Abstract
Dyskeratosis congenita (DC) is an inherited disorder that implicates defects in the biology of telomeres, which are maintained by telomerase, a ribonucleoprotein with reverse transcriptase activity. Like all H/ACA RNAs, the H/ACA domain of nascent human telomerase RNA (hTR) forms a pre-RNP with H/ACA proteins NAF1, dyskerin, NOP10, and NHP2 in vivo. To assess the pre-RNP assembly of hTR mutants that poorly accumulate in vivo, we developed an in vitro system that uses components of human origin. Pre-RNPs were reconstituted with synthetic 32P-labeled RNAs and 35S-labeled proteins produced in rabbit reticulocyte lysate, and immunoprecipitations were carried out to analyze RNP formation. We show that human NAF1 cannot bind directly to the H/ACA domain of hTR, and requires the core trimer dyskerin-NOP10-NHP2 to be efficiently incorporated into the pre-RNP. This order of assembly seems common to H/ACA RNAs since it was observed with snoRNA ACA36 and scaRNA U92, which are predicted to guide pseudouridylation of 18S rRNA and U2 snRNA, respectively. However, the processing H/ACA snoRNA U17 did not conform to this rule, as NAF1 alone was able to bind it. We also provide the first evidence that DC-related mutations of hTR C408G and Δ378-451 severely impair pre-RNP assembly. Integrity of boxes H and ACA of hTR are also crucial for pre-RNP assembly, while the CAB box is dispensable. Our results offer new insights into the defects caused by some mutations located in the H/ACA domain of hTR.
Keywords: telomerase biogenesis, human telomerase RNA, dyskeratosis congenita, H/ACA pre-RNPs, NAF1
INTRODUCTION
Dyskeratosis congenita (DC) is an inherited disorder characterized by a variety of phenotypes, and afflicted subjects usually die of bone marrow failure. DC patients show signs of premature aging, and they are more susceptible to develop cancers. There is evidence that all DC patients have some defects in telomere biology, and that those defects affect the renewing capabilities of hematopoietic stem cells. Furthermore, all DC causal mutations identified to date are found in telomerase components or in telomere stabilizing components (Hockemeyer et al. 2008; Savage et al. 2008; Vulliamy and Dokal 2008; Vulliamy et al. 2008).
Telomerase is a ribonucleoprotein with reverse transcriptase activity responsible for de novo synthesis and maintenance of telomeric DNA, the chromosomal termini (Collins 2006). In vertebrates, the 5′ half of telomerase RNA forms a conserved pseudoknot structure that harbors the telomere template, and the 3′ half evolved as an H/ACA-like domain (Mitchell et al. 1999; Chen and Greider 2004). The H/ACA domain is structurally similar to a large family of small nucleolar RNAs (snoRNAs) and small Cajal body-specific RNAs (scaRNAs). All H/ACA RNAs share a common fold consisting of a hairpin followed by a single-stranded hinge containing box H (consensus sequence ANANNA, where N is any nucleotide), another hairpin, and a single-stranded tail with the ACA box situated 3 nucleotides (nt) from the 3′ end (Balakin et al. 1996; Ganot et al. 1997). Most H/ACA snoRNAs act as guides for the pseudouridylation of rRNA and localize to nucleoli, while scaRNAs guide the pseudouridylation of spliceosomal small nuclear RNAs and localize to Cajal bodies. The sno/scaRNAs families also include another RNA class characterized by two conserved motifs called boxes C and D; most C/D sno/scaRNAs act as guides for the site-specific 2′-O-methylation of target RNAs (Kiss 2004; Matera et al. 2007).
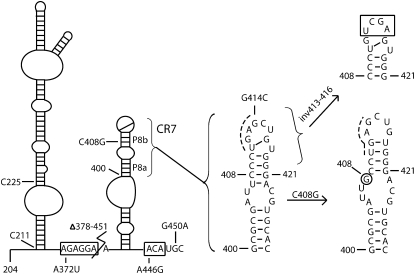
In humans, all H/ACA sno/scaRNAs are encoded within introns of pre-mRNAs and the processed mature RNAs are devoid of a 5′ cap structure (Kiss et al. 2004). In contrast, human telomerase RNA (hTR) is transcribed by RNA polymerase II (pol II) from its own promoter and the mature RNA (451 nt) bears a TMG-cap at the 5′ end. Although the H/ACA domain of hTR does not guide pseudouridylation, this domain is essential for hTR stability and accumulation in vivo (Collins 2006). Within this domain, the terminal loop of the conserved region 7 (CR7) harbors a Cajal body retention signal (CAB box) that is shared by H/ACA scaRNAs, classifying hTR as a scaRNA rather then a snoRNA (Fig. 1). The consensus sequence of the CAB box is ugAG, where the first two residues are varying and the last two are highly conserved (Richard et al. 2003; Jady et al. 2004). The CAB box allows Cajal body localization, but it is not involved in 3′ end formation of the RNA, indicating that the two processes are functionally independent (Theimer et al. 2007). The CR7 also contains key features required for RNP assembly, such as the base pairing between U411 and G417, and an unpaired U418 in the terminal loop. In addition, conservation of base pairing and the internal loop of the P8 stem are also essential for 3′ end processing of hTR (Fu and Collins 2003).
FIGURE 1.
Schematic secondary structure of the H/ACA domain of hTR. Residues are numbered according to full-length hTR. Conserved motifs H and ACA are boxed. Sequence changes made in this study are indicated; they correspond to mutations in conserved box H (A372U), box ACA (A446G), and the CAB box highlighted by a dashed line (G414C and inv413–416), as well as disease-related mutations C408G, G450A, and Δ378–451 deletion. CR7 is enlarged to better show its normal structure, which is altered in the DC mutant C408G.
Human H/ACA RNPs share a common set of core proteins named GAR1, NHP2, NOP10, and dyskerin, a pseudouridine synthase that catalyzes the pseudouridylation reaction (Matera et al. 2007). GAR1 is a late assembly component that is present only in mature, functional RNPs, and it is not detected at the transcription site of an H/ACA snoRNA (Darzacq et al. 2006). In contrast, dyskerin, NHP2, and NOP10 are early assembly components and they interact with nascent H/ACA snoRNA at the transcription site (Darzacq et al. 2006). Formation of these immature and inactive RNPs (or pre-RNPs) requires the assistance of the nuclear assembly factor-1 (NAF1) at the transcription site (Ballarino et al. 2005; Yang et al. 2005; Darzacq et al. 2006; Hoareau-Aveilla et al. 2006). Unlike GAR1, NAF1 is not present in mature H/ACA RNPs and it is not detected in nucleoli and Cajal bodies, the compartments where H/ACA RNPs function (Hoareau-Aveilla et al. 2006; Darzacq et al. 2006). It has been proposed that NAF1 must be replaced by GAR1 to make functional RNPs, but the exchange mechanism remains elusive. In yeast, depletion of Naf1p, Nhp2p, Nop10p, or Cbf5p (the homolog of dyskerin) causes a decrease in levels of H/ACA snoRNAs (Dez et al. 2002, and references therein). Naf1p interacts with the pol II machinery and could be responsible for coupling transcription with pre-RNP formation (Fatica et al. 2002; Ballarino et al. 2005; Yang et al. 2005). These functional interactions are likely conserved in humans because human NAF1 can complement a yeast strain depleted of Naf1p (Hoareau-Aveilla et al. 2006). Dyskerin was also found associated with an unspliced nascent H/ACA snoRNA, which requires the presence of NAF1 to accumulate, just as hTR does (Richard et al. 2006; Hoareau-Aveilla et al. 2006).
Some mutations found in the H/ACA domain of hTR are associated with the autosomal dominant form of DC. One mutation leading to DC is the C408G tranversion, which is located in helix P8b of CR7 (see Fig. 1); this mutation alters the structure of CR7, resulting in the loss of proper processing signals (Theimer et al. 2003, 2007). Another mutation in hTR leading to DC is the deletion of nucleotides 378–451 (Δ378–451), which removes the entire 3′ stem, positioning box H at the 3′ extremity. Accumulation of mutants C408G and Δ378–451 is highly reduced in vivo, as shown by Northern hybridization and RT-PCR experiments. However, normal telomerase activity was obtained for both mutants in an in vitro system, or when RNAs were overexpressed in hTR negative cells, indicating that these two DC mutations affect the biogenesis of telomerase rather than its activity. Other mutations in hTR are related to aplasic anemia (AA), such as the G450A mutant that was found to accumulate near wild-type level in vivo (Vulliamy et al. 2001; Fu and Collins 2003; Marrone et al. 2004; Ly et al. 2005).
NAF1, dyskerin, NOP10, and NHP2 are protein components of H/ACA pre-RNPs. We have developed an in vitro system to test their interaction with the H/ACA domain of hTR and its mutant derivatives. Here we show that NAF1 cannot bind the H/ACA domain of hTR by itself, and requires the presence of the trimer dyskerin-NOP10-NHP2 to be efficiently incorporated into the pre-RNP. We also show that pre-RNP assembly is impaired by some DC-related mutations.
RESULTS AND DISCUSSION
To assess the interactions between human H/ACA proteins, as well as RNP formation of the H/ACA domain of hTR, we used 35S-labeled proteins produced in rabbit reticulocyte lysate (RRL) and 32P-labeled RNA fragments of hTR transcribed in vitro, and we carried out a series of immunoprecipitation (IP) experiments against dyskerin or NAF1.
The precursor complex of human H/ACA proteins assembles correctly in RRL
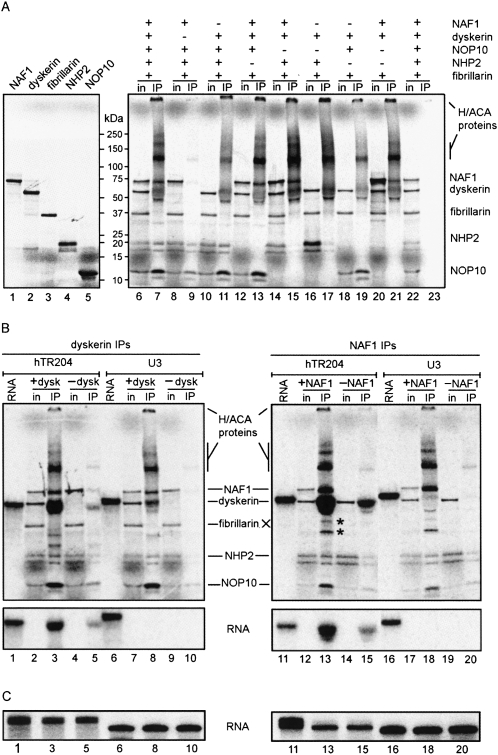
We first analyzed the expression level of each protein-encoding plasmid individually in RRL; the proteins were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE) (Fig. 2A, lanes 1-5). Fibrillarin, a protein associated with box C/D sno/scaRNAs, was used as a negative control for IPs. Human NOP10 failed to accumulate in RRL (data not shown). The RRL is known to work best with proteins in the range of 15-100 kDa, while smaller proteins like human NOP10 may be targeted for degradation by an ubiquitin-dependent pathway (TenHarmsel 2004). To circumvent this problem the HA tag was inserted at the N-terminus of human NOP10; because HA-NOP10 accumulated well in RRL (Fig. 2A, lane 5), we used this construct for all experiments and we hereafter refer to it as NOP10.
FIGURE 2.
The tetramer NAF1-dyskerin-NOP10-NHP2 assembles specifically with hTR204. (A) Individual proteins were translated in vitro in the presence of 35S-labeled methionine and analyzed by PAGE (left). For dyskerin IPs (right), 35S-labeled proteins were cotranslated; the presence or absence of each protein in the RRL mixture is indicated by (+) or (−), respectively. Input of translated proteins (in) and immunoprecipitated products (IP) were separated by PAGE. Molecular weight markers (kDa) are shown between the two panels. Control IP with beads alone is shown (lanes 22,23). Note that dyskerin recovered from IPs gives a fuzzy signal. High molecular weight multimers of H/ACA proteins are also observed. (B) Dyskerin IPs (left) and NAF1 IPs (right) of RRL mixtures containing 35S-labeled proteins and 32P-labeled hTR204 (lanes 1-5,11-15) or U3 (lanes 6-10,16-20) RNAs were analyzed by PAGE. Because RNAs co-migrate with dyskerin in these protein gels, input lanes (in) did not contain the 32P-labeled RNAs to better discern the 35S-labeled proteins. IPs with RRL mixtures lacking either dyskerin (-dysk) or NAF1 (-NAF1) were used as negative controls. Labeled hTR204 (lanes 1,11) and U3 (lanes 6,16) RNAs not incubated in RRL are shown. Bands marked by an asterisk (*) correspond to degradation products of dyskerin/NAF1, not to fibrillarin, which was omitted form NAF1 IPs (right). (Bottom) The 32P signal from the protein gels presented in the top panels; they were obtained by blocking the 35S signal with a transparency. (C) Labeled RNAs from input (lanes 1,6,11,16) and IP supernatants (lanes 3,5,8,10,13,15,18,20) were analyzed separately on 8% denaturing polyacrylamide gels. Lanes are numbered in accordance with the corresponding IPs shown in (B).
We first examined if the order of assembly of human H/ACA proteins occurs as previously reported in other heterologous systems (Wang and Meier 2004; Darzacq et al. 2006); we carried out IPs against dyskerin on RRL mixtures containing all co-translated proteins, or different combinations of those (Fig. 2A, lanes 6-23). IPs showed that all four H/ACA proteins (dyskerin, NOP10, NHP2, and NAF1) can form a tetramer, and that the C/D protein fibrillarin was not co-immunoprecipitated (Fig. 2A, lane 7). In the absence of NAF1 the core trimer is readily formed (Fig. 2A, lane 11), indicating that NAF1 is not required for its assembly. Formation of this core trimer is mediated by NOP10 because omission of NOP10 in the RRL impairs NHP2 co-immunoprecipitation with dyskerin, regardless of the presence of NAF1 (Fig. 2A, lanes 15,17). Conversely, NOP10 was co-immunoprecipitated with dyskerin from an RRL mixture lacking NHP2 (Fig. 2A, lane 19). Co-immunoprecipitation of dyskerin and NAF1 in the absence of NHP2 and/or NOP10 indicated that NAF1 interacts directly with dyskerin (Fig. 2A, lanes 13,15,21). These data suggest that the precursor complex formed by human proteins dyskerin, NOP10, NHP2, and NAF1 can be assembled in vitro in the same way as previously reported (Wang and Meier 2004; Darzacq et al. 2006). The weak IP background found with a mixture lacking dyskerin is likely caused by low levels of endogenous H/ACA proteins present in RRL (see Fig. 2A, lane 9). Indeed, this background was not observed in control IPs with beads alone (Fig. 2A, lane 23) nor with anti-myc monoclonal antibody (data not shown). It was not possible to determine the molar ratio of co-immunoprecipitated proteins because H/ACA protein complexes of high molecular weight were seen in every IP. Disruption of these protein complexes with various harsher conditions prior to PAGE failed (data not shown), suggesting that H/ACA proteins can form highly stable complexes, in line with previous observations (Henras et al. 2004; Leulliot et al. 2007).
H/ACA proteins of the pre-RNP assemble specifically on the H/ACA domain of hTR
To test the ability of the tetramer complex to assemble with the H/ACA domain of hTR, we used a radiolabeled RNA fragment encompassing residues 204–451 of hTR (hTR204) (see Fig. 1). When RRL mixtures containing all proteins were subjected to dyskerin or NAF1 IPs, hTR204 co-immunoprecipitated efficiently (Fig. 2B, bottom, lanes 3,13, respectively). Specificity of this interaction was confirmed using the box C/D snoRNA U3 as negative control; U3 does not bind H/ACA proteins and it was not co-immunoprecipitated in our assays (Fig. 2B, lanes 8,18). Thus, H/ACA pre-RNP assembly occurs specifically in our RRL system. RNAs from IP supernatants were analyzed separately on 8% denaturing polyacrylamide gels; there were no differences in RNA quantities or integrity that could have explained the reduced RNA levels in the IPs (Fig. 2C). The background of hTR204 seen in the IPs carried out in absence of either dyskerin (Fig. 2B, lane 5) or NAF1 (Fig. 2B, lane 15) correlates with the low background level of H/ACA proteins in RRL, as shown in the top panels of Figure 2B (lanes 5,10,15,20) and in Figure 2A (lane 9). We also carried out dyskerin IPs and NAF1 IPs with RRL mixtures containing only labeled hTR204 (no translated protein) and obtained the same background levels of hTR204, however, control IPs made with anti-NAF1 beads incubated with labeled RNA alone (no RRL present) showed virtually no background (data not shown). These control experiments revealed that RRL contains traces of H/ACA proteins that can interact together (see Fig. 2A) and with the radiolabeled RNA, thus producing a low level background in our experiments.
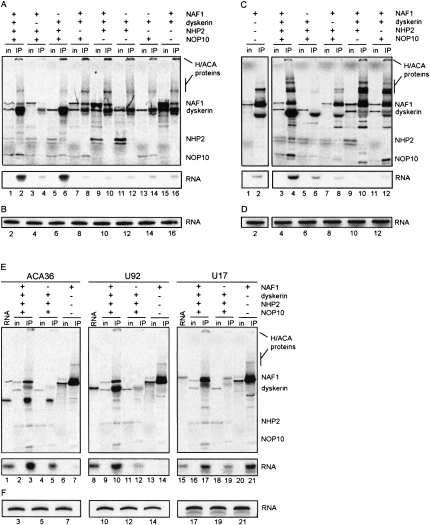
NAF1 assembly on the H/ACA domain of hTR is mediated by the core trimer
A series of dyskerin IPs indicated that the core trimer dyskerin-NOP10-NHP2 is necessary and sufficient to bind hTR204 in RRL (see Fig. 3A), in accordance with the report of Wang and Meier (2004) for the rat counterparts. However, NAF1 was not included in these previous experiments, and here we further found that NAF1 could be co-immunoprecipitated with hTR204 and dyskerin only when NOP10 and NHP2 were present (Fig. 3A). These results suggest that NAF1 requires the core trimer to be assembled on the H/ACA domain of hTR. Because NAF1 interacts with dyskerin in absence of RNA (Fig. 2A), dyskerin IPs did not clearly demonstrate the association of NAF1 with hTR204. To clarify this point, we carried out NAF1 IPs with radiolabeled hTR204 incubated in RRL mixtures bearing all proteins or different combinations of those (Fig. 3C). We observed that NAF1 cannot bind directly to hTR204 (Fig. 3C, lanes 1,2), however, the RNA was efficiently co-immunoprecipitated with NAF1 when all the other H/ACA proteins were present in the translation mixture (Fig. 3C, lanes 3,4). The omission of any protein that forms the core trimer dyskerin-NOP10-NHP2 resulted in a RNA signal weaker or equal to the background level obtained in the absence of NAF1 (Fig. 3C, bottom, lanes 5-12). RNAs from IP supernatants were analyzed separately, and there was no sign of degradation to explain the reduced RNA levels in IPs (Fig. 3B,D). Thus, NAF1 cannot bind directly to hTR204 and needs to associate with the core trimer to be incorporated in the RNP. This finding is supported by the lack of conserved electrostatic positive patch in NAF1 that could define an RNA-binding motif (Leulliot et al. 2007). Our data differ from gel retardation experiments showing that bacterially produced yeast Naf1p binds directly to snR36 snoRNA (Fatica et al. 2002). To determine if our results were specific to hTR, we tested different H/ACA RNAs in the RRL assay. We used the human homolog of yeast snR36, ACA36, which is predicted to guide pseudouridylation of 18S rRNA (Kiss et al. 2004), U92, a human scaRNA predicted to guide pseudouridylation of U2 snRNA (Darzacq et al. 2002), and U17, a human snoRNA involved in early pre-rRNA processing reactions (Atzorn et al. 2004, and references therein). As expected, ACA36, U92, and U17 co-immunoprecipitated with NAF1 when incubated in a mixture containing all the H/ACA proteins (Fig. 3E, bottom, lanes 3,10,17). However, when these RNAs were incubated with NAF1 alone, only U17 co-immunoprecipitated with NAF1 (Fig. 3E, lane 21), in marked contrast with ACA36 and U92, which were virtually absent from immunoprecipitates (Fig. 3E, bottom, lanes 7,14; the 32P signal for ACA36 and U92 RNAs was equal or lower than the background level seen in the IPs carried out with the mixture lacking NAF1). Identical results were obtained with human snoRNA ACA36B, a paralog of ACA36 (data not shown). Analysis of RNAs in IP supernatants ruled out the possibility that absence of RNAs in immunoprecipitates was due to degradation (Fig. 3F). Overall, our data suggest that direct binding of human NAF1 to H/ACA RNAs is not a common theme, and U17 could be the exception that proves the rule. We also noted that NAF1 IPs with HeLa cell extracts co-immunoprecipitated much more U17 than other H/ACA sno/scaRNAs (Hoareau-Aveilla et al. 2006). Intron-encoded U17 is unique in that it has no pseudouridylation target and it is one of the few snoRNAs that participate in pre-rRNA processing reactions. Moreover, U17 has structural features not found in other H/ACA snoRNAs, and it has been proposed that it might interact with U17-specific proteins (Dragon et al. 2000; Atzorn et al. 2004). One possible explanation for our results is that these features allow a distinct order of assembly of the U17 pre-RNP. Alternatively, it is possible that an unidentified U17-specific protein present in the RRL could mediate the interaction between U17 and NAF1.
FIGURE 3.
Binding of NAF1 to hTR204 requires the trimer dyskerin/NOP10/NHP2. (A,C) RRL mixtures containing 35S-labeled proteins and 32P-labeled hTR204 were subjected to (A) dyskerin IP or (C) NAF1 IP and analyzed by (top) PAGE. The presence or absence of each protein in the RRL mixtures is indicated by (+) or (−), respectively. Input lanes (in) did not contain the 32P-labeled hTR204. (C, left,right) Come from the same gel exposition. (Bottom) Only the 32P signal of the protein gel (see legend to Figure 2B for details). (B,D) Labeled hTR204 recovered from supernatants of dyskerin or NAF1 IPs (see A and C, respectively) was analyzed on 8% denaturing polyacrylamide gels. Lanes are numbered in accordance with the corresponding dyskerin and NAF1 IPs shown in (A) and (C), respectively. (E) RRL mixtures containing 35S-labeled proteins and 32P-labeled ACA36, U92, or U17 were subjected to NAF1 IP and analyzed by (top) PAGE. The presence or absence of each protein in the RRL mixtures is indicated by (+) or (−), respectively. Input lanes (in) did not contain the radiolabeled RNAs. Labeled ACA36 (lane 1), U92 (lane 8), and U17 (lane 15) RNAs not incubated in RRL are shown. (Bottom) Only the 32P signal of the protein gels presented in top panels. (F) Labeled ACA36, U92, or U17 recovered from supernatants of NAF1 IPs shown in (E) were analyzed on 8% denaturing polyacrylamide gels. Lanes are numbered in accordance with the corresponding IPs shown in (E).
At the level of protein-protein interactions, we did not observe any direct interaction between NAF1 and NHP2 in our system. As demonstrated in Figure 2, endogenous rabbit H/ACA proteins are present in RRL, and this could explain why trace amounts of in vitro translated yeast Nhp2p were pulled down with GST-Naf1p (Fatica et al. 2002), and why small amounts of human NAF1 co-immunoprecipitated with rat HA-NOP10 in the presence of rat NHP2 (Darzacq et al. 2006).
Taken together, our data indicate that the core trimer dyskerin-NOP10-NHP2 is necessary and sufficient to bind the H/ACA domain of hTR, and that NAF1 requires the core trimer to be incorporated in the pre-RNP; these results are in agreement with in vivo studies showing that all four proteins were localized at the transcription site of an H/ACA snoRNA, and the observation that knocking down dyskerin impaired localization of NAF1 at the transcription site (Darzacq et al. 2006).
The Cajal body localization signal is not required for pre-RNP assembly
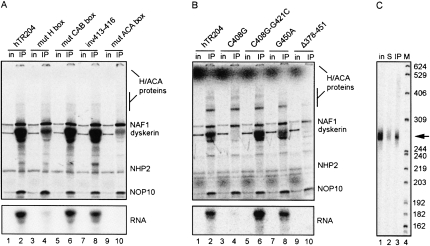
We next assessed pre-RNP assembly of hTR204 in the context of a mutated H box (AGAGGA→UGAGGA), a mutated ACA box (ACA→GCA), a mutated CAB box (UGAG→UGAC) or inverted nucleotides 413–416 (inv413–416) of the CR7 terminal loop encompassing the last two nucleotides of the CAB box (UGAGCU→UGUCGA). This inversion maintains the U411-G417 base pair, and the G418 bulge that are required for processing and accumulation of the H/ACA domain in vivo (Theimer et al. 2007). Since our experiments indicated that NAF1 requires the trimer dyskerin-NOP10-NHP2 to be efficiently assembled on the H/ACA domain of hTR (see Fig. 3C), we decided to carry out NAF1 IPs on RRL mixtures containing all proteins incubated with wild-type hTR204 or its mutant derivatives. Mutations in box H or box ACA severely impaired RNP formation (Fig. 4A, bottom, lanes 4,10, respectively). As mutants of boxes H or ACA do not accumulate in vivo (Mitchell et al. 1999), these results provide the first evidence linking accumulation defects with impaired or inefficient pre-RNP assembly. In contrast with mutations in conserved motifs H and ACA, the mutated CAB box and the inversion of the CR7 terminal loop (inv413-416) did not affect pre-RNP assembly (Fig. 4A, bottom, lanes 6,8), consistent with the in vivo study showing that H/ACA processing and accumulation of hTR are independent of the CAB box element (Theimer et al. 2007). RNAs from IP supernatants were analyzed on denaturing polyacrylamide gels; there was no RNA degradation that could account for the reduced RNA levels in the IPs (data not shown).
FIGURE 4.
Some mutations in hTR204 impair pre-RNP assembly. (A) PAGE analysis of NAF1 IPs conducted on RRL mixtures containing 35S-labeled proteins and 32P-labeled hTR204 or its derivatives bearing mutations in conserved motifs (see text for details). Input lanes (in) did not contain the 32P-labeled RNAs. (Bottom) Only the 32P signal of the protein gel (see legend to Figure 2B for details). (B) NAF1 IPs conducted and analyzed as in (A) with 32P-labeled hTR204 or its derivatives bearing disease-related mutations C408G, G450A, and Δ378-451. The double mutant C408G-G412C restores base pairing (see text for details). (C) 32P-labeled hTR204 (arrow) recovered from a NAF1 IP (lane 3) and the IP supernatant (lane 2) was analyzed on an 8% sequencing gel. Untreated 32P-labeled hTR204 (in) was run in parallel (lane 1). Sizes of 32P-labeled DNA markers (lane 4) are indicated in nt on the right.
DC mutations in the H/ACA domain of hTR impair pre-RNP assembly
A number of disease-causing mutations map to the H/ACA domain of hTR. To determine if some of these mutations affect pre-RNP assembly, we analyzed hTR H/ACA pre-RNP assembly of DC-related mutants C408G and the 3′ stem deletion (Δ378–451) in our RRL system, as well as the AA-related mutation G450A (see Fig. 1). The G450A mutant showed no detectable defects for RNP assembly (Fig. 4B, bottom, lane 8). Such result was expected because the G450A mutant accumulates normally in vivo (Ly et al. 2005), suggesting that the first steps of telomerase biogenesis occur correctly with this mutant. In contrast, mutations C408G and Δ378–451 greatly impaired RNP assembly (Fig. 4B, bottom, lanes 4,10). Note that mutant Δ378-451 corresponds to the 5′ hairpin followed by box H of the H/ACA domain of hTR, a region that apparently lacks essential elements required for RNP formation. In contrast, a fragment encompassing only the 3′ hairpin followed by box ACA formed an RNP in RRL and could be immunoprecipitated (data not shown), in accordance with previous studies showing that this region of hTR contains critical assembly elements (Dragon et al. 2000). The defect caused by the C408G point mutation, which disrupts the CR7 terminal helix and loop structure (Theimer et al. 2003, 2007), could be specifically overcome by the compensatory mutation G421C (Fig. 4B, bottom, lane 6) that restores the 408:421 base pairing and the structure of the CR7, as well as the activity of telomerase (Ly et al. 2005). Again, RNAs from all IP supernatants were analyzed separately on denaturing polyacrylamide gels; there were no differences in RNA quantities or integrity that could have explained the reduced RNA levels in the IPs (data not shown).
Our results are in accordance with the study of Theimer et al. (2007) who showed that the C408G mutant localized in dot-like structures presumed to be transcription sites of hTR, as if the mutant could not be processed into mature RNPs. However, when Theimer et al. (2007) expressed only the H/ACA domain portion encompassing nt 210–451 from a β-globin intron, the C408G mutant RNA got processed up to C225 at its 5′ end, and it formed RNPs that localized in Cajal bodies. In this case, 5′ end processing of the fragment promotes an alternative structure of the 5′ stem that may be assembled differently than in the context of full-length hTR. We verified the integrity of the hTR204 fragment in our RRL system: RNAs were recovered from NAF1 IPs and analyzed on a sequencing gel. Trimming of hTR204 RNA up to C225 would be easily visualized on a sequencing gel, as the processed fragment would migrate faster than the input RNA. As shown in Figure 4C (cf. lanes 1,2 and 3), hTR204 is not processed in our system. Therefore, this difference in 5′ end processing of the H/ACA domain could account for the RNP assembly/accumulation discrepancy between our study and the study of Theimer et al. (2007). It is not known if the 5′ processed H/ACA domain of hTR mutant C408G expressed from the β-globin intron can be co-immunoprecipitated with H/ACA proteins, or if alternative RNPs are formed in that context. As the Cajal body retention signal is functionally distinct from the processing elements (Theimer et al. 2007), such alternative RNPs could still be imported to Cajal bodies. Another explanation could reside in the pol II machinery that may differ in composition, or have components that favor pre-RNP assembly of intronic sno/scaRNAs versus hTR.
The in vitro experiments reported here nicely complement in vivo studies. Indeed, they present more mechanistic details on the order of interactions between components of a complex that is difficult to study in vivo. More specifically, we showed that some DC-related mutations in the H/ACA domain of hTR impair assembly of the pre-RNP. Overall, our data provide a first glimpse as to why these mutations can cause hTR insufficiency in vivo, leading directly to DC and eventually bone marrow failure.
MATERIALS AND METHODS
DNA constructs
The sequence corresponding to the open reading frame (ORF) of NAF1 was amplified by PCR from a cDNA library derived from a human Namalwa (Burkitt lymphoma) cell line (Strubin et al. 1995) with oligonucleotides NAF1-F (5′-GGGGGATCCACCATGGAGGTAGTGGAGGCCGC-3′) and NAF1-R (5′-TGGGAAGCTTCTAATAGTAAGGTCCAAAATGAGA-3′). The ORF encoding fibrillarin was amplified similarly with oligonucleotides fib-F (5′-GGGGAATTCACCATGAAGCCAGGATTCAGTC-3′) and fib-R (5′-GGGGAATTCTCAGTTCTTCACCTTGGG-3′). The PCR products were cloned into BamHI-HindIII sites and EcoRI site of pcDNA3.1(−) (Invitrogen) to generate plasmids pNAF1 and pFib, which respectively encode untagged NAF1 and fibrillarin. The cDNAs encoding human dyskerin and NHP2 were subcloned from pcDNA-DKC1 and pcDNA-hNHP2 (Pogacic et al. 2000) into the EcoRI site of pcDNA3.1(−) to generate pDKC1 and pNHP2, which encode untagged dyskerin and NHP2, respectively. The ORF of NOP10 was subcloned from pcDNA-hNOP10 (Pogacic et al. 2000) into the EcoRI site of pGADT7 (Clontech) to generate pGAD-NOP10. In vitro transcription/translation of this construct with TNT T7-Quick (see below) produces an N-terminal HA-tagged NOP10 that does not contain the activation domain located upstream of the T7 RNA polymerase promoter of pGADT7. The H/ACA domain of human telomerase RNA was amplified by PCR from phTR206 (Dragon et al. 2000) using oligonucleotide hTR204-F 5′-CCCAAGCTTTAATACGACTCACTATAGGGTACCGGGGACCTGCGGCGGGT-3′ harboring a T7 RNA polymerase promoter (bold letters) flanked by HindIII and KpnI restriction sites, and oligonucleotide hTR204-R (5′-CGGAATTCTGCGCATGTGTGAGCCGAG-3′), which contains an EcoRI site at its 5′ end. The PCR product was digested by HindIII and EcoRI, and inserted into pUC19 to yield phTR204. Site-directed mutagenesis (Higuchi et al. 1988) was performed on phTR204 to generate the mutant derivatives A372U (mutated H box), C408G, C408G-G421C, G414C (mutated CAB box) and inverted nucleotides 413–416 (inv413–416) of the CR7 domain. Single PCR amplification was used to obtain mutants A446G (mutated ACA box), G450A and Δ378–451. The sequence of oligonucleotides used for site-directed mutagenesis are available on request. All PCR products of mutagenized hTR204 where then cleaved with StuI and EcoRI, and swapped with the corresponding fragment of wild-type phTR204. The cDNA encoding human U17a snoRNA was PCR amplified from pHU17(E-B) (Dragon et al. 2000) with oligonucleotides hU17-F (5′-CCCAAGCCTTCCAACGTGGATACACC-3′) and hU17-R (5′-CGGAATTCGGCTGTTTCCTGCATGG-3′) and cloned into restriction sites HindIII and EcoRI of pBluescript SK(−) to yield pBS-U17. The cDNA encoding H/ACA snoRNA ACA36 (Kiss et al. 2004; Lestrade and Weber 2006) was amplified from HeLa genomic DNA with oligonucleotides ACA36-F (5′-AAATTAATACGACTCACTATAGGGAATTCCAAAGTGTTGAGTTCAGTCC-3′; the T7 RNA polymerase promoter sequence is in bold letters) and ACA36-R (5′-GGGAAGCTTTTTGTAGGCTGGGAGGCA-3′), and cloned into restriction sites SmaI and HindIII of pUC19 to generate pACA36. The cDNA encoding human scaRNA U92 (Darzacq et al. 2002; Lestrade and Weber 2006) was amplified similarly with oligonucleotides hU92-F (5′-CCGGAATTCTGGGAGGCTGATACACAAATTG-3′) and hU92-R (5′-GCCTCTAGAATCTGTCTGCCCCGTATCTGTC-3′) and cloned into EcoRI and XbaI restriction sites of pBluescript SK(−) to yield pBS-U92. The integrity of all constructs was verified by automated sequencing at the McGill University and Génome Québec Innovation Center. The plasmid encoding human U3 snoRNA has been described previously (Baserga et al. 1991).
In vitro RNA synthesis
All plasmids used for in vitro transcription of 32P-labeled RNAs were first linearized with the appropriate enzyme (EcoRI for U17 and hTR constructs, HindIII for ACA36, XbaI for U92, and RsaI for U3). Radiolabeled RNAs were synthesized with T7 RNA polymerase (Ambion) essentially as described (Dragon et al. 2000), and purified on 8% denaturing polyacrylamide gels.
In vitro protein synthesis
Proteins were synthesized with the TNT T7-Quick coupled Transcription/Translation System (Promega). Individual or mixed templates were incubated with 20 μL of rabbit reticulocyte lysate and 11 μCi of [35S]-EXPRE35S35S Protein Labeling Mix (PerkinElmer) in a final volume of 25 μL for 90 min at 30°C. The TNT mixtures were then clarified by centrifugation at 14,000g for 10 min at room temperature; a 1 μL aliquot was retrieved for protein analysis by PAGE before proceeding with IPs or RNP assembly (see below).
RNP assembly
Clarified TNT mixtures (24 μL) were incubated with 100,000 cpm of radiolabeled RNAs for 30 min at 30°C, and centrifuged for 10 min at 14,000g before proceeding with IPs.
Immunoprecipitations
Mixtures were diluted 20 times in IP buffer containing 20 mM HEPES (pH 7.9), 150 mM NaCl, 2 mM MgCl2, 0.1% NP-40 and Complete EDTA-free protease inhibitor cocktail (Roche), and incubated on a nutator at room temperature for 1 h with 25 μL of protein A-agarose beads (Roche) previously saturated with antibodies raised against human dyskerin (H-300, Santa Cruz Biotechnology) or human NAF1 (Hoareau-Aveilla et al. 2006). The beads were then washed 5 times with 1 mL of IP buffer, and elution was achieved by heating the beads 3 min at 90°C in 25 μL of XT sample buffer containing reducing agent (Bio-Rad). Eluates and the 1-μL aliquots of TNT reactions (see in vitro protein synthesis) were fractionated on 4%-12% gradient Criterion XT bis-tris gels (Bio-Rad). Gels were fixed in a 50% methanol, 10% acetic acid, and 10% glycerol solution, dried on Whatman chromatography paper, exposed to a phosphor screen and revealed with a Molecular Imager FX (Bio-Rad). In order to visualize only the 32P-labeled RNA signal from the Criterion gels, a transparency was used to block the 35S-methionine signal that emanates from the proteins in the gels (Wang and Meier 2004). To check for possible RNA degradation, supernatant of IPs were extracted with phenol-chloroform and analyzed on 8% denaturing polyacrylamide gels. To evaluate RNA processing in RRL, radiolabeled hTR204 from a NAF1 IP (carried out in the presence of NAF1, dyskerin, NOP10, and NHP2) and the IP supernatant were extracted with phenol-chloroform, precipitated in ethanol, and fractionated a 8% polyacrylamide sequencing gel together with the input RNA alone. For DNA size marker, MspI-digested pBR322 was terminally labeled with the Klenow fragment of DNA polymerase I (NEB).
ACKNOWLEDGMENTS
We are indebted to Yves Henry, Patrick Matthias, and Vanda Pogacic for the gift of materials, and Chantal Autexier for critical reading of the manuscript. This work was supported by grants from the Banting Research Foundation, the Fonds de la recherche en santé du Québec (FRSQ), the Canadian Institutes for Health Research, and a Chercheur-boursier award of the FRSQ to F.D.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1354009.
REFERENCES
- Atzorn V., Fragapane P., Kiss T. U17/snR30 is a ubiquitous snoRNA with two conserved sequence motifs essential for 18S rRNA production. Mol. Cell. Biol. 2004;24:1769–1778. doi: 10.1128/MCB.24.4.1769-1778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin A.G., Smith L., Fournier M.J. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- Ballarino M., Morlando M., Pagano F., Fatica A., Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 2005;25:5396–5403. doi: 10.1128/MCB.25.13.5396-5403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baserga S.J., Yang X.D., Steitz J.A. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.L., Greider C.W. Telomerase RNA structure and function: Implications for dyskeratosis congenita. Trends Biochem. Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat. Rev. Mol. Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Jady B.E., Verheggen C., Kiss A.M., Bertrand E., Kiss T. Cajal body-specific small nuclear RNAs: A novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X., Kittur N., Roy S., Shav-Tal Y., Singer R.H., Meier U.T. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J. Cell Biol. 2006;173:207–218. doi: 10.1083/jcb.200601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C., Noaillac-Depeyre J., Caizergues-Ferrer M., Henry Y. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol. Cell. Biol. 2002;22:7053–7065. doi: 10.1128/MCB.22.20.7053-7065.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F., Pogacic V., Filipowicz W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 2000;20:3037–3048. doi: 10.1128/mcb.20.9.3037-3048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Dlakic M., Tollervey D. Naf1 p is a box H/ACA snoRNP assembly factor. RNA. 2002;8:1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Fu D., Collins K. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell. 2003;11:1361–1372. doi: 10.1016/s1097-2765(03)00196-5. [DOI] [PubMed] [Google Scholar]
- Ganot P., Caizergues-Ferrer M., Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- Henras A.K., Capeyrou R., Henry Y., Caizergues-Ferrer M. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA. 2004;10:1704–1712. doi: 10.1261/rna.7770604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R.K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau-Aveilla C., Bonoli M., Caizergues-Ferrer M., Henry Y. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA. 2006;12:832–840. doi: 10.1261/rna.2344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Palm W., Wang R.C., Couto S.S., de Lange T. Engineered telomere degradation models dyskeratosis congenita. Genes & Dev. 2008;22:1773–1785. doi: 10.1101/gad.1679208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady B.E., Bertrand E., Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- Kiss A.M., Jady B.E., Bertrand E., Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 2004;24:5797–5807. doi: 10.1128/MCB.24.13.5797-5807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade L., Weber M.J. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulliot N., Godin K.S., Hoareau-Aveilla C., Quevillon-Cheruel S., Varani G., Henry Y., Van Tilbeurgh H. The box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. J. Mol. Biol. 2007;371:1338–1353. doi: 10.1016/j.jmb.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Ly H., Calado R.T., Allard P., Baerlocher G.M., Lansdorp P.M., Young N.S., Parslow T.G. Functional characterization of telomerase RNA variants found in patients with hematologic disorders. Blood. 2005;105:2332–2339. doi: 10.1182/blood-2004-09-3659. [DOI] [PubMed] [Google Scholar]
- Marrone A., Stevens D., Vulliamy T., Dokal I., Mason P.J. Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood. 2004;104:3936–3942. doi: 10.1182/blood-2004-05-1829. [DOI] [PubMed] [Google Scholar]
- Matera A.G., Terns R.M., Terns M.P. Noncoding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Mitchell J.R., Cheng J., Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacic V., Dragon F., Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P., Darzacq X., Bertrand E., Jady B.E., Verheggen C., Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:4283–4293. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P., Kiss A.M., Darzacq X., Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol. Cell. Biol. 2006;26:2540–2549. doi: 10.1128/MCB.26.7.2540-2549.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage S.A., Giri N., Baerlocher G.M., Orr N., Lansdorp P.M., Alter B.P. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am. J. Hum. Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubin M., Newell J.W., Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- TenHarmsel A. TNT® rabbit reticulocyte lysate systems-Easy protein expression. Promega Notes. 2004;88:24–26. [Google Scholar]
- Theimer C.A., Finger L.D., Trantirek L., Feigon J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer C.A., Jady B.E., Chim N., Richard P., Breece K.E., Kiss T., Feigon J. Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol. Cell. 2007;27:869–881. doi: 10.1016/j.molcel.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Vulliamy T.J., Dokal I. Dyskeratosis congenita: The diverse clinical presentation of mutations in the telomerase complex. Biochimie. 2008;90:122–130. doi: 10.1016/j.biochi.2007.07.017. [DOI] [PubMed] [Google Scholar]
- Vulliamy T., Beswick R., Kirwan M., Marrone A., Digweed M., Walne A., Dokal I. Mutations in the telomerase component NHP2 cause the premature ageing syndrome dyskeratosis congenita. Proc. Natl. Acad. Sci. 2008;105:8073–8078. doi: 10.1073/pnas.0800042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy T., Marrone A., Goldman F., Dearlove A., Bessler M., Mason P.J., Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- Wang C., Meier U.T. Architecture and assembly of mammalian H/ACA small nucleolar and telomerase ribonucleoproteins. EMBO J. 2004;23:1857–1867. doi: 10.1038/sj.emboj.7600181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P.K., Hoareau C., Froment C., Monsarrat B., Henry Y., Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol. Cell. Biol. 2005;25:3295–3304. doi: 10.1128/MCB.25.8.3295-3304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]