Abstract
Localizing specific components in three-dimensional reconstructions of protein complexes visualized in an electron microscope increases the scientific value of those structures. Subunits are often identified within the complex by labeling; however, unless the label produces directly visible features, it must be detected by computational comparison with unlabeled complex. To bypass this step, we generated a cloneable tag from the actin-nucleating protein Spire that produces a directly visible “pointer” to the subunit after actin polymerization. We have used this new label to identify the intron of the C complex spliceosome to its small domain by fusing the 10 kDa Spire moiety to the affinity label that binds recombinant stem loops in the pre-mRNA substrate and assembling an actin filament on the particle.
Keywords: spliceosome, RNA, actin, single particle, electron microscopy, Spire
INTRODUCTION
Localizing individual components in single particle electron microscopy (EM) reconstructions of large protein complexes is a major challenge in interpreting these structures. Identifying the position of key components can provide anchor points to facilitate docking of atomic resolution coordinates into molecular envelopes determined by single particle EM. Knowing where components are may also shed light on the possible role of those components within a complex. Current methods for labeling single particle EM reconstructions include adding gold label, adding antibodies (with or without gold conjugates), adding mass by recombinant protein fusions, or deletion mapping of domains (Jurica et al. 2002; Wolf et al. 2003; Kim et al. 2004; Stroupe et al. 2006).
Each of these methods has unique advantages and disadvantages. Covalently added 5 nm gold clusters can be seen directly on the specimen to provide information about subunit positions at moderate (2 nm) resolution. However, the target requires an attachment site, such as a single, surface-accessible cysteine to achieve specific labeling. Furthermore, labeling is usually much less than 100%, and the use of gold labels often leads to non-specific labeling and aggregation of label and/or sample. Monoclonal antibodies, on the other hand, do not require the engineering of specific attachment sites. They are exogenously added and can be used to provide tags of different sizes. For example, antibody tags can be directly visualized in micrographs by coupling them to gold clusters, or their size can be increased by adding a secondary antibody. Similar to gold labels, labeling is usually substoichiometric, even with excess antibody added, which may require another purification step to remove unbound labels. Also, unless an additional heavy atom label is used, an antibody label cannot be localized easily in raw images of heterogeneous samples. In this case, averaging over many labeled particles is required to localize the label. Labeling can also be achieved by adding or deleting domains of subunits. This is appealing because these modifications are stoichiometric without additional purification. However, additions or deletions may affect sample integrity, and averaging may be required to localize the label in larger and/or heterogeneous particles.
The ideal label would have high affinity, specific binding, low background, be adjustable in size, and be directly and unambiguously identifiable in raw micrographs to avoid the need for image averaging. This last criterion is of particular importance for reconstructions of large structures whose resolution is limited by compositional heterogeneity, such as the C complex spliceosome (Jurica et al. 2004). The spliceosome is a highly dynamic assembly of RNAs and proteins that removes intervening non-coding regions (introns) from the coding regions (exons) of nascent pre-mRNA transcripts. Various spliceosome assemblies have molecular masses of several megadaltons, increasing the challenge to their structure determination (Jurica and Moore 2003).
C complex has a diameter of about 20 nm and three discernable domains (Jurica et al. 2004). The largest domain is believed to be the U5 snRNP, one major component that accounts for about 1 MDa of mass (Sander et al. 2006). This hypothesis further places Prp19 in the arm domain and places SF3b, which is part of the U2 snRNP and interacts with the intron, within the smallest domain (Jurica et al. 2004). Recent labeling of the pre-mRNA 5′ and 3′ exons using a dimeric, donut-shaped protein fused to a sequence-specific RNA-binding protein shows that the exons fall at the junction of the three domains (Alcid and Jurica 2008). The location of the lariat-intron remains to be determined to complete the map of the substrate pre-mRNA within C complex.
RESULTS AND DISCUSSION
We have generated an EM label suitable for use with such massive complexes: a novel, cloneable tag from the Drosophila melanogaster actin nucleating protein Spire (Quinlan et al. 2005). Nucleating assembly of an actin filament (F-actin) makes the label directly visible in an image. Spire initiates barbed-end filament growth by binding and scaffolding actin monomers (G-actin) using its four tandem Wiskott-Aldrich Syndrome protein (WASp) homology 2 (WH2) domains (SpireABCD) (Quinlan et al. 2005). The two C-terminal WH2 repeats (SpireCD) plus intervening loop, comprising 10 kDa, are sufficient to nucleate actin polymerization.
As proof of concept for this label, we created a fusion of the DNA repair enzyme Thermatoga maritima RuvB, a 37 kDa protein that assembles into homohexamers (Putnam et al. 2001), with SpireCD joined to its N-terminus through an eight amino acid linker. In a standard actin polymerization assay that monitors the change in fluorescence as pyrene-labeled actin monomers (G-actin) assemble into F-actin (Moseley and Goode 2005), SpireCD-RuvB weakly nucleated F-actin growth (data not shown). In images of labeled SpireCD-RuvB, single actin filaments are associated with RuvB hexamers (Fig. 1) and there are few free actin filaments that are likely due to monomeric RuvB. The width of the filament with respect to the hexamer is equivalent to about one or two 37 kDa RuvB subunits, suggesting that the F-actin label could localize subunits of about 50 kDa to a position of about 5 nm within a larger complex (Fig. 1).
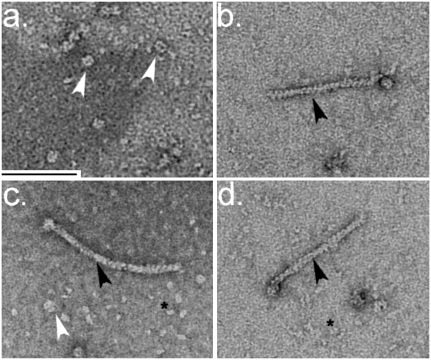
FIGURE 1.
SpireCD-RuvB with F-actin labels. In the background, SpireCD-RuvB hexamers without actin filaments (white arrow head), as well as monomeric actin (asterisk), are visible. The scale bar represents a length of 100 nm. Images were collected on a 1k × 1k AMT detector on an FEI Morgani microscope operating at 80 keV and 36,000 times magnification. (a) SpireCD-RuvB hexamers (white arrowheads); (b–d) SpireCD-RuvB with single actin filaments (black arrowheads).
Next, we purified C complex spliceosomes as described (Jurica et al. 2002), but with one change. We used a modified RNA-affinity tag with five tandem moieties: SpireCD, an eight amino acid linker, maltose binding protein (MBP), MS2 coat protein, and a six-histidine peptide. SpireCD, the linker, and the six-histidine tag add 11 kDa in mass to the traditional MS2:MBP RNA affinity tag used in C complex purification. The MS2 coat protein moiety binds to one of three tandem MS2 stem loops encoded upstream of the branchpoint within the intron of the pre-mRNA splicing substrate. To form the labeled complex, we mixed purified C complex with G-actin, quenched filament growth with cytochalasin B, and stabilized the filament with phalloidin. To visualize the resulting complexes in the EM, we prepared negatively stained continuous carbon grids using fresh 1% uranyl formate.
C complex spliceosomes associated with individual actin filaments could be readily observed in EM images (Fig. 2). This particle binds to the carbon substrate such that the majority of the particles adopt a single orientation (Fig. 2a; Jurica et al. 2002), allowing us to identify this representative view in our raw images (Fig. 2b–f). The filament appeared to be attached to the spliceosome at the small domain, which has previously been proposed to contain the intron-interacting U2 snRNP component SF3b (Jurica et al. 2004).
FIGURE 2.
Images of C complex spliceosomes with F-actin labels on the intron. (a) Class average of the dominant view (“preferred orientation”) of C complex spliceosomes. The class average was calculated using Spider (Frank et al. 1996) with raw data from Jurica et al. (2002). Domains are labeled to indicate their approximate locations. (b–f) Individual images of preferred-orientation C complex spliceosomes labeled with a single actin filament marking the small domain through the Spire tag attached to the mRNA substrate's intron. Particles were stained with 1% uranyl formate on continuous carbon films and imaged at 59,000 times magnification on a Technai F30 operating at 200 keV. White arrows mark the spliceosome and black arrows mark the F-actin. The scale bar is 10 nm.
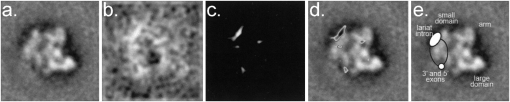
To show that the filament junction with the particle is localized to the small domain, we selected 1576 negatively stained particles from 885 images collected at 59,000 times magnification on an FEI Techani F30 operating at 200 kV. From these raw data, we could easily identify particles with the preferred orientation (Fig. 2b–f), which we aligned with the expected class average (Fig. 2a). The resulting average of the 100 particles that aligned with the highest cross correlation coefficient (Fig. 3a), and its difference map (Fig. 3b,c) with the unlabeled complex (Fig. 2a), support our observation from the raw data that the label tags the small domain of C complex (Fig. 3d). Although the filament is not visible in the average, the additional mass that is localized at the small domain is clearly seen in the single 4-sigma peak in the difference map of the labeled and unlabeled averages (Fig. 3d). This 4-sigma peak is significantly stronger and broader than the other positive and negative peaks, the strongest of which are 3-sigma. Its broad shape can be attributed either to the three sequential MS2 loops in the substrate RNA, which are separated by 13 nucleotides (nt) that can span a maximum distance of 12 nm, or to the filament itself. This analysis shows that our label can be located both in raw and averaged images.
FIGURE 3.
(a) Average of 100 labeled particles, which best align and correlate with the unlabeled C complex. (b) Difference map of the labeled minus the unlabeled class averages filtered at 4 nm resolution. (c) The difference map contoured at 2.5-sigma threshold. (d) The superposition of (c) on (a), in which the difference is colored in grayscale according to the intensity of the peak, where white is about 2.5-sigma and black is about 4-sigma. (e) Class average of the unlabeled C complex, as in Figure 2a. The location of the exons is marked with a small circle and the location of the intron with an oval. The putative area where the RNA joining these elements could run is demarked with the black lines.
Understanding where the pre-mRNA substrate is within the massive C complex spliceosome has been challenging owing to a lack of techniques that are appropriate for labeling such difficult samples. By comparing our result with those from a recently developed RNA label that has been used to tag the two exons (Alcid and Jurica 2008), we show that the intron lariat (120 nt in length not including the stem loops) sits about 3 nm from the free 5′ and intron-joined 3′ exons in the small domain near its junction with the arm domain (Fig. 3). The three main elements of the pre-mRNA substrate (the two exons and the intron-lariat) appear to fall at the periphery of the complex (Figs. 2a, 3). The RNA strand that joins the 3′ exon to the lariat could run either around the surface of the molecule or through the cavity that is found between the small and large domains (Fig. 3e).
In both RuvB and C complex splicesomes, the low background, extreme specificity, and visually identifiable mass of the F-actin label show that it fulfills our requirements for an easy-to-use EM label. Additionally, the length of the actin filament “pointer” is easy to adjust by adding cytochalasin B earlier or later to quench growth. Filament growth rate is directly proportional to the concentration of available actin monomers, so that at 0.5 μM monomeric actin in the presence of Mg2+, monomers are added to the barbed end at a rate of about 6 per second (about 20 nm of polymer length) (Pollard 1986). For visualizing a filament on a small protein or protein complex, short filaments might be desirable and therefore one might use a low actin concentration (0.5–1 μM) and short polymerization time (10–30 sec). For a larger complex, one might use a higher actin concentration (1–3 μM) and longer polymerization times (30–120 sec).
Additional benefits include that the tag is small during expression and purification, and thus less likely to interfere with these steps. The small size of the tag means it is less disruptive to the complex of interest if it falls on the surface; however, if it were to be buried, the G-actin could not interact with Spire and, therefore, there would be no actin nucleation. Successful actin nucleation can be detected, for example, by a simple solution scattering experiment (Moseley and Goode 2005). It should be noted that SpireCD is not as efficient an actin-assembly promoting factor (Quinlan et al. 2005) (data not shown) as either Arp2/3 complex or formins (Pollard 2007). This could be an advantage when expressing recombinant protein-Spire fusions in a eukaryotic expression system where efficient actin nucleation might interfere with cellular functions. Although the size and flexible tethering of the filament can interfere with alignment (data not shown), the filament can be accurately identified in the raw images so averaging is not needed to interpret the labeled particles.
In summary, we have tested the F-actin label on two different samples suitable for negative-stain single particle EM. First, we labeled the N-terminus of RuvB, showing that single, untangled actin filaments could be readily assembled from SpireCD fused to individual RuvB molecules and localizing a mass of ∼50 kDa to within ∼5 nm. Second, we labeled the intron of the C complex spliceosome and localized the pre-mRNA intron to the small domain of C complex, not far from the junction of its three domains where the exons are believed to reside. This result shows that the three main elements of the substrate pre-mRNA in C complex (the 5′ exon, the intron lariat, and the 3′ exon) are at the periphery of the spliceosome, poised for the second step of catalysis and intron release.
MATERIALS AND METHODS
To produce SpireCD-RuvB proteins with nucleated actin filaments, we incubated 0.5 μM gel-filtered monomeric rabbit muscle actin purified as described (Spudich and Watt 1971) with 4 μM SpireCD-RuvB for 10 min and then initiated filament growth with the addition of KCl and MgCl2. Filament growth was quenched at intervals between 10 and 120 sec with cytochalasin B. Filaments were stabilized with phalloidin before being diluted 1:10 into filament-stabilizing buffer (F-buffer). Then, samples were added to continuous carbon films made hydrophilic by a 2-min glow discharge, washed with F-buffer (50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.2 mM CaCl2, 0.7 mM ATP, 0.2 mM DTT, 10 mM tris at pH 7.5), and stained with fresh 2% uranyl acetate (Baker Chemical). No filaments were found in images of control experiments with monomeric actin but no SpireCD-RuvB. Single filaments are associated with RuvB hexamers in images of labeled SpireCD-RuvB, with few free actin filaments that likely arise from monomeric RuvB molecules.
We purified the recombinantly expressed C complex spliceosome RNA affinity tag according to the published protocol but adding an additional nickel affinity step (Jurica et al. 2002). By using each of the three affinity tags for purification we ensured that all three of the functions (actin binding, amylose binding, and RNA binding) were intact.
To facilitate actin filament formation on the spliceosome, we added 0.5–2 mM G-actin monomers to purified C complex after diluting the concentrated monomers 1:5 into water. C complex has a concentration of less than 1 pM after purification, however the vast excess of G-actin does not add significant background because of its small size relative to the large spliceosome. C complex purification buffer contains 200 mM KCl, which is sufficient to facilitate F-actin nucleation. It was not necessary to add excess Mg+2 or ATP, despite the presence of 5 mM EDTA in the C complex buffer. Filament growth was quenched as described above after 30–120 sec. The samples were immediately applied to hydrophilic continuous carbon grids and stained with 1% uranyl formate (SPI) at pH 5.0.
Images of labeled C complex were acquired at 1.5 μm defocus on a Technai F30 operating at 200 kV using Serial EM (Mastronarde 2005) to drive semiautomated data collected on a 4k × 4k Gatan CCD camera. Particles were picked using Ximdisp (Smith 1999) and all other image analysis was performed using Spider (Frank et al. 1996).
ACKNOWLEDGMENTS
We thank Omar Quintero-Monson, Amy Grace DuPage, Kyoko Okada, and Christopher Gould, for help with the actin polymerization assays and for supplying G-actin. The Spire clone was generously provided by Dyche Mullens and the tmRuvB clone by John Tainer. We also thank David DeRosier, Michael Rigney, and Melissa Jurica for helpful discussion. This work was supported in part by grants from the National Institutes of Health (GM063691 and GM083137) and the March of Dimes.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1313609.
REFERENCES
- Alcid E.A., Jurica M.S. A protein-based EM label for RNA identifies the location of exons in spliceosomes. Nat. Struct. Mol. Biol. 2008;15:213–215. doi: 10.1038/nsmb.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Licklider L.J., Gygi S.R., Grigorieff N., Moore M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica M.S., Moore M.J. Pre-mRNA splicing: Awash in a sea of proteins. Mol. Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Jurica M.S., Sousa D., Moore M.J., Grigorieff N. Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat. Struct. Mol. Biol. 2004;11:265–269. doi: 10.1038/nsmb728. [DOI] [PubMed] [Google Scholar]
- Kim L.A., Furst J., Gutierrez D., Butler M.H., Xu S., Goldstein S.A., Grigorieff N. Three-dimensional structure of I(to); Kv4.2-KChIP2 ion channels by electron microscopy at 21 Å resolution. Neuron. 2004;41:513–519. doi: 10.1016/s0896-6273(04)00050-9. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Moseley J.B., Goode B.L. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Putnam C.D., Clancy S.B., Tsuruta H., Gonzalez S., Wetmur J.G., Tainer J.A. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 2001;311:297–310. doi: 10.1006/jmbi.2001.4852. [DOI] [PubMed] [Google Scholar]
- Quinlan M.E., Heuser J.E., Kerkhoff E., Mullins R.D. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Sander B., Golas M.M., Makarov E.M., Brahms H., Kastner B., Luhrmann R., Stark H. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol. Cell. 2006;24:267–278. doi: 10.1016/j.molcel.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Smith J.M. Ximdisp—A visualization tool to aid structure determination from electron microscope images. J. Struct. Biol. 1999;125:223–228. doi: 10.1006/jsbi.1998.4073. [DOI] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin–troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Stroupe M.E., Tange T.O., Thomas D.R., Moore M.J., Grigorieff N. The three-dimensional arcitecture of the EJC core. J. Mol. Biol. 2006;360:743–749. doi: 10.1016/j.jmb.2006.05.049. [DOI] [PubMed] [Google Scholar]
- Wolf M., Eberhart A., Glossmann H., Striessnig J., Grigorieff N. Visualization of the domain structure of an L-type Ca2+ channel using electron cryo-microscopy. J. Mol. Biol. 2003;332:171–182. doi: 10.1016/s0022-2836(03)00899-4. [DOI] [PubMed] [Google Scholar]