Abstract
Codon recognition by aminoacyl-tRNA on the ribosome triggers a process leading to GTP hydrolysis by elongation factor Tu (EF-Tu) and release of aminoacyl-tRNA into the A site of the ribosome. The nature of this signal is largely unknown. Here, we present genetic evidence that a specific set of direct interactions between ribosomal protein S12 and aminoacyl-tRNA, together with contacts between S12 and 16S rRNA, provide a pathway for the signaling of codon recognition to EF-Tu. Three novel amino acid substitutions, H76R, R37C, and K53E in Thermus thermophilus ribosomal protein S12, confer resistance to streptomycin. The streptomycin-resistance phenotypes of H76R, R37C, and K53E are all abolished by the mutation A375T in EF-Tu. A375T confers resistance to kirromycin, an antibiotic freezing EF-Tu in a GTPase activated state. H76 contacts aminoacyl-tRNA in ternary complex with EF-Tu and GTP, while R37 and K53 are involved in the conformational transition of the 30S subunit occurring upon codon recognition. We propose that codon recognition and domain closure of the 30S subunit are signaled through aminoacyl-tRNA to EF-Tu via these S12 residues.
Keywords: decoding, EF-Tu, ribosomal protein S12, ribosome, streptomycin
INTRODUCTION
The discrimination between cognate and near-cognate aminoacyl-tRNAs by the ribosome is determined by the interplay among multiple components of the translation apparatus (for reviews, see Ogle and Ramakrishnan 2005; Rodnina et al. 2005). This process is initiated by the delivery of aminoacyl-tRNA to the ribosome as part of a ternary complex with elongation factor Tu (EF-Tu) and GTP (Fig. 1A). In an initial recognition step, cognate codon–anticodon pairing is signaled to EF-Tu, resulting in GTPase activation and GTP hydrolysis. This induces EF-Tu to undergo a conformational rearrangement leading to dissociation of EF-Tu•GDP from the ribosome as the acceptor end of aminoacyl-tRNA moves into the 50S subunit A-site where it can participate in peptide bond formation. Near-cognate aminoacyl-tRNA is rejected in a proofreading step subsequent to GTP hydrolysis and release of EF-Tu. While recent structural studies have provided an atomic-resolution view of codon recognition (Ogle et al. 2001, 2002), the mechanism by which this event is signaled to EF-Tu to stimulate GTP hydrolysis is a major unresolved issue in our understanding of protein synthesis.
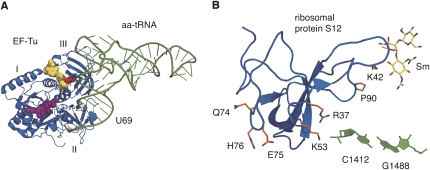
FIGURE 1.
(A) The EF-Tu•Phe-tRNAPhe•GTP ternary complex with kirromycin bound (PDB file 1OB2). The QEH triplet of ribosomal protein S12 contacts Phe-tRNAPhe (green) in the vicinity of residue U69. EF-Tu is colored blue, with A375 shown as red spheres. Kirromycin is shown as yellow spheres and the GTP analog, phosphoaminophosphonic acid-guanylate ester, is shown as purple spheres. (B) Structure of T. thermophilus ribosomal protein S12 as observed in the context of the 30S ribosomal subunit (PDB file 1FJG; Carter et al. 2000), showing the positions of streptomycin (yellow sticks) and residues altered in mutants (orange sticks), K42, K53, P90, and H76. Also shown as orange sticks are Q74 and E75, which together with H76 protrude into the intersubunit cavity. K53 makes direct contact with C1412 in 16S rRNA helix 44 (green). Both images were rendered with PyMol (DeLano 2002).
X-ray crystallographic studies of the Thermus thermophilus 30S ribosomal subunit show the existence of an induced fit mechanism of substrate binding, with the subunit undergoing a transition from an open to a closed conformation upon codon recognition (Ogle et al. 2001, 2002; Ogle and Ramakrishnan 2005). Large-scale movements of the head and shoulder of the subunit during this “domain closure” entail the formation of contacts between ribosomal protein S12 and 16S rRNA in the decoding center, and the concomitant breaking of contacts between ribosomal proteins S4 and S5 on the far side of the subunit. Cryo-electron microscopic (cryo-EM) reconstructions of Escherichia coli 70S ribosomes associated with ternary complex (Stark et al. 2002; Valle et al. 2003) show the ribosome to be in the closed conformation when ternary complex is bound.
Discrimination between cognate and near-cognate ternary complexes results in part from the greater ability of cognate complexes to induce domain closure and thereby stimulate GTP hydrolysis (Ogle et al. 2002). Pre-steady-state kinetic experiments indicate that the rates of GTP hydrolysis differ for cognate and near-cognate tRNAs by at least two orders of magnitude (Rodnina et al. 2005), and antibiotics such as streptomycin or mutations conferring streptomycin resistance impact both decoding accuracy and GTP hydrolysis rates (Bilgin et al. 1992; Gromadski and Rodnina 2004). Streptomycin causes misreading by forming contacts with both S12 and 16S rRNA (Carter et al. 2000) and shifting the conformational equilibrium of the 30S subunit toward domain closure, but in a conformation that differs from that attained through cognate codon recognition (Ogle et al. 2003). In doing so it uncouples GTP hydrolysis from codon recognition by inhibiting GTPase activation by cognate complexes and stimulating GTPase activation by near-cognate complexes, thus destroying the ribosome's ability to discriminate between the two (Gromadski and Rodnina 2004). Thus, a link exists between domain closure and GTP hydrolysis, both of which are key elements of the decoding mechanism of the ribosome. There should exist mutations that impact both 30S subunit domain closure and the signaling of codon recognition to EF-Tu by ribosomal protein S12, and such mutations should confer resistance to streptomycin.
Here we describe streptomycin-resistance mutations in ribosomal protein S12 at sites distant from the streptomycin-binding site and positions of other previously identified resistance mutations (Carter et al. 2000; Gregory et al. 2001; Sharma et al. 2007). One of these substitutions, H76R, is located in a QEH triplet consisting of residues Q74, E75, and H76, which has been interpreted in low-resolution cryo-EM maps as a protrusion contacting the aminoacyl-tRNA acceptor helix at or near residue U69 (Stark et al. 2002; Valle et al. 2003). As this contact occurs exclusively in the initial recognition step and is lost upon release of EF-Tu and movement of the acceptor helix into the peptidyltransferase center on the 50S subunit, it is a potential candidate for involvement in signaling codon recognition to EF-Tu and activating GTP hydrolysis. We also describe two other amino acid substitutions, R37C and K53E, in ribosomal protein S12 at residues involved in domain closure. Further, we find that streptomycin resistance conferred by each of these three substitutions is completely abolished by a kirromycin-resistance mutation in EF-Tu, establishing a functional relationship between the S12–ternary complex interaction and GTP hydrolysis during the initial recognition phase of tRNA selection. These observations suggest the existence of a signal relay, involving ribosomal protein S12, whose function is to stimulate GTPase activation of EF-Tu upon cognate codon recognition and 30S subunit domain closure. We propose that the interactions involving these S12 residues serve to place the ternary complex on the ribosome in a GTPase-activated state.
RESULTS AND DISCUSSION
Amino acid substitution H76R in ribosomal protein S12 of drug-dependent mutants
Among our collection of T. thermophilus mutants selected for antibiotic-resistance phenotypes, we discovered a paromomycin-dependent mutant bearing two amino acid substitutions, P90L and H76R, in ribosomal protein S12. The P90L mutation on its own has been identified previously as a streptomycin- or paromomycin-dependence mutation in a number of organisms including E. coli (Kurland et al. 1996) and T. thermophilus (Gregory et al. 2001; Carr et al. 2005). Such “ancillary” mutations in combination with P90L have been observed previously in E. coli (Timms and Bridges 1993) but their phenotypes have never been examined separately from the primary drug-dependence mutation. They do not appear to influence the phenotype of the P90L substitution, as the double mutants behave indistinguishably from mutants bearing the P90L substitution alone. This is consistent with the distance of H76 from both P90 (24 Å) and the streptomycin binding site (32 Å) (Carter et al. 2000). Thus, the reason for the appearance of ancillary mutations is unknown.
H76 is part of a loop motif embedded within the β-barrel domain of S12, consisting of the triplet sequence QEH (Fig. 1B). The QEH sequence is highly conserved among bacteria, while the motif is DEH among archaea. These residues are more variable among the eukarya, although E75 is highly conserved, as is an acidic residue at position 74. Prompted by the proximity of H76 to the acceptor helix of aminoacyl-tRNA in cryo-EM reconstructions and the lack of prior information regarding the phenotype of the H76R substitution, we examined it on its own, independent of the P90L mutation.
Streptomycin-resistance conferred by H76 substitutions in ribosomal protein S12
We used a gene replacement technique (Carr et al. 2005) to reconstruct H76R as well as three other single amino acid substitutions, H76A, H76E, and H76K. This technique does not involve selection for streptomycin resistance, and it allows us to recover mutant alleles independent of their phenotypes, e.g., streptomycin sensitive. H76R, H76A, and H76K produced moderate increases in doubling time (58, 62, and 57 min, respectively, compared with 49 min for wild type). H76E proved to be severely debilitating (doubling time 109 min) and genetically unstable, with faster growing derivatives readily appearing on plates. One possible explanation for the more severe phenotype of H76E is that replacement of histidine with a negatively charged glutamic acid creates electrostatic repulsion with the negatively charged phosphate backbone of aminoacyl-tRNA.
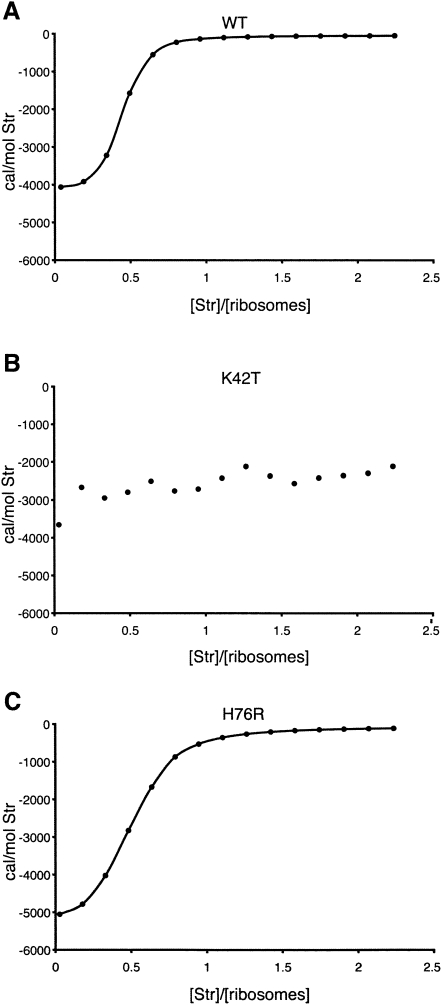
H76R, H76A, and H76K produce high-levels of streptomycin-resistance (Table 1). Amino acid substitutions can produce resistance either directly by perturbing drug binding or indirectly by avoiding the action of the antibiotic when it is bound. To distinguish between these mechanisms, we directly assessed the effect of H76R on streptomycin binding using isothermal titration calorimetry (ITC). ITC measures the heat liberated by intermolecular binding interactions, and while the small energy released upon streptomycin binding to the ribosome (due to the large mass difference between the ribosome and streptomycin) precludes the assignment of precise binding affinities, this method is sufficiently sensitive to distinguish between binding and its absence. As indicated in Figure 2, ribosomes bearing H76R bind streptomycin in a manner similar to wild-type ribosomes, consistent with the large distance (32 Å) of H76 from streptomycin observed in the 30S subunit-streptomycin co-crystal structure (Carter et al. 2000). In contrast, ribosomes with K42T do not bind streptomycin, as indicated by the random distribution of data points. This is expected, as K42 directly contacts streptomycin (Carter et al. 2000). These results exclude the possibility that substitutions at H76 act by perturbing the drug-binding site through a global conformational alteration of S12 and suggest instead that they confer resistance indirectly through influencing the interaction of S12 with ternary complex.
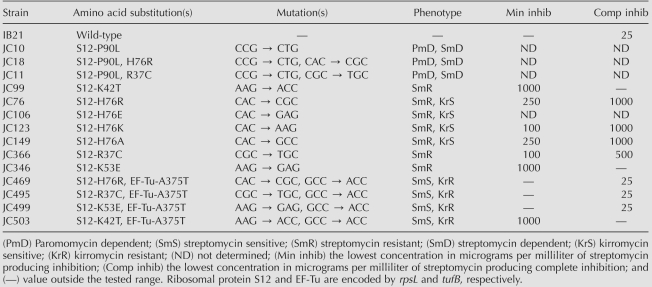
TABLE 1.
Ribosomal protein S12 and EF-Tu amino acid substitutions and their associated phenotypes.
FIGURE 2.
Binding of streptomycin to ribosomes analyzed by isothermal titration calorimetry (ITC). (A) Wild-type ribosomes. (B) Ribosomes bearing the S12–K42T substitution. (C) Ribosomes bearing the S12–H76R substitution.
Suppression of streptomycin-resistance phenotype of H76R by an amino acid substitution in EF-Tu
In order to detect a genetic interaction between ribosomal protein S12 and ternary complex we searched for EF-Tu mutations that would interfere with the phenotype generated by H76R. The antibiotic kirromycin freezes ternary complex on the ribosome after a single round of GTP hydrolysis, and mutants resistant to kirromycin have amino acid substitutions in EF-Tu (Parmeggiani and Nissen 2006). We, therefore, selected spontaneous kirromycin-resistant derivatives of the H76R mutant and screened these for ones that had lost streptomycin resistance.
The tufA and tufB genes encoding EF-Tu from kirromycin-resistant, streptomycin-sensitive mutants were then individually amplified by PCR and sequenced. The rpsL gene encoding ribosomal protein S12 was also sequenced to exclude reversion of the H76R allele. One mutant, designated JC469, was found to have a wild-type tufA and a mutant tufB producing an A375T amino acid substitution in EF-Tu. This mutant displayed streptomycin sensitivity similar to wild type (Fig. 3). A375T was previously identified as the E. coli tufAR mutation (Duisterwinkel et al. 1981) and the Salmonella typhimurium tufA8 mutation (Tubulekas et al. 1991).
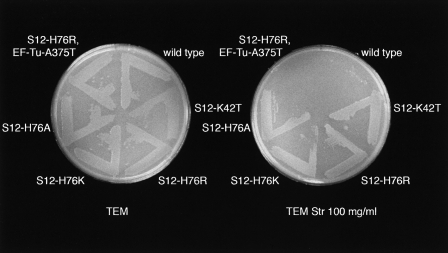
FIGURE 3.
Antibiotic-resistance phenotypes produced by amino acid substitutions at H76 of ribosomal protein S12, and the suppression of streptomycin resistance by the EF-Tu-A375T mutation. Wild-type and mutant T. thermophilus IB-21 were streaked for single colonies on TEM medium with (A) no drug and (B) 100 μg/mL streptomycin. The amino acid substitutions are indicated. The S12-K42T mutation confers streptomycin resistance by abrogating streptomycin binding to the ribosome.
Suppression of streptomycin-resistance phenotypes of R37C and K53E by EF-Tu-A375T
In S. typhimurium, EF-Tu-A375T abolishes the streptomycin-resistance phenotype of an R53L substitution in ribosomal protein S12 (Tubulekas et al. 1991). The equivalent residue in T. thermophilus S12, K53, makes direct contact with the backbone of 16S rRNA residue C1412 during domain closure (Ogle et al. 2002), and a T. thermophilus K53E substitution conferring streptomycin resistance has been identified (Carter 2002; S.T. Gregory and A.E. Dahlberg, unpubl.). K53 is 21 Å from the streptomycin binding site (Carter et al. 2000), and, rather than interfering with drug binding, K53E is likely to interfere with domain closure due to the clash of the negatively charged glutamate side chain with the negatively charged phosphates of 16S rRNA helix 44. Another S12 residue, R37, situated between the two β-strands containing the QEH triplet and K53, makes van der Waals contact with K53, suggesting that substitutions at this position could cause streptomycin resistance indirectly by influencing the K53–16S rRNA contact. From our collection of mutants, we identified a streptomycin-dependent P90L R37C double mutant and found that a reconstructed R37C single mutant is streptomycin resistant.
Introduction of the EF-Tu-A375T mutation into the T. thermophilus R37C and K53E single mutant strains resulted in complete loss of streptomycin resistance; in contrast, this EF-Tu mutation had no effect on the streptomycin-resistance phenotype of K42T (Table 1). This result is consistent with the observations that, in S. typhimurium, A375T only slightly reduced the streptomycin-resistance levels of K42T and K42N, but completely abolished the resistance conferred by the R53L substitution (Tubulekas et al. 1991).
While the exact nature of the mechanism by which EF-Tu-A375T suppresses K53E and H76R is not known, it is worth recalling that in E. coli and S. typhimurium A375T produces an error-prone EF-Tu, a phenotype opposite that of hyperaccurate S12 mutations such as R53L (Tubulekas et al. 1991). As hyperaccurate S12 mutants have reduced GTP hydrolysis rates (Bilgin et al. 1992), we might expect the A375T EF-Tu to exhibit higher rates of GTP hydrolysis. The location of A375T in the cleft between domains I and III of EF-Tu suggests that it could do so by influencing the relative movement of these domains that occurs upon GTP hydrolysis (Berchtold et al. 1993). Interestingly, A375T or other substitutions in the cleft lower the binding affinity of EF-Tu for aminoacyl-tRNA (Sam et al. 1985; Vorstenbosch et al. 2000), perhaps reflecting an enhanced ability to release aminoacyl-tRNA into the A site after GTP hydrolysis.
Structural changes in ternary complex, domain closure, and GTPase activation
Two major structural rearrangements in cognate ternary complex occur upon ribosome binding—a shift in the relative position of aminoacyl-tRNA acceptor helix with respect to EF-Tu and a deformation of the anticodon helix (Valle et al. 2003). These rearrangements are dependent on domain closure as well as on multiple contacts between the ternary complex, and both subunits of the ribosome and may serve to place the ternary complex into a high-energy, GTPase-active conformation. As suggested previously (Valle et al. 2003), the shift of aminoacyl-tRNA relative to EF-Tu may serve as the direct signal for GTPase activation upon domain closure. Conversely, by failing to induce domain closure, near-cognate complexes are unable to facilitate this aminoacyl-tRNA shift and are rejected prior to GTPase activation. The role of S12 in this process may be to induce this shift of the aminoacyl-tRNA acceptor helix as S12 moves toward 16S rRNA helix 44 during domain closure. As EF-Tu is anchored on the 50S subunit, the shift of the aminoacyl-tRNA acceptor helix could facilitate interdomain movements of the factor, leading to a GTPase activated conformation.
One corollary of this model is that the structural integrity of aminoacyl-tRNA is essential for proper GTPase activation. Indeed, GTPase activation is abolished when aminoacyl-tRNA is replaced with two equivalent fragments, an anticodon stem–loop analog and an aminoacylated acceptor helix (Piepenburg et al. 2000). Conversely, a mutation expected to facilitate tRNA deformation, the G24A substitution in tRNATrp (Hirsh 1971), accelerates the forward rate constant for GTPase activation (Cochella and Green 2005). Given the position of G24 at or near the site of tRNA bending observed by cryo-EM, the suppressor tRNA could be more readily deformed, facilitating the shift of the acceptor helix relative to EF-Tu.
A role for ribosomal protein S12 in signaling EF-Tu during decoding
Cryo-EM reconstructions have led to speculation that other EF-Tu-ribosome contacts participate in GTPase activation (Valle et al. 2003). Our genetic data directly implicate ribosomal protein S12 in signaling codon recognition to EF-Tu during decoding of mRNA and suggest that it may do so by signaling the transition of the 30S subunit from the open to the closed form. In this view, the repositioning of S12 that occurs upon domain closure directly facilitates the shift in position of aminoacyl-tRNA relative to EF-Tu, thereby stimulating GTPase activation. One of the key elements in this process is the H76 region of S12. A more detailed description of this process will require a synthesis of genetic, structural, and kinetic approaches.
MATERIALS AND METHODS
Genetic methods
All mutants are derived from the Icelandic strain of T. thermophilus, IB-21 (ATCC 43815; Kristjansson et al. 1986). The S12-K53E strain was derived by transformation of IB-21 with DNA from a T. thermophilus HB8 mutant, kindly provided by Andrew Carter and Venki Ramakrishnan, MRC, Cambridge, UK. Growth was performed in ATCC medium 1598 (thermus-enhanced medium [TEM]) at 72°C. Streptomycin sulfate and paromomycin sulfate were purchased from Sigma Chemical. Kirromycin was a generous gift of Eric Cundliffe, University of Leicester, UK, and of Michael O'Connor, University of Missouri at Kansas City.
Spontaneous mutants were selected by plating ∼109 cells onto TEM plates containing antibiotic. Drug-dependent mutants were isolated on 20 μg/mL paromomycin, and kirromycin-resistant mutants were selected on 25 μg/mL kirromycin. Individual mutants were purified by restreaking serially twice for single colonies and were not subjected to antibiotic selection subsequent to the initial isolation. Antibiotic-resistance phenotypes were screened using a disc assay as previously described (Gregory et al. 2005) or by streaking for single colonies on plates containing closely spaced concentrations of antibiotic. Spontaneous kirromycin-resistant mutants of T. thermophilus were isolated on 25 μg/mL kirromycin, and 150 individual colonies were screened for loss of streptomycin resistance by toothpicking onto antibiotic-free TEM plates and TEM plates containing 100 μg/mL streptomycin sulfate. Streptomycin-sensitive mutants were purified by restreaking and then rescreened for kirromycin resistance. Transformation of T. thermophilus IB-21 was performed with plasmid or chromosomal DNA according to the method of Koyama (Koyama et al. 1986).
Site-directed mutagenesis of rpsL was performed by PCR amplification from chromosomal DNA in two steps. For example, for H76R, the N-terminal portion was amplified with TthS12PCRI (5′-GAAGGGATCCGTGGCACTGCCG-3′), containing a BamHI site, and TthS12PCRIV (5′-GACCACCGAGCGCTCCTGAAGG-3′); the C-terminal portion was amplified with TthS12PCRIII (5′-CCTTCAGGAGCGCTCGGTGGTC-3′) and TthS12PCRII (5′-GCTCACAAGCTTCTTGGCCGC-3′), containing a HindIII site. These PCR products were used as templates in fusion PCR with the primer pair TthS12PCRI and TthS12PCRII. This PCR product was then digested with BamHI/ HindIII and cloned into BamHI-HindIII-digested pUC19. The H76A, H76E, and H76K mutagenesis was conducted in the same way using the pairs TthS12PCRI and TthS12PCRII; the internal mutagenesis primers were as follows: H76A with TthS12H76A-1 (5′-CCTTCAGGAGGCCTCGGTGGTC-3′) and TthS12H76A-2 (5′-GACCACCGAGGCCTCCTGAAGG-3′); H76E with TthS12H76E-1 (5′-CCTTCAGGAGGAGTCGGTGGTC-3′) and TthS12H76E-2 (5′-GACCACCGACTCCTCCTGAAGG-3′); and H76K with TthS12H76K-1 (5′-CCTTCAGGAGAAGTCGGTGGTC-3′) and TthS12H76K-2 (5′-GACCACCGACTTCTCCTGAAGG-3′). The genes encoding EF-Tu, tufA and tufB, were individually amplified from genomic DNA by PCR using either primer pair Tth tufA-1 (5′-CCAAGCAGGTCCAGGAGAAGCTCATC-3′) and Tth tufA-2 (5′-GGCCGAGGCGTCCAGGGTCTTGTG-3′), or primer pair Tth tufB-1 (5′-GACTGCCCTGGAGCCACCCAGG-3′) and Tth tufB-2 (5′-CCGGACCTCGCTGGCCATCTCGG-3′). Sequencing of PCR products was performed using primers Tth tufAB-1 (5′-GGAGAACGAGTGGGTGGACAAG-3′) or Tth tufAB-2 (5′-GGAATGTACTCGTCAATCGCGTCC-3′).
Isothermal titration calorimetry
ITC was conducted at MicroCal, LLC. Purified 70S ribosomes were sent at a concentration of 3 μM in the following buffer against which they were dialyzed: 10 mM HEPES•KOH (pH 7.6), 10 mM MgCl2, 50 mM NH4Cl, 5 mM β-mercaptoethanol. Streptomycin was resuspended in the excess dialysis buffer at 60 μM concentration. Ribosomes and streptomycin were shipped and stored at 4°C. Binding was conducted at 37°C with the following technical details provided by MicroCal: A negative control lacking ribosomes showed small and constant heat effects as expected. A 300 μL syringe was used for all samples. For experimental samples, 60 μM streptomycin was titrated into 2.8 μM 70S in this manner: 10 μL per injection (except a 2 μL first injection); 3.5 min per injection; 20 sec injection duration; 2 sec filter; 280 rpm stirring; 1.428 mL cell volume. Data were fitted using Microsoft Excel.
ACKNOWLEDGMENTS
We thank Andrew Carter and Venki Ramakrishnan for providing their K53E mutant prior to publication, Eric Cundliffe and Michael O'Connor for providing kirromycin, Michael O'Connor, James Dahlberg, and Elsebet Lund for discussions, and Judith Nathanson for help with the figures. This work was supported by grant GM19756 from the U.S. National Institutes of Health to A.E.D. This work is dedicated to the memory of David Carr.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1355709.
REFERENCES
- Berchtold H., Reshetnikova L., Reiser C.O., Schirmer N.K., Sprinzl M., Hilgenfeld R. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- Bilgin N., Claesens F., Pahverk H., Ehrenberg M. Kinetic properties of Escherichia coli ribosomes with altered forms of S12. J. Mol. Biol. 1992;224:1011–1027. doi: 10.1016/0022-2836(92)90466-w. [DOI] [PubMed] [Google Scholar]
- Carr J.F., Gregory S.T., Dahlberg A.E. Severity of the streptomycin resistance and streptomycin dependence phenotypes of ribosomal protein S12 of Thermus thermophilus depends on the identity of highly conserved amino acid residues. J. Bacteriol. 2005;18:3548–3550. doi: 10.1128/JB.187.10.3548-3550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P. University of Cambridge; Cambridge, UK: 2002. “Structural studies of the 30S ribosomal subunit.”. Ph.D. thesis, [Google Scholar]
- Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Cochella L., Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. The PyMol molecular graphics system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Duisterwinkel F.J., de Graaf J.M., Kraal B., Bosch L. A kirromycin resistant elongation factor EF-Tu from Escherichia coli contains a threonine instead of an alanine in position 375. FEBS Lett. 1981;131:89–93. doi: 10.1016/0014-5793(81)80894-0. [DOI] [PubMed] [Google Scholar]
- Gregory S.T., Cate J.H.D., Dahlberg A.E. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus . J. Mol. Biol. 2001;309:333–338. doi: 10.1006/jmbi.2001.4676. [DOI] [PubMed] [Google Scholar]
- Gregory S.T., Carr J.F., Rodriguez-Correa D., Dahlberg A.E. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus . J. Bacteriol. 2005;187:4804–4812. doi: 10.1128/JB.187.14.4804-4812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski K.B., Rodnina M.V. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 2004;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J. Mol. Biol. 1971;58:439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Koyama Y., Hoshino T., Tomizuka N., Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjansson J.K., Hreggvidsson G.O., Alfredsson G.A. Isolation of halotolerant Thermus spp. from submarine hot springs in Iceland. Appl. Environ. Microbiol. 1986;52:1313–1316. doi: 10.1128/aem.52.6.1313-1316.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland C.G., Hughes D., Ehrenberg M. Limitations of translational accuracy. In: Neidhardt F.C., et al., editors. Escherichia coli and Salmonella: Cellular and molecular biology. American Society for Microbiology; Washington, DC: 1996. pp. 979–1004. [Google Scholar]
- Ogle J.M., Ramakrishnan V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Brodersen D.E., Clemons W.M., Jr, Tarry M.J., Carter A.P., Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Murphy F.V., IV, Tarry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ogle J.M., Carter A.P., Ramakrishnan V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Parmeggiani A., Nissen P. Elongation factor Tu-targeted antibiotics: Four different structures, two mechanisms of action. FEBS Lett. 2006;580:4576–4581. doi: 10.1016/j.febslet.2006.07.039. [DOI] [PubMed] [Google Scholar]
- Piepenburg O., Pape T., Pleiss J.A., Wintermeyer W., Uhlenbeck O.C., Rodnina M.V. Intact aminoacyl-tRNA is required to trigger GTP hydrolysis by elongation factor Tu on the ribosome. Biochemistry. 2000;39:1734–1738. doi: 10.1021/bi992331y. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V., Gromadski K.B., Kothe U., Wieden H.J. Recognition and selection of tRNA in translation. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Sam T., Pingoud A., Bosch L. Mutant species of EF-Tu, altered at position 375, exhibit a reduced affinity for aminoacylated transfer-RNAs. FEBS Lett. 1985;185:51–56. doi: 10.1016/0014-5793(85)80739-0. [DOI] [PubMed] [Google Scholar]
- Sharma D., Cukras A.R., Rogers E.J., Southworth D.R., Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J. Mol. Biol. 2007;374:1065–1076. doi: 10.1016/j.jmb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Rodnina M.V., Wieden H.J., Zemlin F., Wintermeyer W., van Heel M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- Timms A.R., Bridges B.A. Double, independent mutational events in the rpsL gene of Escherichia coli: An example of hypermutability? Mol. Microbiol. 1993;9:335–342. doi: 10.1111/j.1365-2958.1993.tb01694.x. [DOI] [PubMed] [Google Scholar]
- Tubulekas I., Buckingham R.H., Hughes D. Mutant ribosomes can generate dominant kirromycin resistance. J. Bacteriol. 1991;173:3635–3643. doi: 10.1128/jb.173.12.3635-3643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Li W., Stagg S.M., Sengupta J., Nielsen R.C., Nissen P., Harvey S.C., Ehrenberg M., Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Vorstenbosch E.L., Potapov A.P., de Graaf J.M., Kraal B. The effect of mutations in EF-Tu on its affinity for tRNA as measured by two novel and independent methods of general applicability. J. Biochem. Biophys. Methods. 2000;42:1–14. doi: 10.1016/s0165-022x(99)00032-9. [DOI] [PubMed] [Google Scholar]