAs as cofractionation with the CPEB complex would suggest. We note, however, that tethering of Xiwi1 or Xili to the 3′-UTR of firefly luciferase mRNA in the MS2-tethered assay showed no significant effect on expression of the reporter mRNA in Xenopus oocytes as compared with Xp54, a well-characterized translational repressor (Minshall and Standart 2004; Minshall et al. 2007; data not shown). These observations confirm and extend those of Pillai et al. (2004), who examined the effect of tethered Hiwi on reporter RNA in HeLa cells, which lack an endogenous Piwi/piRNA pathway.
There are conflicting reports as to the subcellular localization of Piwi proteins. In Drosophila, while Piwi is distributed between the nucleus and cytoplasm (Cox et al. 2000; Megosh et al. 2006; Saito et al. 2006), Aub and Ago3 are predominantly cytoplasmic as well as localizing in the germline granule called nuage (Harris and Macdonald 2001; Brennecke et al. 2007; Gunawardane et al. 2007), leading to the speculation of compartmentalized function of the proteins. In particular, Piwi would be responsible for the nuclear import of piRNAs and mediate epigenetic events (Klattenhoff and Theurkauf 2008). In mammals, Miwi and Mili are localized in cytoplasmic granules, when present in ovary and testis (Grivna et al. 2006; Houwing et al. 2007; Aravin et al. 2008; Kuramochi-Miyagawa et al. 2008), while Miwi2 in testis is nuclear (Aravin et al. 2008), as are the human Piwi-like proteins in HEK 293 cells (Sugimoto et al. 2007). For Piwi proteins to fulfill their epigenetic role, they presumably at least temporarily reside in the nucleus, while the current model of piRNA biogenesis would require their presence in the cytoplasm.
Our data support the notion that Piwi protein family members may shuttle between the nucleus and the cytoplasm. One previous report has described the presence of NESs and NLSs in members of the human Piwi protein family; however, the investigators noted nuclear localization of all four PIWIL proteins, while only two contain consensus NLS sequences (Sugimoto et al. 2007). We note that the presence of the NLS in Xiwi orthologs is conserved among all vertebrates and that, although Xili proteins do not appear to contain NLSs, they possess multiple NES sequences. While the localization of the human proteins was observed for ectopically expressed tagged constructs in HEK293T cells in which there is no evidence for expression of any of the proteins, our observations are for endogenous protein in the Xenopus oocyte. Interestingly, Ziwi is cytoplasmic and undetectable in the oocyte nucleus by immunohistochemistry, so perhaps the residence time in this cellular compartment is very transient (Houwing et al. 2007), while Zili has a dynamic nucleocytoplasmic distribution in oocytes, concentrating in the nucleus as oogenesis progresses (Houwing et al. 2008). In this regard, our data are not in agreement with those in zebrafish, as we find Xiwi1 but not Xili in the nuclear fraction of stage VI oocytes. Altogether, it is intriguing to note the similarities, and differences, in the number of and localization of Piwi proteins in vertebrate germline cells, possibly reflecting their relatively rapid evolution.
In conclusion, our observations extend the repertoire of Piwi proteins found in vertebrate female germlines, underscoring the conservation and importance of the cellular mechanisms they are involved in.
MATERIALS AND METHODS
Plasmid constructs
The X. tropicalis Xiwi1 open reading frame (ORF) was amplified by PCR (Pwo polymerase; Roche Applied Sciences) from IMAGE clone number 7687655 and inserted into the BamHI site of the pET15b vector. The clone contained a 2-nt deletion at positions 1280–1281 of the coding sequence. The missing nucleotides (AT) were reinserted by site-directed mutagenesis using the Stratagene QuikChange Mutagenesis kit. The Xili ORF was amplified by PCR from IMAGE clone number 7676520, which contained the full-length coding sequence of the protein, and inserted into the NdeI site of the pET15b vector. pET-Xiwi_Stop and pET-Xili_Stop were generated by introduction of termination codons (TAA and TAG, respectively) at Q359 and K360, respectively, using the QuikChange Mutagenesis kit.
Recombinant protein expression
Full-length Xiwi1 and Xili, as well as truncated versions of both proteins, Xiwi_Stop and Xili_Stop, were expressed in BL21-Rosetta (for Xiwi and Xiwi_Stop) and BL21-Star (Xili and Xili_Stop). Full-length recombinant protein was subsequently used to analyze reactivity of Xiwi1 and Xili antisera, while the Xili-Stop truncated protein was used for purification of the Xili antiserum.
Antibody preparation and purification
The Xiwi1 antibody was raised against the peptide RGGRRRDFHDLGIN, and the Xili antibody was raised against the peptide SDTTQRDPSDSRNV (Sigma-Genosys). The Xiwi1 antibody was used in all experiments as crude serum. The Xili antibody was purified using 10 μg of blotted recombinant N-terminally truncated Xili protein (Xili_Stop), as previously described (Minshall and Standart 2004).
Xenopus oocyte, embryo, and testis lysate preparation
Isolation, staging, handling, lysate preparation, and enucleation of Xenopus laevis oocytes was performed as previously described (Minshall et al. 2007). X. tropicalis testes were homogenized in RIPA buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS, 1× protease inhibitors [Roche Applied Sciences]).
Western blotting
SDS-PAGE and Western blotting were performed as described before (Minshall and Standart 2004; Minshall et al. 2007) on 10% and 15% gels. The primary antibodies used were Xiwi1 (1:1000) (this study); Xili (1:100) (this study); CPEB (1:2000) (Wilczynska et al. 2005); PARN (1:2000) (Copeland and Wormington 2001); eIF4E (1:10000) (Minshall et al. 2007); Xp54 (1:1000) (Minshall and Standart 2004); actin (1:1000, A2066; Sigma); and MIWI (1:1000) (Deng and Lin 2002).
Cap-binding assay
Stage III/IV Xenopus oocyte lysate (80 μL) was applied to 20 μL of packed m7GTP Sepharose beads (Amersham Biosciences) equilibrated in 750 μL of HKE buffer (50 mM HEPES [NaOH] at pH 7.4, 150 mM KCl, 0.5 mM EDTA, 0.5 mM EGTA, 0.1% [v/v] β-mercaptoethanol, 1× Complete EDTA-free protease inhibitors [Roche Applied Science]) and incubated for 1 h with agitation at 4°C and then washed with HKE buffer. Proteins were eluted from the resin with 0.1 mM GTP and then with 70 μM m7GpppG in HKE buffer and finally with 1× SDS loading buffer.
Immunoprecipitation of Xiwi1 complexes and small RNA extraction
Xiwi1 antiserum (25 μL) was bound to 10 μL of protein A-Sepharose beads (GE Healthcare) equilibrated in 1 mL of NET buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA at pH 8.0, 0.25% gelatin, 0.02% NaN3) supplemented with 1 mM DTT, 80 U/mL RNase inhibitor for 6 h at 4°C with agitation. Precleared X. tropicalis oocyte lysate (40 μL; stage V/VI) was incubated with serum-bound beads for 4 h at 4°C in NET buffer. Beads were washed three times in NET buffer supplemented with NaCl to a final concentration of 500 mM. RNA was extracted from bound beads with 250 μL of TNES (0.1 M Tris at pH 7.5, 0.3 M NaCl, 5 mM EDTA, 2% SDS) containing 200 μg/mL proteinase K for 30 min at 50°C, followed by phenol/chloroform extraction and ethanol precipitation in the presence of glycogen. Total RNA was extracted from whole oocyte lysate in the same fashion and further purified using the mirVana kit (Ambion) according to the manufacturer's instructions to enrich for small RNAs.
5′-End labeling and electrophoresis of immunoprecipitated RNA
RNA extracted from immunoprecipitated complexes was dephosphorylated using calf intestinal phosphatase (NEB) followed by 5′-end labeling with [γ-32P]ATP using T4 polynucleotide kinase (NEB) according to the manufacturer's instructions. Unincorporated nucleotides were removed from labeled RNA by purification on an Illustra G-25 microspin column (GE Lifesciences).
ACKNOWLEDGMENTS
We are grateful to Rachel Allison for the gel filtration Western blots. Staged embryo samples were a generous gift from Francis von Horck. We thank Jim Smith and David Simpson for help and materials, Haifan Lin for the Miwi antibody, and Mike Gilchrist for assistance with the Gurdon Institute EST database and helpful discussions. This work was supported by the Biotechnology and Biological Sciences Research Council.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1422509.
REFERENCES
- Allison R., Czaplinski K., Git A., Adegbenro E., Stennard F., Houliston E., Standart N. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA. 2004;10:1751–1763. doi: 10.1261/rna.7450204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A.A., Bourc'his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes & Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Hannon G.J., Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachdanandam R., Bourc'his D., Scaefer C., Pezic D., Toth K.F., Bestor T., Hannon G.J. A piRNA pathway primed by individual transposons is linked to de novo methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista P.J., Ruby J.G., Claycomb J.M., Chiang R., Fahlgren N., Kasschau K.D., Chaves D.A., Gu W., Vasale J.J., Duan S., et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans . Mol. Cell. 2008;31:67–78. doi: 10.1016/j.molcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila . Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B., Findley S.D., Jiang L., Liu L., Yin H., Dus M., Zhou P., Elgin S.C., Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes & Dev. 2007;21:2300–2311. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Cerutti L., Mian N., Bateman A. Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Copeland P.R., Wormington M. The mechanism and regulation of deadenylation: Identification and characterisation of Xenopus PARN. RNA. 2001;7:875–886. doi: 10.1017/s1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D.N., Chao A., Baker J., Chang L., Qiao D., Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D.N., Chao A., Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Das P.P., Bagijn M.P., Goldstein L.D., Woolford J.R., Lehrbach N.J., Sapetschnig A., Buhecha H.R., Gilchrist M.J., Howe K.L., Stark R., et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Farazi T.A., Juranek S.A., Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Gilchrist M.J., Zorn A.M., Voigt J., Smith J.C., Papalopulu N., Amaya E. Defining a large set of full-length clones from a Xenopus tropicalis EST project. Dev. Biol. 2004;271:498–516. doi: 10.1016/j.ydbio.2004.04.023. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Grivna S.T., Pyhtila B., Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila . Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Harris A.N., Macdonald P.M. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development. 2001;128:2823–2832. doi: 10.1242/dev.128.14.2823. [DOI] [PubMed] [Google Scholar]
- Hock J., Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Houwing S., Berezikov E., Ketting R.F. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulo N., Bairoch A., Bulliard V., Cerutti L., Cuche B.A., de Castro E., Lachaize C., Langendijk-Genevaux P.S., Sigrist C.J. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Simard M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Kalmykova A.I., Klenov M.S., Gvozdev V.A. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Res. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C., Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Yomogida K., Kuroiwa A., Tadokoro Y., Fujita Y., Sato M., Matsuda Y., Nakano T. Two mouse Piwi-related genes: Miwi and Mili. Mech. Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Kimura T., Ijiri T.W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T.W., et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Guttinger S. Leucine-rich nuclear-export signals: Born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Ladomery M., Wade E., Sommerville J. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 1997;25:965–973. doi: 10.1093/nar/25.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Seto A.G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D.P., Kingston R.E. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Lin H., Spradling A.C. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Megosh H.B., Cox D.N., Campbell C., Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Minshall N., Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N., Reiter M.H., Weil D., Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biol. Chem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- Nair R., Carter P., Rost B. NLSdb: Database of nuclear localization signals. Nucleic Acids Res. 2003;31:397–399. doi: 10.1093/nar/gkg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K.M., Saito K., Mori T., Kawamura Y., Nagami-Okada T., Inagaki S., Siomi H., Siomi M.C. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–1922. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L., Meister G. Argonaute proteins: Mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Artus C.G., Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao D., Zeeman A.M., Deng W., Looijenga L.H., Lin H. Molecular characterization of Hiwi, a human member of the Piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–3999. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- Ro S., Song R., Park C., Zheng H., Sanders K.M., Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Nishida K.M., Mori T., Kawamura Y., Miyoshi K., Nagami T., Siomi H., Siomi M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarot E., Payen-Groschene G., Bucheton A., Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G. Mechanisms of translational control in early development. Curr. Opin. Genet. Dev. 1996;6:555–561. doi: 10.1016/s0959-437x(96)80083-9. [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Kage H., Aki N., Sano A., Kitagawa H., Nagase T., Yatomi Y., Ohishi N., Takai D. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem. Biophys. Res. Commun. 2007;359:497–502. doi: 10.1016/j.bbrc.2007.05.136. [DOI] [PubMed] [Google Scholar]
- Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.H., Lee T.C., Weeraratne S.D., Korzh V., Lim T.M., Gong Z. Ziwi, the zebrafish homologue of the Drosophila Piwi: co-localization with Vasa at the embryonic genital ridge and gonad-specific expression in the adults. Gene Expr. Patterns. 2002;2:257–260. doi: 10.1016/s1567-133x(02)00052-2. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between Dcp1 bodies and stress granules. J. Cell Sci. 2005;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- Yin H., Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster . Nature. 2007;450:304–308. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]