Abstract
The exit (E) site has been implicated in several ribosomal activities, including translocation, decoding, and maintenance of the translational reading frame. Here, we target the 30S subunit E site by introducing a deletion in rpsG that truncates the β-hairpin of ribosomal protein S7. This mutation (S7ΔR77–Y84) increases both −1 and +1 frameshifting but does not increase miscoding, providing evidence that the 30S E site plays a specific role in frame maintenance. Mutation S7ΔR77–Y84 also stimulates +1 programmed frameshifting during prfB′-lacZ translation in many synthetic contexts. However, no effect is seen when the E codon of the frameshift site corresponds to those found in nature, suggesting that E-tRNA release does not normally limit the rate of prfB frameshifting. Ribosomes containing S7ΔR77–Y84 exhibit an elevated rate of spontaneous reverse translocation and an increased K 1/2 for E-tRNA. These effects are of similar magnitude, suggesting that both result from destabilization of E-tRNA. Finally, this mutation of the 30S E site does not inhibit EF-G-dependent translocation, consistent with a primary role for the 50S E site in the mechanism.
Keywords: ribosome, tRNA, S7, frameshift, translocation
INTRODUCTION
During translation, the tRNAs pass through three distinct sites of the ribosome: the aminoacyl (A) site, the peptidyl (P) site, and the exit (E) site. Aminoacyl-tRNA enters the A site with the help of elongation factor Tu (EF-Tu) in a multistep selection process involving GTP hydrolysis and governed by cognate codon–anticodon interaction. Once aminoacyl-tRNA is in the A site, the nascent peptide is transferred from the P-site tRNA to the A-site tRNA, thereby lengthening the peptide by one residue. The tRNAs are then translocated to their adjacent ribosomal sites in a reaction catalyzed by elongation factor G (EF-G), which hydrolyzes GTP in the process. This results in a posttranslocation complex containing deacylated tRNA in the E site, peptidyl-tRNA in the P site, and a vacant A site. To this complex, EF-Tu delivers aminoacyl-tRNA to the A site in the next round of elongation.
A question of considerable interest is how the E site contributes to specific steps in translation elongation. Several studies point to the importance of the E site in the mechanism of translocation. In one study, a number of tRNA derivatives were prepared, each having a different modification of the 3′-terminal adenosine (A76) (Lill et al. 1988, 1989). Ribosomal complexes containing these 3′-modified tRNAs in the P site exhibited decreased rates of translocation. These decreased rates correlated with defects in E-site binding, strongly suggesting that interaction between the 50S E site and the 3′ end of the P-site tRNA is critical for translocation. Similar conclusions were drawn from a study in which 2′ substitutions were systematically engineered into the acceptor arm of the P-site substrate (Feinberg and Joseph 2001). O-methyl, deoxy, fluoro, and amino substitutions of the 2′ hydroxyl group at positions 71 and 76 of tRNA inhibited translocation, as did 2′-O-methyl substitution at positions 66 and 70. These regions of the tRNA contact rRNA in the 50S E site, further implicating this site in the mechanism of translocation. Mutation of 23S rRNA nucleotide C2394, which interacts with A76 of tRNA in the 50S E site, decreased the rate of translocation substantially, providing direct evidence for involvement of the 50S E site in the mechanism of translocation (Walker et al. 2008). Together these studies provide strong evidence that movement of tRNA into the P/E site is integral to the mechanism of translocation.
It has been proposed that another critical role for the E site is in maintenance of the translational reading frame, dependent on codon–anticodon pairing (Wilson and Nierhaus 2006). Some of the evidence supporting this model comes from analysis of a programmed +1 frameshift required for expression of the RF2 gene (prfB) (Márquez et al. 2004; Sanders and Curran 2007). Because frameshifting competes with UGA-directed termination, this programmed frameshift enables RF2 to negatively regulate its expression in Escherichia coli and many other bacteria. Efficient frameshifting depends on a Shine–Dalgarno (SD)-like element just upstream of the frameshift site. It has been suggested that base-pairing between this SD element and the 3′ end of 16S rRNA destabilizes the E-site tRNA (E-tRNA) and thereby promotes high-level frameshifting. Consistent with this model, in vitro studies have shown that frameshifting correlates with the dissociation of tRNA from the E site (Márquez et al. 2004). Using model mRNAs that lack a SD element, it was also shown that an E-tRNA could stabilize the mRNA register in the ribosome. In vivo studies of prfB′-lacZ expression also suggest that E-tRNA plays a role in maintenance of the reading frame (Sanders and Curran 2007). An extensive mutagenesis of the E codon at the frameshift site revealed that the efficiency of frameshifting is inversely correlated with the predicted stability of codon–anticodon interaction in the E site. These data are consistent with the idea that +1 frameshifting in prfB depends on dissociation of tRNA from the E site.
It is generally agreed that codon–anticodon pairing can occur in the E site. However, the kinetic stability of E-tRNA and the degree to which codon–anticodon pairing contributes to E-tRNA binding have been controversial (Semenkov et al. 1996; Nierhaus et al. 1997). When the first X-ray crystal structures of ribosomes containing E-tRNA were solved, the position of the E codon was incompatible with Watson–Crick pairing to the tRNA (Yusupov et al. 2001; Yusupova et al. 2001; Korostelev et al. 2006; Selmer et al. 2006). A caveat to these observations was that the E-tRNA was noncognate, which raised questions about the biological relevance of the E-codon position. More recently, structures of complexes believed to resemble post-initiation-stage ribosomes have been solved (Yusupova et al. 2006; Jenner et al. 2007). In these complexes, the ribose–phosphate backbones of the E-site codon and anticodon were positioned to form an A-helix, consistent with codon–anticodon pairing in the E site during elongation. Another conclusion that can be drawn from these studies is that the SD–anti-SD (ASD) helix and its relative orientation impact the positioning of the E codon and its potential to pair with an E-site tRNA.
The E site has also been implicated in the fidelity of aminoacyl-tRNA selection (Wilson and Nierhaus 2006). It has been proposed that noncognate ternary complexes are excluded from the A site when the E site is occupied. Support for this model comes from experiments in which mixtures of cognate and noncognate ternary complexes (EF-Tu·GTP·Phe-tRNA and EF-Tu·GTP·Asp-tRNA, respectively) were added to ribosomes programmed with polyU and containing P-site N-acetyl-Phe-tRNA (Geigenmüller and Nierhaus 1990). Analysis of the dipeptides formed indicated that substantial miscoding occurred unless excess deacylated tRNAPhe was added prior to the mix of ternary complexes. It was suggested that deacylated tRNAPhe binding to the E site allosterically influenced the A site, making decoding more stringent. This interpretation was in line with other biochemical evidence for allosteric coupling between the E and A sites (Spahn and Nierhaus 1998).
In this study, we analyze the effects of truncating the β-hairpin of ribosomal protein S7 (ΔR77–Y84) on ribosome function. We find that this 30S E-site mutation stimulates −1 and +1 frameshifting but does not increase the frequency of misreading or stop codon readthrough. In addition, we show that this 30S E-site mutation fails to inhibit EF-G-dependent translocation, in sharp contrast to the effects of mutations in the 50S E site. These data provide direct evidence that the E site plays a role in reading frame maintenance in vivo and suggest that the function of the E site in translocation is largely confined to the 50S subunit.
RESULTS
Construction of an E. coli strain with a mutated 30S E site
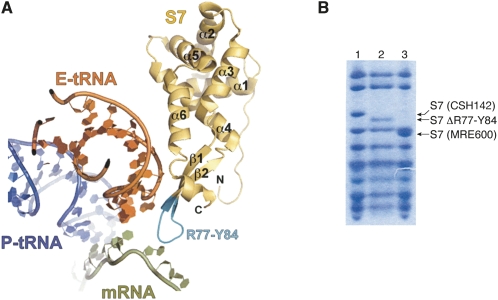
Using the method of Church and coworkers (Link et al. 1997), the region of rpsG encoding residues R77–Y84 of S7 was replaced with two glycine codons on the chromosome of E. coli. This mutation (rpsGΔR77–Y84) effectively truncates the β hairpin element of S7, removing several residues positioned to interact with the anticodon region of E-tRNA and/or the E codon (Fig. 1A). Positive isolates were easily identified by PCR amplification of the central portion of rpsG because the mutation shortens the PCR product by 18 base pairs. The mutant strain, named KLF3027, was verified in two ways. First, the PCR product amplified from KLF3027 was sequenced and found to contain the expected mutation. Second, 30S subunits were isolated from KLF3027, its parental strain CSH142, and strain MRE600, and the ribosomal proteins were resolved by SDS-PAGE (Fig. 1B). S7 from KLF3027 migrated faster than S7 from CSH142, consistent with the engineered mutation. In addition, there was no indication that the mutant protein was underrepresented in KLF3027 subunits or that the wild-type S7 protein was additionally present. As expected, S7 from MRE600 migrated faster than S7 from either CSH142 or KLF3027, because the latter E. coli K strains uniquely encode 23 additional residues at the 3′ end of rpsG (Reinbolt et al. 1978; Johanson and Hughes 1992).
FIGURE 1.
Truncation of the β-hairpin element of ribosomal protein S7 in E. coli. (A) Shown is the position of S7 (gold), E-site tRNA (orange), P-site tRNA (blue), and mRNA (green) based on the 4.5 Å X-ray crystal structure of a ribosomal complex believed to correspond to the post-initiation stage of translation (Jenner et al. 2007). For clarity, the rRNA and other proteins were computationally removed. In this work, the region of rpsG encoding residues R77–Y84 of S7 (cyan) was replaced by two glycine codons on the chromosome of E. coli. This mutation effectively truncates the β-hairpin element of S7, removing residues positioned to interact with tRNA and/or mRNA in the 30S E site. (B) Tricine SDS-PAGE analysis (Schägger and von Jagow 1987) indicated that S7 from strain KLF3027 (lane 2) was smaller than S7 from its parent strain CSH142 (lane 1), consistent with the engineered deletion. For comparison, 30S subunit proteins from MRE600 were also subjected to electrophoresis (lane 3). S7 from MRE600 lacks the 23 amino acid extension characteristic of E. coli K strains.
Although strain KLF3027 was readily isolated, mutation rpsGΔR77–Y84 conferred an obvious growth defect. This mutation decreased the growth rate of E. coli in LB broth from 1.9 ± 0.03 doublings/h to 1.4 ± 0.01 doublings/h at 37°C and from 1.4 ± 0.01 doublings/h to 1.1 ± 0.02 doublings/h at 30°C (data not shown).
It was shown previously that S7 lacking the entire β-hairpin element (ΔE73–E89) was efficiently incorporated into 30S subunits, and those subunits were detected in polysome fractions (Fredrick et al. 2000). Thus, we tried to engineer the corresponding chromosomal deletion using the same approach for allelic replacement. However, our efforts proved unsuccessful (data not shown), suggesting that E. coli can only tolerate partial removal of the β-hairpin of S7.
Mutation rpsGΔR77–Y84 increases the efficiency of frameshifting
To test whether this 30S E site mutation affected the fidelity of translation during elongation, we employed a dual luciferase reporter system described previously (Kramer and Farabaugh 2007). In this system, Renilla luciferase (R-luc) and firefly luciferase (F-luc) are encoded from a single open reading frame. Because these luciferases use different substrates, the activities of R-luc and F-luc can be independently measured. The ability of ribosomal mutations to suppress missense, nonsense, or frameshift mutations in the downstream F-luc gene (or the intergenic linker region) can be assessed by measuring the activity of F-luc relative to R-luc.
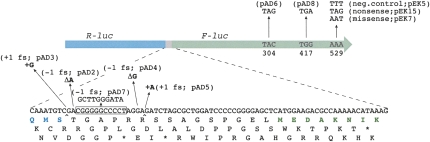
In this work, a number of dual luciferase constructs were used (Fig. 2), some of which were described previously (Kramer and Farabaugh 2007). Five different plasmids were used to measure frameshift suppression. Plasmids pAD2 and pAD4 each contain a single base-pair deletion in the linker region, pAD3 and pAD5 each contain a single base-pair insertion, and pAD7 contains a −1 frameshift shown previously to be suppressed by repairing of peptidyl-tRNAGly on an upstream overlapping codon (O'Connor et al. 1997). Plasmid pEK7, which contains AAT in place of AAA at codon 529 of F-luc, was used to measure missense suppression. Codon 529 corresponds to a critical active-site lysine residue of F-luc; thus Lys-tRNA must misread AAU in this case to generate functional F-luc protein. Plasmids pAD6, pAD8, and pEK15 each contain a premature stop codon in F-luc and were used to measure nonsense suppression. Plasmid pEK5, which contains the noncognate phenylalanine codon at 529, served as a negative control for these experiments. Strains containing this plasmid define the background level of F-luc activity, since UUU cannot form any canonical base pairs with the anticodon of Lys-tRNA (Kramer and Farabaugh 2007).
FIGURE 2.
Dual luciferase reporter constructs used to analyze the effects of ribosomal mutations on the fidelity of translation elongation. In this system, a gene fusion between R-luc (blue) and F-luc (green) allows the efficiency of missense, nonsense, or frameshift suppression to be measured. To measure missense and nonsense suppression, base-pair changes in the F-luc coding region (as indicated) were employed. To measure frameshift suppression, single base-pair insertions and deletions were introduced in the intergenic linker region (pAD2–5; as indicated), as was a −1 frameshift known to be suppressed by slippage of peptidyl-tRNAGly (pAD7) (O'Connor et al. 1997).
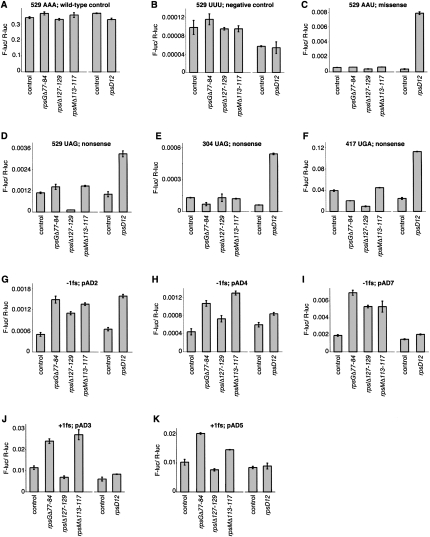
We found that mutation rpsGΔR77–Y84 increased the efficiency of both −1 and +1 frameshifting (Fig. 3). This E-site mutation increased the relative F-luc activity in strains carrying pAD2 (−1 fs), pAD4 (−1 fs), and pAD7 (−1 fs) by two- to fourfold and in strains carrying pAD3 (+1 fs) and pAD5 (+1 fs) by twofold (Fig. 3G–K). Mutation rpsGΔR77–Y84 failed to increase miscoding of AAU, UAG, or UGA codons (Fig. 3C–F). These data suggest that truncation of the β-hairpin of S7 in the 30S E site confers a specific defect in reading frame maintenance.
FIGURE 3.
Effects of 30S subunit mutations on the fidelity of translation elongation. Each panel compares the activity of F-luc relative to that of R-luc (F-luc/R-luc) in various genetic backgrounds. Reporter constructs contained no mutation (A), a mutation of the catalytic Lys codon 529 of F-luc as indicated (B–D), an amber mutation at codon 304 of F-luc (E), an opal mutation at codon 417 of F-luc (F), or a frameshift mutation in the intergenic linker region as indicated (G–K). Isogenic strains harboring mutations rpsGΔR77–Y84, rpsIΔS127–R129, and rpsMΔK113-K117 were derived from CSH142 [control; Δ(gpt-lac)5 ara-600 relA1 spoT1 thi-1] and are shown on the left-hand side of each panel. Strains lacking [control; Δ(gpt-lac)5 ara argE(Am) gyrA(NalR ) rpoB(RifR ) thi-1] or containing mutation rpsD12 were also analyzed and are shown on the right-hand side of each panel. Data represent mean±SEM of three or more independent experiments.
As a positive control, we analyzed the effects of rpsD12 in parallel (Andersson et al. 1982; Dahlgren and Rydén-Aulin 2000). This well-characterized ram mutation in S4 increased misreading of AAU by 20-fold, increased stop codon readthrough by three- to ninefold, and increased −1 frameshifting in certain contexts. These effects of rpsD12 are in line with previous studies and confirm that the missense and nonsense mutations in these constructs can be suppressed by error-prone ribosomes.
Effects of 30S P-site mutations on elongation fidelity
It is clear that codon–anticodon interactions in the 30S P site are critical for maintenance of the reading frame. With this in mind, we tested the effects of two previously described 30S P-site mutations on elongation fidelity and found that both influence frame maintenance. These mutations, rpsIΔS127–R129 and rpsMΔK113–K117, truncate the C-terminal “tails” of ribosomal proteins S9 and S13, respectively (Hoang et al. 2004). Like rpsGΔR77–Y84, rpsMΔK113–K117 increased both −1 and +1 frameshifting by two- to threefold, while rpsIΔS127–R129 increased −1 but not +1 frameshifting. In addition, rpsIΔS127–R129 reduced misreading of UAG (by Lys-tRNA) at codon 529 by ninefold and reduced readthrough of UGA at codon 417 by fourfold. However, no restrictive phenotype was observed with UAG as codon 304, suggesting that the ability of this S9 deletion to decrease misreading of stop codons depends on the context.
Mutation rpsGΔR77–Y84 can increase programmed +1 frameshifting
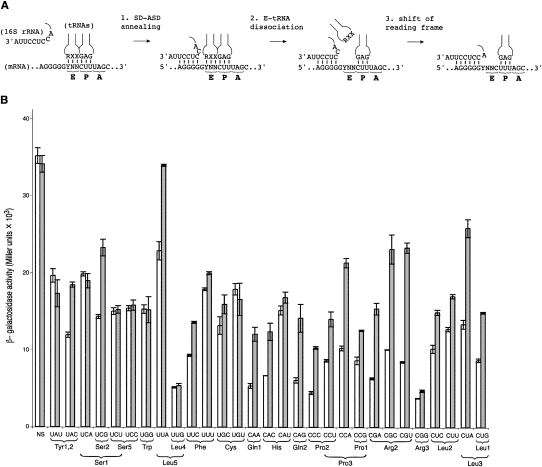
To further investigate the role of the 30S E site in frame maintenance, we tested the effects of mutation rpsGΔR77–Y84 on expression of various prfB′-lacZ reporter constructs (Fig. 4), which differ in the identity of the E codon at the frameshift site (Sanders and Curran 2007). For about two thirds of the mRNAs tested, mutation rpsGΔR77–Y84 increased the efficiency of programmed frameshifting significantly. In the other cases, including the mRNA with the wild-type E codon UAU, no effect was observed. There was no obvious relationship between increased frameshifting and the tRNA isoacceptor predicted to occupy the E site prior to the frameshift event. Furthermore, the ability of rpsGΔR77–Y84 to increase frameshifting did not strictly depend on the identity of any particular nucleotide of the E codon. However, in the six cases where rpsGΔR77–Y84 clearly failed to confer an effect (i.e., UAU, UCA, UCU, UCC, UGG, and UUG), the first nucleotide of the E codon was U.
FIGURE 4.
Mutation rpsGΔR77–Y84 increases +1 programmed frameshifting in a context-dependent manner. (A) Proposed model depicting the major steps involved in the mechanism of prfB programmed frameshifting (Márquez et al. 2004). When the elongating ribosome encounters the in-frame stop codon (normally UGA), annealing between the SD-like sequence in the mRNA and the ASD of the 16S rRNA occurs (step 1), which promotes E-tRNA release (step 2) and repairing of peptidyl-tRNALeu with UUU in the +1 frame (step 3). It has been suggested that the SD–ASD helix extends to include the first nucleotide of the E codon (denoted Y) (Márquez et al. 2004), but structural studies indicate that at least 4 nt of mRNA are necessary to span the distance between the P site and the SD–ASD helix (Yusupova et al. 2006; Jenner et al. 2007; Kaminishi et al. 2007; Korostelev et al. 2007). (B) Plasmids containing prfB′-lacZ with various E codons at the frameshift site (as indicated) were transformed into CSH142 (control; white bars) and KLF3027 (rpsGΔR77–Y84; gray bars), and the level of β-galactosidase activity in the transformants was determined. Relevant tRNA isoacceptor(s) are indicated below each E codon (Dong et al. 1996). In one construct (no shift [NS]), the frameshift of prfB was removed, placing lacZ in frame with the start codon, thus no shift is required for translation of lacZ. Data represent mean±SEM of three or more independent experiments.
Mutation rpsGΔR77–Y84 does not affect regulation of the str operon
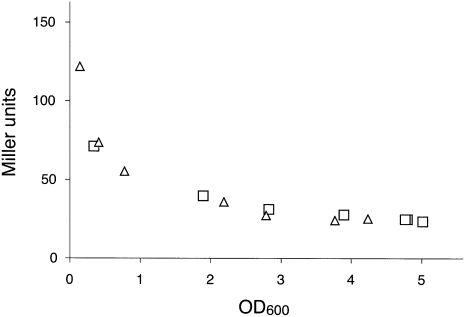
Besides being a component of the ribosome, S7 is a translational feedback repressor of the str operon (Saito et al. 1994; Saito and Nomura 1994), which contains rpsL (S12), rpsG (S7), fusA (EF-G), and tufA (one of the two genes encoding EF-Tu). S7 negatively regulates each product of the operon by binding to a structured region of the mRNA between the rpsL and rpsG reading frames. Hence, we considered the possibility that mutation rpsGΔR77–Y84 conferred its phenotypes indirectly by altering regulation of the str operon. To test this possibility, we cloned the 5′ portion of the operon upstream of lacZ, generating an rpsG′-lacZ translational fusion, and moved this fusion onto the chromosome of E. coli. Expression of rpsG′-lacZ was then compared in control and rpsGΔR77–Y84 cells by assaying β-galactosidase activity at various times throughout batch growth (Fig. 5). No difference was observed, indicating that residues R77–Y84 are unimportant for the regulatory function of S7. These data suggest that mutation rpsGΔR77–Y84 decreases frame maintenance directly by altering the 30S E site rather than indirectly by altering regulation of the str operon.
FIGURE 5.
Mutation rpsGΔR77–Y84 does not affect regulation of the str operon. A fragment of DNA containing the promoter, rpsL, and the first 15 codons of rpsG was cloned upstream of lacZ, such that rpsG′ and lacZ were in frame. This translational fusion was moved as part of a recombinant λ prophage onto the chromosome of E. coli lacking (△) or containing mutation rpsGΔR77–Y84 (□). β-galactosidase activity in the resulting lysogens was assayed throughout batch growth in LB media.
Effects of S7ΔR77–Y84 on spontaneous reverse translocation
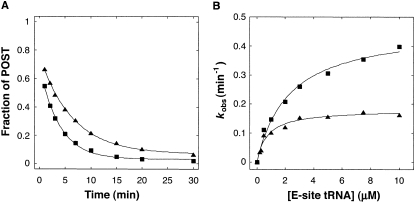
In certain ribosomal complexes, the pre-translocation (PRE) state is more stable than the post-translocation (POST) state; thus movement of the codon–anticodon helices occurs spontaneously in the reverse direction (Shoji et al. 2006; Konevega et al. 2007). To further characterize E-site mutation S7ΔR77–Y84, we studied its effects on spontaneous reverse translocation in vitro (Fig. 6). We purified ribosomes from strains CSH142 (control) and KLF3027 (rpsGΔR77–Y84), incubated them with message m292 and N-acetyl-Val-tRNA to bind the P site, and then added tRNA cognate for the E codon (tRNAfMet) to initiation reverse translocation. Apparent rates of reverse translocation were measured at several concentrations of E-tRNA, which allowed us to determine the maximal rate of reverse translocation (k rev) and the apparent affinity of tRNA for the E site (i.e., K 1/2) (Fig. 6B). Mutation S7ΔR77–Y84 increased both k rev and K 1/2 by threefold. These effects are consistent with destabilization of E-tRNA by mutation S7ΔR77–Y84.
FIGURE 6.
Effects of S7ΔR77–Y84 on spontaneous reverse translocation. (A) A representative experiment measuring reverse translocation. Ribosomes (0.7 μM) without (▲) or with mutation S7ΔR77–Y84 (■) were incubated with message m292 (0.5 μM) and N-acetyl-Val-tRNAVal (1 μM) to bind the P site; complexes were then diluted by 10-fold (to 0.05 μM) in the presence of tRNA cognate for the E codon (tRNAfMet; 3 μM in this example), and the fraction of ribosomes remaining in the posttranslocation state (POST) as a function of time was determined by toeprinting. The data were fit to a single exponential function to obtain apparent rates (k app). For each k app value obtained, the standard error was less than 10%. (B) For control (▲) and mutant (■) ribosomes, the apparent rate of reverse translocation was determined at several concentrations of E-site tRNAfMet. The data were fit to the equation k app = ([E-tRNA]·k rev) / ([E-tRNA] + K 1/2) to estimate k rev, the maximal rate of reverse translocation (control, 0.18 min−1; mutant, 0.47 min−1), and K 1/2, the concentration of E-tRNA at which half-maximal rate was observed (control, 0.74 μM; mutant, 2.3 μM).
Effects of S7ΔR77–Y84 on EF-G-dependent translocation
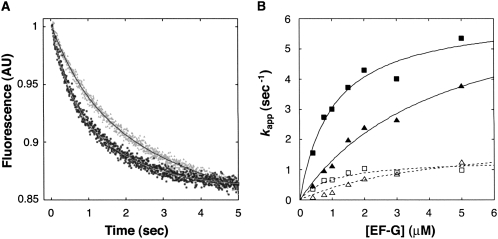
We also investigated the effects of S7ΔR77–Y84 on EF-G-dependent translocation (Fig. 7), using a fluorescence stopped-flow assay (Studer et al. 2003). PRE complexes were made by incubating ribosomes with 3′-pyrene-labeled mRNA and tRNAfMet to bind the P site and then adding N-acetyl-Val-tRNAVal to bind the A site. PRE complexes were rapidly mixed with EF-G·GTP, and decreased pyrene fluorescence resulting from mRNA movement was observed. For both control and mutant ribosomes, the decrease in fluorescence intensity over time did not fit a single-exponential function, but instead fit a double-exponential function. This phenomenon was also seen using ribosomes isolated from E. coli MRE600 (data not shown) and has been observed previously (Peske et al. 2004; Walker et al. 2008; S. Joseph, pers. comm.). The two phases may reflect two populations of ribosomes undergoing translocation at different rates, although this hypothesis remains to be proven. Amplitudes for the fast and slow processes were similar and were unaffected by S7ΔR77–Y84. We determined apparent rates at various concentrations of EF-G and used the k app1 values to estimate k trans, the maximal rate of translocation, and K 1/2, the concentration of EF-G at which half-maximal rate was observed. Based on this analysis, mutation S7ΔR77–Y84 did not affect k trans but decreased K 1/2 by threefold.
FIGURE 7.
Effects of S7ΔR77–Y84 on EF-G-dependent translocation. (A) A representative experiment measuring EF-G-dependent translocation. Pretranslocation (PRE) complexes were formed by incubating control (▲) and mutant (■) ribosomes with pyrene-labeled mRNA and tRNAfMet to bind the P site, and then adding N-acetyl-Val-tRNAVal to bind the A site. Each PRE complex (0.2 μM) was then rapidly mixed with EF-G·GTP (0.75 μM in this example) in a stopped-flow spectrometer, and the change in fluorescence (reflecting mRNA movement) was monitored as a function of time. In each case, the data were fit to a double exponential function, from which apparent rate constants for the fast and slow processes were obtained. (B) Apparent rate constants (k app) for the fast (closed symbols) and slow (open symbols) processes were plotted versus EF-G concentration for control (▲,△) and mutant ribosomes (■,□). Data for the fast process were fit to the equation k app1 = ([EF-G]·k trans) / ([EF-G] + K 1/2) to yield k trans, the maximal rate of translocation (control, 8.3 sec−1; mutant, 9.1 sec−1), and K 1/2, the concentration of EF-G at which half-maximal rate was observed (control, 4.0 μM; mutant, 1.2 μM).
DISCUSSION
The 30S E site contributes to maintenance of the translational reading frame
Several studies support the idea that codon–anticodon interaction in the E site contributes to frame maintenance (Márquez et al. 2004; Trimble et al. 2004; Jenner et al. 2007; Sanders and Curran 2007), but direct in vivo evidence for this hypothesis has been scant. Here, we generated a deletion in rpsG that truncates the β-hairpin element in ribosomal protein S7, removing several residues predicted to interact with tRNA and/or mRNA in the 30S E site of the ribosome. This mutation, S7ΔR77–Y84, increases −1 and +1 frameshifting in an Rluc-Fluc reporter system and promotes +1 programmed frameshifting during translation of prfB′-lacZ in many synthetic contexts. The defects conferred by this E-site mutation seem specific to frame maintenance, because there was no evidence for suppression of a missense mutation or three nonsense mutations in F-luc. These nonsense and missense mutations were suppressed by the ram mutation rpsD12, demonstrating that misreading could be detected using these constructs. Regulation of the str operon was uninfluenced by mutation S7ΔR77–Y84, arguing against the possibility that its effects on translation are conferred indirectly. Together these data provide strong evidence that the 30S E site contributes primarily to reading frame maintenance.
Recent X-ray structures suggest that, during elongation, the β-hairpin of S7 is in position to contact not only the anticodon loop of tRNA but also nucleotides of mRNA immediately upstream of the E codon (i.e., positions −4 and −5) (Yusupova et al. 2006; Jenner et al. 2007). Based on these structures, one can envision two ways in which S7ΔR77–Y84 may increase the frequency of frameshifting. S7ΔR77–Y84 may stimulate dissociation of E-tRNA, thereby increasing the probability that E and A sites are simultaneously unoccupied. In this situation, the elongating ribosome is presumably more susceptible to a frameshift because a single codon–anticodon helix holds the mRNA register. Another possibility is that S7ΔR77–Y84 destabilizes mRNA more directly. This mutation removes ribosomal contacts to mRNA nucleotides just 5′ of the E codon, contacts that appear to stabilize the conformation of the E codon and its ability to pair to tRNA. Without these contacts, the elongating ribosome may be more susceptible to a frameshift even before tRNA dissociates from the E site.
The ability of S7ΔR77–Y84 to increase programmed +1 frameshifting in prfB depends on the context
We found that S7ΔR77–Y84 increases programmed +1 frameshifting during translation of many but not all of the prfB′-lacZ constructs tested, which differ only in the sequence of the E codon at the frameshift site. The molecular basis for this striking context dependence remains unclear. The particular species of tRNA in the E site does not seem to govern whether S7ΔR77–Y84 increases the frequency of programmed +1 frameshifting. For example, S7ΔR77–Y84 confers an effect when the E codon is UUA but not UUG, despite that both are recognized by tRNALeu5, UCG but not UCA, despite that both are recognized by tRNASer1, and UAC but not UAU, despite that both are recognized by tRNATyr1 and tRNATyr2 (which are nearly identical molecules). It seems instead that the sequence of the E codon holds more relevance for the observed context dependence. An alignment of prfB frameshift sites from many bacteria reveals that the first and third nucleotides of the E codon are most often U (∼90% conservation), while the middle nucleotide is anything but U (Baranov et al. 2002). Of the six constructs where S7ΔR77–Y84 clearly fails to confer an effect (i.e., UAU, UCA, UCU, UCC, UGG, UUG, and UGU), three contain an E codon that is well represented in naturally occurring prfB frameshift sites (i.e., UAU, UCU, and UGU).
A general model has been proposed to describe the molecular mechanism of programmed +1 frameshifting of prfB (Fig. 4A; Márquez et al. 2004). Once the elongating ribosome pauses at the stop codon (normally UGA), initial annealing between the ASD of 16S rRNA and the SD-like sequence 5′-AGGGGG-3′ of the mRNA occurs (step 1). Extension of this SD–ASD helix destabilizes E-tRNA, causing it to dissociate (step 2). Then, the +1 shift of the reading frame occurs, an event promoted by the short suboptimal distance between SD–ASD helix and the P codon (step 3). For the majority of prfB′-lacZ constructs tested, mutation S7ΔR77–Y84 stimulates +1 frameshifting. The simplest interpretation of these data is that, by destabilizing the E-site codon–anticodon helix, S7ΔR77–Y84 increases the rate of step 2, which contributes (at least in these cases) to the overall rate of programmed frameshifting. A similar explanation has been provided to explain the inverse correlation between the efficiency of prfB frameshifting and the predicted stability of the E-site codon–anticodon helix (Sanders and Curran 2007). However, as mentioned above, six constructs were clearly unaffected by the E-site mutation. Among these constructs, the naturally occurring E codons of the frameshift site (UAU, UCU, and UGU) are represented (Baranov et al. 2002). The fact that S7ΔR77–Y84 confers no effect suggests that, in these cases, either (1) S7ΔR77–Y84 fails to increase the rate of step 2 or (2) the overall rate of programmed frameshifting is limited by a step other than step 2. In our view, the former explanation seems less likely. It seems improbable that an engineered mutation in S7 would destabilize most codon–anticodon helices in the E site except for a small subset that includes those naturally involved in the frameshift mechanism. It seems more likely that E-tRNA release does not normally limit the rate of prfB programmed frameshifting. More experiments will be needed to investigate this hypothesis further.
Effects of P-site mutations on translation elongation
Removal of the C-terminal tails of S9 and S13, which contact P-tRNA, also conferred defects in frame maintenance. S13Δ113–117 increased both −1 and +1 frameshifting while S9Δ127–129 increased only −1 frameshifting. These in vivo phenotypes do not correlate with the effects of these mutations on the stability of tRNA for the P site. Mutation S9Δ127–129 increases the dissociation rate (k off) of several tRNAs (tRNAPhe, tRNAVal, tRNATyr, and tRNAMet) from the P site whereas S13Δ113–117 does not (S. Shoji and K. Fredrick, unpubl.), in line with earlier equilibrium binding and toeprinting experiments (Hoang et al. 2004). In other words, increased frameshifting conferred by S13Δ113–117 cannot be attributed to destabilization of the P-site tRNA per se. Interestingly, the C-terminal residues of S13 interact with P-tRNA near the junction of the ASL stem and D stem (Berk et al. 2006; Korostelev et al. 2006; Selmer et al. 2006). Cryo-EM studies indicate that this region of tRNA is particularly flexible, as bending of tRNA at this location has been observed in various complexes (Valle et al. 2002, 2003a; Li and Frank 2007). We hypothesize that the C terminus of S13 braces the coaxially stacked ASL and D helices to prevent bending of the anticodon arm, thereby maintaining optimal codon–anticodon interactions. Precedence for this idea comes from the cryo-EM reconstruction of a ribosomal complex stalled at a −1 programmed frameshift site (Namy et al. 2006). In this structure, which is believed to be a snapshot of the complex just before the frameshift event, the P-tRNA is obviously bent near the junction of the ASL and D stems.
To our surprise, mutation S9Δ127–129 restricted stop codon readthrough by ninefold in one case (AAA 529 UAG) and fourfold in another (UGG 417 UGA). These data suggest that tRNA–ribosome interactions in the 30S subunit P site somehow affect termination frequency. The molecular basis of this phenomenon remains unclear.
Mutation S7ΔR77–Y84 may stimulate reverse translocation by destabilizing the POST state of the ribosome
The rate constant of spontaneous reverse translocation (k rev) in wild-type ribosomes containing tRNAfMet and N-acetyl-Val-tRNAVal is 0.18 min−1. Using this rate constant, the free energy barrier (ΔG‡) for the reaction can be calculated as 91 kJ/mol from the equation ΔG‡ = −RT in [(k rev·h)/ (k B·T)], where R is the gas constant, T is temperature in degrees K, h is Planck's constant, and k B is Boltzmann's constant. In ribosomes harboring S7ΔR77–Y84, k rev is approximately threefold larger (0.47 min−1), which indicates a decrease in the free energy barrier by 2.5 kJ/mol (i.e., ΔΔG‡ = − 2.5 kJ/mol). Mutation S7ΔR77–Y84 also increases K 1/2 for E-site tRNA by threefold. Recent single-molecule FRET experiments that directly monitored tRNA dissociation from the E/E site (Fei et al. 2008) support earlier evidence that E-tRNA is bound in a kinetically labile fashion (Robertson et al. 1986; Robertson and Wintermeyer 1987; Semenkov et al. 1996). Those data predict that the rate of tRNA dissociation from the E site in our experiments is at least 20 times faster than k rev, and thus K 1/2 should approximate the equilibrium dissociation constant (K D) for E-site tRNA quite well. Because the free energy of E-tRNA binding contributes to the stability to the POST (ground) state, destabilization of the POST (ground) state caused by the mutation can be approximated as ΔΔG° = −RT in (K 1/2, control / K 1/2, mutant) = 2.9 kJ/mol. This estimate of ΔΔG° is similar to and may account for ΔΔG‡. In other words, these data are consistent with the idea that S7ΔR77–Y84 decreases the energy barrier for reverse translocation solely by destabilizing tRNA in the E site of the POST (ground) state.
Mutation S7ΔR77–Y84 does not affect the rate of EF-G-dependent translocation
Mutation S7ΔR77–Y84 has no effect on the maximal rate of EF-G-dependent translocation, suggesting that the 30S E site plays little or no role in the mechanism. In contrast, mutation C2394A of the 50S subunit E site decreases k trans by eightfold (Walker et al. 2008). These differential effects can be interpreted in the framework of the hybrid-state model of translocation. While mutation C2394A is predicted to inhibit formation of the P/E complex (Walker et al. 2008), an important intermediate in the pathway, mutation S7ΔR77–Y84 is not.
Interestingly, S7ΔR77–Y84 enhances the apparent affinity of EF-G for the PRE complex (i.e., decreases K 1/2) by threefold. This effect also contrasts that of C2394A, which increases K 1/2 of EF-G by >20-fold (Walker et al. 2008). Cryo-EM and FRET studies suggest that movement of tRNA into the P/E site is coupled to an intersubunit rotation (termed ratchet-subunit rotation or RSR) (Valle et al. 2003b; Gao et al. 2004; Ermolenko et al. 2007a,b; Cornish et al. 2008), and these events may regulate the interaction of EF-G with the ribosome (Zavialov and Ehrenberg 2003). While it remains unclear how S7ΔR77–Y84 decreases K 1/2 for EF-G, it is tempting to speculate that the mutation somehow stabilizes the RSR conformation of the PRE complex and thereby confers its effect.
Concluding remarks
In this study, the 30S E site of the ribosome was targeted by truncation of the β-hairpin of S7. This mutation reduced the growth rate of E. coli and stimulated frameshifting but not miscoding. These data support the idea that codon–anticodon pairing in the E site helps maintain the reading frame during elongation. We also found that mutation S7ΔR77–Y84 increased programmed frameshifting in prfB in many synthetic contexts. However, no effect was seen when the sequence of E codon of the frameshift site corresponded to those found in nature. These data suggest that E-tRNA release can limit the rate of programmed frameshifting but may not normally do so during prfB translation. Finally, we showed that mutation of the 30S E site does not inhibit EF-G-catalyzed translocation, implying that the importance of the E site in the mechanism is primarily confined to the 50S subunit.
MATERIALS AND METHODS
Bacterial strains and plasmids
Mutation rpsGΔR77–Y84 was introduced into the genome of E. coli strain CSH142 [F- ara Δ(gpt-lac)5] using a method described previously (Link et al. 1997). A DNA fragment containing the 5′ portion of the str operon (including the promoter, rpsL, rpsG, and part of fusA) was amplified from using oligonucleotides #133 (5′-GAAGATCTCTCGAGGCGTCTGCTCAGTGAAGGAGACG-3′) and #135 (5′-GAAGATCTGAAGGTCAGGTTACCAACAAACGG-3′), digested with XhoI and BglII, and cloned into the compatible SalI and BamHI sites of vector pKO3 to generate pKF280. QuikChange mutagenesis (Stratagene) with oligonucleotides #523 (5′-GACTGTAGAAGTTAAGTCTGGTGGTCAGGTACCAGTTGAAGTC-3′) and #524 (5′-GACTTCAACTGGTACCTGACCACCAGACTTAACTTCTACAGTC-3′) was then used to replace codons 77–84 of rpsG with two glycine codons, generating pEH6. Plasmid pEH6 was used to move mutation rpsGΔR77–Y84 onto the chromosome of CSH142 as described (Link et al. 1997), generating strain KLF3027.
Strains used in Figure 5 to determine whether truncation of the β-hairpin affected the ability of S7 to act as a translational repressor were made as follows. A DNA fragment containing the 5′ portion of the str operon (including the promoter, rpsL, and part of rpsG) was amplified using oligonucleotides #840 (5′-GAATTCTTGCCACCTTTTCGGCAT–3′) and #49 (5′-CATGGATTTATCACCGGCTTTACGAGCAGCTTC-3′), digested with EcoRI and BamHI, and cloned into the EcoRI and BamHI sites of pRS552 (Simons et al. 1987). The resulting Pstr-rpsLrpsG′-lacZ fusion was transferred to λRS45 by homologous recombination in vivo, and KanR lysogens of CSH142 and KLF3027 harboring the recombinant λ prophage were selected (Simons et al. 1987). For both lysogens, the presence of a single-copy prophage was confirmed by PCR (Powell et al. 1994).
To generate plasmids pAD2-pAD8, the linker region of pEK4 (Kramer and Farabaugh 2007) was mutagenized using the QuikChange method (Stratagene). Although the first codon of the F-luc reading frame in pEK4 is ATG, mutation of this codon to CTG has no effect on the level of F-luc activity (data not shown), indicating that this AUG is not used as an internal translation initiation site.
Dual luciferase assay
Cells were grown at 37°C to mid-logarithmic phase in LB medium supplemented with ampicillin (100 μg/mL), pelleted, resuspended in 200 μL lysis buffer (1 mg/mL lysozyme, 10 mM Tris-HCl at pH 8.0, 1 mM EDTA), incubated in ice for 10 min, and flash frozen in liquid nitrogen. The samples were then thawed on ice, and 5 μL of extract from each was assayed for firefly and Renilla luciferase activities using the Dual-Luciferase Reporter Assay System (Promega). For each reaction, luminescence (Relative Light Units) was collected over a 10-sec interval using a Wallac 1420 Victor-3 luminometer (Perkin Elmer). Standardized F-luc activity was calculated as the ratio of the F-Luc to R-Luc activity.
β-galactosidase assay
An overnight culture of each strain grown in LB supplemented with chloramphenicol (20 μg/mL) was diluted 1000-fold into fresh media and grown at 37°C for 9 h. The cells were pelleted, washed once in Z buffer (100 mM sodium phosphate at pH 7.0, 10 mM KCl, 10 mM MgSO4), and assayed for β-galactosidase activity as described (Abdi and Fredrick 2005).
Spontaneous reverse translocation
The rate of spontaneous reverse translocation was measured by toeprinting essentially as described (Shoji et al. 2006). Message m292 (0.5 μM) with [32P]-labeled primer annealed at the 3′ end was incubated with 0.7 μM ribosomes and 1 μM N-acetyl-Val-tRNAVal at 37°C for 20 min in polymix buffer (Ehrenberg et al. 1990) to form the P-site binding complex. Reverse translocation was initiated by diluting the complex 10-fold (to 0.05 μM) into polymix buffer containing tRNAfMet (cognate for the E codon; various concentrations) at t = 0, and the reaction was incubated at 37°C. At several time points, a 2-μL aliquot was removed, immediately added to 10 μL of prewarmed extension mix, and further incubated at 37°C for 2 min. Data were plotted and fit to a single exponential function using KaleidaGraph (Synergy Software) to obtain apparent rates (k app). For each k app value obtained, the standard error was less than 10%. The k app values were plotted versus concentration of tRNAfMet and fit to a hyperbolic function as described (Shoji et al. 2006).
EF-G-dependent translocation
The rate of EF-G-dependent translocation was measured by stopped-flow spectroscopy (Studer et al. 2003). Ribosomes (1 μM) were incubated with 1.5 μM m625 [5′-AAGGAAAUAAAAAUGGUAUA(2′-py-U)-3′; py, pyrene] and 1.5 μM tRNAfMet at 37°C for 20 min in polymix buffer to form the P-site binding complex. N-acetyl-Val-tRNAVal (1.5 μM) was then added and incubated an additional 10 min to fill the A site. The resulting PRE complex was diluted fivefold (to 0.2 μM) with polymix buffer just before an experiment. EF-G·GTP was formed by incubating EF-G (various concentrations) with GTP (2 μM) at 37°C for 10 min. Equal volumes of the PRE complex and EF-G·GTP were rapidly mixed at 37°C in an SX18-MV stopped-flow spectrometer (Applied Photophysics). For each experiment, data from at least five shots were averaged and then fit to a double exponential function by using KaleidaGraph (Synergy Software). Apparent rate constants obtained from the curve fitting were plotted versus EF-G concentration and fit to a hyperbolic function as described (Walker et al. 2008).
ACKNOWLEDGMENTS
We thank E. Kramer and P. Farabaugh for providing the dual-luciferase constructs and technical advice, J. Curran for providing the prfB′-lacZ constructs, and M. Rydén-Aulin for providing E. coli strains Xac (rpsD+) and A112 (rpsD12). This work was supported by NIH grant GM072528.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1320109.
REFERENCES
- Abdi N.M., Fredrick K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli . RNA. 2005;11:1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson D.I., Bohman K., Isaksson L.A., Kurland C.G. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli . Mol. Gen. Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- Baranov P.V., Gesteland R.F., Atkins J.F. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk V., Zhang W., Pai R.D., Cate J.H.D. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish P.V., Ermolenko D.N., Noller H.F., Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren A., Rydén-Aulin M. A novel mutation in ribosomal protein S4 that affects the function of a mutated RF1. Biochimie. 2000;82:683–691. doi: 10.1016/s0300-9084(00)01160-3. [DOI] [PubMed] [Google Scholar]
- Dong H., Nillson L., Kurland C.G. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Ehrenberg M., Bilgin N., Kurland C.G. Design and use of a fast and accurate in vitrotranslation system. In: Spedding G., editor. Ribosomes and protein synthesis—A practical approach. IRL Press; Oxford, UK: 1990. pp. 101–129. [Google Scholar]
- Ermolenko D.N., Majumdar Z.K., Hickerson R.P., Spiegel P.C., Clegg R.M., Noller H.F. Observation of intersubunit movement of the ribosome in solution using FRET. J. Mol. Biol. 2007a;370:530–540. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Ermolenko D.N., Spiegel P.C., Majumdar Z.K., Hickerson R.P., Clegg R.M., Noller H.F. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 2007b;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- Fei J., Kosuri P., MacDougall D.D., Gonzalez R.L. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Feinberg J., Joseph S. Identification of molecular interactions between P site tRNA and the ribosome essential for translocation. Proc. Natl. Acad. Sci. 2001;98:11120–11125. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K., Dunny G.M., Noller H.F. Tagging ribosomal protein S7 allows rapid identification of mutants defective in assembly and function of 30 S subunits. J. Mol. Biol. 2000;298:379–394. doi: 10.1006/jmbi.2000.3563. [DOI] [PubMed] [Google Scholar]
- Gao H., Valle M., Ehrenberg M., Frank J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J. Struct. Biol. 2004;147:283–290. doi: 10.1016/j.jsb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Geigenmüller U., Nierhaus K.H. Significance of the third tRNA binding site, the E site, on E. coli ribosomes for the accuracy of translation: An occupied E site prevents the binding of noncognate aminoacyl-tRNA to the A site. EMBO J. 1990;9:4527–4533. doi: 10.1002/j.1460-2075.1990.tb07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang L., Fredrick K., Noller H.F. Creating ribosomes with an all-RNA 30S subunit. Proc. Natl. Acad. Sci. 2004;101:12439–12443. doi: 10.1073/pnas.0405227101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L., Rees B., Yusupov M., Yusupova G. Messenger RNA conformations in the ribosomal E site revealed by X-ray crystallography. EMBO Rep. 2007;8:846–858. doi: 10.1038/sj.embor.7401044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U., Hughes D. Comparison of the complete sequence of the str operon in Salmonella typhimurium and Escherichia coli . Gene. 1992;120:93–98. doi: 10.1016/0378-1119(92)90014-g. [DOI] [PubMed] [Google Scholar]
- Kaminishi T., Wilson D.N., Takemoto C., Harms J.M., Kawazoe M., Schluenzen F., Hanawa-Suetsugu K., Shirouzu M., Fucini P., Yokoyama S. A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine–Dalgarno interaction. Structure. 2007;15:289–297. doi: 10.1016/j.str.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Konevega A.L., Fischer N., Semenkov Y.P., Stark H., Wintermeyer W., Rodnina M.V. Spontaneous reverse movement of mRNA-bound tRNA through the ribosome. Nat. Struct. Mol. Biol. 2007;14:318–324. doi: 10.1038/nsmb1221. [DOI] [PubMed] [Google Scholar]
- Korostelev A., Trakhanov S., Laurberg M., Noller H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1–13. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Korostelev A., Trakhanov S., Asahara H., Laurberg M., Lancaster L., Noller H.F. Interactions and dynamics of the Shine–Dalgarno helix in the 70S ribosome. Proc. Natl. Acad. Sci. 2007;104:16840–16843. doi: 10.1073/pnas.0707850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.B., Farabaugh P.J. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Frank J. Transfer RNA in the hybrid P/E state: Correlating molecular dynamics simulations with cryo-EM data. Proc. Natl. Acad. Sci. 2007;104:16540–16545. doi: 10.1073/pnas.0708094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Lepier A., Schwagele F., Sprinzl M., Vogt H., Wintermeyer W. Specific recognition of the 3′-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J. Mol. Biol. 1988;203:699–705. doi: 10.1016/0022-2836(88)90203-3. [DOI] [PubMed] [Google Scholar]
- Lill R., Robertson J.M., Wintermeyer W. Binding of the 3′ terminus of tRNA to 23S rRNA in the ribosomal exit site actively promotes translocation. EMBO J. 1989;8:3933–3938. doi: 10.1002/j.1460-2075.1989.tb08574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link A.J., Phillips D., Church G.M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. J. Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez V., Wilson D.N., Tate W.P., Triana-Alonso F., Nierhaus K.H. Maintaining the ribosomal reading frame: The influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. doi: 10.1016/j.cell.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Namy O., Moran S.J., Stuart D.I., Gilbert R.J.C., Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus K.H., Junemann R., Spahn C.M.T. Are the current three-site models valid descriptions of the ribosomal elongation cycle? Proc. Natl. Acad. Sci. 1997;94:10499–10500. doi: 10.1073/pnas.94.20.10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor M., Thomas C.L., Zimmermann R.A., Dahlberg A.E. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F., Savelsbergh A., Katunin V.I., Rodnina M.V., Wintermeyer W. Conformational changes of the small ribosomal subunit during elongation factor G-dependent tRNA–mRNA translocation. J. Mol. Biol. 2004;343:1183–1194. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- Powell B.S., Court D.L., Nakamura Y., Rivas M.P., Turnbough C.L. Rapid confirmation of single copy λ prophage integration by PCR. Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbolt J., Tritsch D., Wittmann-Liebold B. The primary structure of ribosomal protein S7 from E. coli strains K and B. FEBS Lett. 1978;91:297–301. doi: 10.1016/0014-5793(78)81196-x. [DOI] [PubMed] [Google Scholar]
- Robertson J.M., Wintermeyer W. Mechanism of ribosomal translocation: tRNA binds transiently to an exit site before leaving the ribosome during translocation. J. Mol. Biol. 1987;196:525–540. doi: 10.1016/0022-2836(87)90030-1. [DOI] [PubMed] [Google Scholar]
- Robertson J.M., Paulsen H., Wintermeyer W. Pre-steady-state kinetics of ribosomal translocation. J. Mol. Biol. 1986;192:351–360. doi: 10.1016/0022-2836(86)90370-0. [DOI] [PubMed] [Google Scholar]
- Saito K., Nomura M. Post-transcriptional regulation of the str operon in Escherichia coli: Structural and mutational analysis of the target site for translational repressor S7. J. Mol. Biol. 1994;235:125–139. doi: 10.1016/s0022-2836(05)80021-x. [DOI] [PubMed] [Google Scholar]
- Saito K., Mattheakis L.C., Nomura M. Post-transcriptional regulation of the str operon in Escherichia coli: Ribosomal protein S7 inhibits coupled translation of S7 but not its independent translation. J. Mol. Biol. 1994;235:111–124. doi: 10.1016/s0022-2836(05)80020-8. [DOI] [PubMed] [Google Scholar]
- Sanders C.L., Curran J.F. Genetic analysis of the E site during RF2 programmed frameshifting. RNA. 2007;13:1483–1491. doi: 10.1261/rna.638707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylimide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Selmer M., Dunham C.M., Murphy F.V., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Semenkov Y.P., Rodnina M.V., Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli . Proc. Natl. Acad. Sci. 1996;93:12183–12188. doi: 10.1073/pnas.93.22.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S., Walker S.E., Fredrick K. Reverse translocation of tRNA in the ribosome. Mol. Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R.W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Spahn C.M.T., Nierhaus K.H. Models of the elongation cycle: An evaluation. Biol. Chem. 1998;379:753–772. doi: 10.1515/bchm.1998.379.7.753. [DOI] [PubMed] [Google Scholar]
- Studer S.M., Feinberg J.S., Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. J. Mol. Biol. 2003;327:369–381. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- Trimble M.J., Minnicus A., Williams K.P. tRNA slippage at the tmRNA resume codon. RNA. 2004;10:805–812. doi: 10.1261/rna.7010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Sengupta J., Swami N.K., Grassucci R.A., Burkhardt N., Nierhaus K.H., Agrawal R.K., Frank J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Li W., Stagg S.M., Sengupta J., Nielsen R.C., Nissen P., Harvey S.C., Ehrenberg M., Frank J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003a;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003b;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Walker S.E., Shoji S., Pan D., Cooperman B.S., Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc. Natl. Acad. Sci. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.N., Nierhaus K.H. The E-site story: The importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell. Mol. Life Sci. 2006;63:2725–2737. doi: 10.1007/s00018-006-6125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov M., Yusupova G., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Yusupova G.Z., Yusupov M., Cate J.H.D., Noller H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Yusupova G., Jenner L., Rees B., Moras D., Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–122. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]