Abstract
Characterization of base substitutions in rRNAs has provided important insights into the mechanism of protein synthesis. Knowledge of the structural effects of such alterations is limited, and could be greatly expanded with the development of a genetic system based on an organism amenable to both genetics and structural biology. Here, we describe the genetic analysis of base substitutions in 16S ribosomal RNA of the extreme thermophile Thermus thermophilus, and an analysis of the conformational effects of these substitutions by structure probing with base-specific modifying agents. Gene replacement methods were used to construct a derivative of strain HB8 carrying a single 16S rRNA gene, allowing the isolation of spontaneous streptomycin-resistant mutants and subsequent genetic mapping of mutations by recombination. The residues altered to give streptomycin resistance reside within the central pseudoknot structure of 16S rRNA comprised of helices 1 and 27, and participate in the U13–U20–A915 base triple, the G21–A914 type II sheared G–A base pair, or the G885–C912 Watson–Crick base pair closing helix 27. Substitutions at any of the three residues engaged in the base triple were found to confer resistance. Results from structure probing of the pseudoknot are consistent with perturbation of RNA conformation by these substitutions, potentially explaining their streptomycin-resistance phenotypes.
Keywords: pseudoknot, rRNA, ribosome, streptomycin resistance, Thermus
INTRODUCTION
Genetic analysis has long been a vital instrument in the effort to dissect the mechanism of protein synthesis, and recent advances in structural biology have placed genetic information into a three-dimensional context. While conformational effects of base or amino acid substitutions can sometimes be predicted by inspection of wild-type ribosome structures, the ability to utilize both genetics and structural biology within a single system would allow a direct assessment of the structural impact of these alterations. Such a synthesis of experimental approaches would incorporate the ability to generate genetically homogeneous populations of mutant ribosomes as well as the establishment of criteria to identify mutants as potential targets worthy of crystallization trials. To this end, we have worked to develop the genetics of ribosomes of the thermophile Thermus thermophilus HB8, the source of a number of ribosome crystal structures, and apply structural biochemistry techniques to recognize structurally informative mutants.
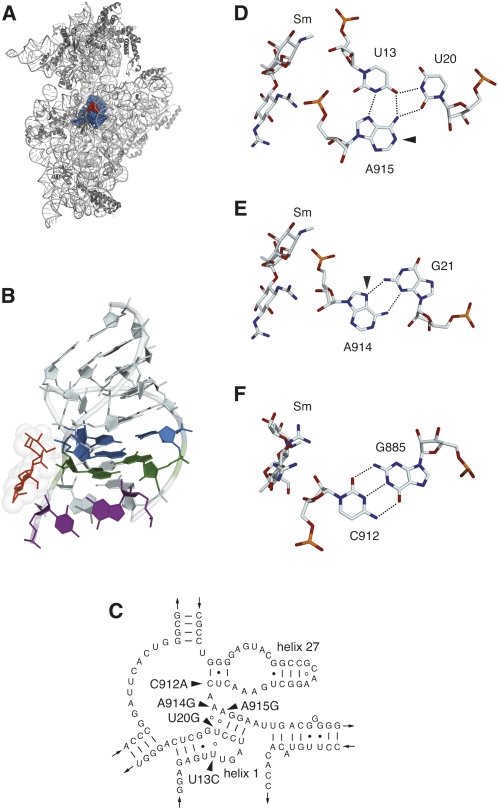
One critical structural element of the ribosome is the central pseudoknot of 16S rRNA, located at the junction of the three major domains of the molecule and composed of parts of helices 1 and 27 (Fig. 1A–C; Cannone et al. 2002). As seen in the crystal structure of the T. thermophilus 30S subunit (Schlüenzen et al. 2000; Wimberly et al. 2000), the central pseudoknot is buried deep within the subunit and makes direct contact with other critical functional components, including 16S rRNA helices 18 and 44 and ribosomal proteins S5 and S12. Genetic studies of Escherichia coli ribosomes have indicated that the structural integrity of the central pseudoknot is important for ribosome function (Poot et al. 1998), and base substitutions in the pseudoknot have been found to interfere with translation initiation (Pinard et al. 1995). A cold-sensitive phenotype attributed to a C23U base substitution in helix 1 indicates that the proper folding of the pseudoknot is important for 30S subunit assembly (Dammel and Noller 1993), consistent with its location in the subunit and with its interaction with multiple structural elements.
FIGURE 1.
(A) Location of the central pseudoknot (blue spheres) and the streptomycin (red) binding site in the T. thermophilus 30S subunit crystal structure (Carter et al. 2000). (B) Structure of the central pseudoknot, showing the U13–A915–U20 base triple (blue), the G21–A915 base pair (green), and the C912–G885 base pair (purple). Streptomycin is shown as red sticks with van der Waals radii indicated. (C) Secondary structure model of the central pseudoknot region of T. thermophilus HB8 16S rRNA (Cannone et al. 2002) modified to reflect the base pairing and base triple arrangements observed in the 30S subunit crystal structure (Wimberly et al. 2000). Sites of base substitutions conferring streptomycin resistance are indicated. (D) The U13–U20–A915 base triple and streptomycin (Sm), showing the hydrogen bonding network (dashed lines) and the site of DMS hyperreactivity (arrowhead) at the N1 of A915. (E) The G21–A914 type II sheared G–A base pair and streptomycin. The arrowhead indicates the N7 of A914 that becomes reactive to DMS in the A914G mutant. (F) The C912–G885 Watson–Crick pair and streptomycin. (A,B,D–F) Generated with PyMol (DeLano 2002) using PDB file 1FJG (Carter et al. 2000).
Examination of the T. thermophilus 30S subunit crystal structure also reveals that the central pseudoknot comprises a significant portion of the binding site for the aminoglycoside antibiotic streptomycin (Carter et al. 2000). Streptomycin causes translational misreading (Davies et al. 1964; Gorini 1974), and a strong case has been made that mistranslation constitutes its primary mode of action (Davis et al. 1986). Crystallographic data indicate that streptomycin-ribosome contacts stabilize a high-affinity substrate binding conformation of the 30S subunit thereby stimulating the erroneous selection of near-cognate aminoacyl-tRNAs (Ogle et al. 2002). Further, by uncoupling codon recognition from GTP hydrolysis by EF-Tu (Gromadski and Rodnina 2004), streptomycin abolishes the contribution of initial recognition and proofreading mechanisms that the ribosome utilizes to discriminate between cognate and near-cognate aminoacyl-tRNAs. The antibiotic makes extensive contact with the 16S rRNA backbone, and base substitutions at any of a number of residues in this vicinity confer resistance (Montandon et al. 1985; Gauthier et al. 1988; Harris et al. 1989; Leclerc et al. 1991; Pinard et al. 1993; Yeh et al. 1994). Streptomycin therefore provides a convenient genetic selection for base substitutions in the drug-binding site, including the central pseudoknot.
Resistance to streptomycin is genetically recessive (Lederberg 1951), and was first found to result from mutation of the rpsL gene encoding ribosomal protein S12 (reviewed by Kurland et al. 1996). The dominance of sensitivity to streptomycin delayed the discovery of resistance mutations in rRNA genes, as rRNA operons are present in multiple copies in most bacteria that are used as experimental organisms. Indeed, the first streptomycin-resistance mutations in 16S rRNA genes were identified in the single rRNA genes of chloroplasts of Euglena gracilis (Montandon et al. 1985) and Chlamydomonas reinhardtii (Gauthier et al. 1988; Harris et al. 1989). Attempts to construct streptomycin-resistance mutations by site-directed mutagenesis in E. coli using multicopy plasmid-encoded rRNA operons have met with some difficulties owing to the recessive nature of the drug-resistance phenotype (Montandon et al. 1985; Frattali et al. 1990). The impediment of mixed ribosome populations can be partly overcome by the use of a secondary mutation allowing phenotypic suppression of wild-type ribosomes (Powers and Noller 1991). Alternatively, the problem of dominance of sensitivity can be bypassed entirely by examining the binding of streptomycin by mutant ribosomes in vitro (Leclerc et al. 1991; Pinard et al. 1993). More recently, streptomycin-resistance mutations have been isolated in a strain of Mycobacterium smegmatis engineered to contain a single rRNA operon and thereby containing homogeneous populations of mutant ribosomes (Springer et al. 2001) and have been characterized in an E. coli strain engineered to express only plasmid-encoded rRNA (Vila-Sanjurjo et al. 2007). Thus, streptomycin resistance could, in principle, permit a direct selection for mutations in the central pseudoknot, provided that the recessive nature of such resistance can be circumvented.
We have previously described antibiotic-resistance mutations in ribosomal protein and rRNA genes of the extremely thermophilic bacterium T. thermophilus (Gregory et al. 2001a, b; Cameron et al. 2004; Carr et al. 2005; Gregory et al. 2005a, b). Our selections for streptomycin-resistant mutants led to the exclusive identification of lesions in rpsL (Gregory et al. 2001a), presumably owing to the presence of two 16S rRNA (rrs) gene copies in this species (Borges and Bergquist 1993). In the present study, we describe the isolation and genetic analysis of streptomycin-resistant mutants of T. thermophilus HB8 bearing single base substitutions in the central pseudoknot. This was made possible by construction of a strain in which one of the two 16S rRNA genes had been inactivated and replaced with a kanamycin-resistance gene. Streptomycin-resistance mutations could be mapped genetically by transformation to the single remaining 16S rRNA gene, relative to other antibiotic-resistance mutations, thus establishing the identified mutations as the cause of resistance. Structural analysis by chemical modification of the streptomycin binding site of mutant ribosomes indicates that these base substitutions confer resistance by sufficiently perturbing the conformation of the central pseudoknot structure so as to abrogate the inhibitory effect of streptomycin. This study also paves the way for future structural analyses of mutant ribosomes, using classical genetic approaches as a starting point.
RESULTS AND DISCUSSION
Construction of a system for genetic analysis of 16S rRNA mutations
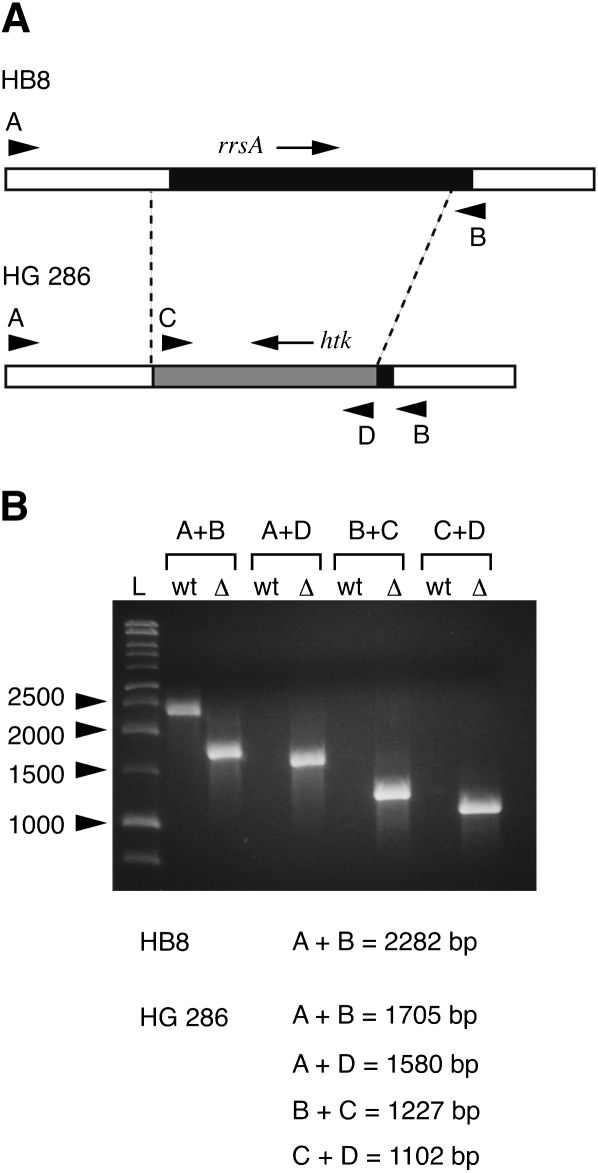
In order to facilitate genetic mapping of mutations by recombination, we constructed a derivative of T. thermophilus HB8 having a single gene encoding 16S rRNA. To construct the null allele, upstream and downstream segments flanking the 16S rRNA gene corresponding to locus 16Sa (spanning positions 131,300–132,803, and hereafter referred to as rrsA) of the unpublished T. thermophilus HB8 genome sequence (Masui et al. 2005; GenBank Accession number AP00826), were amplified by PCR and cloned simultaneously into plasmid pUC18 (see Materials and Methods for details). The htk gene encoding a thermostable kanamycin-adenyltransferase (a kind gift of Hiroyuki Kagamiyama, Osaka Medical College) (Hashimoto et al. 2001) was then inserted into the junction of the upstream and downstream flanking sequences. The final construct (see Fig. 2) contains 475 base pairs of upstream homology and 165 base pairs of downstream homology with the bacterial chromosomal rrsA locus. The resulting deletion was designed to remove the promoter region including the −35 and −10 sequences, the transcription start site, and the entire 16S rRNA coding region except for the 3′ terminal 15–20 nucleotides.
FIGURE 2.
Construction of the ΔrrsA∷ htk1 knockout strain. (A) The 16S rRNA gene rrsA is represented by the black box, with the surrounding chromosomal regions in white. The htk gene is represented by the gray box. Arrows indicate the direction of transcription. Dashed lines indicate the limits of sequence identity between the two chromosomal loci. Arrowheads indicate the positions of primers used for diagnostic PCR analysis; A, primer Tth P16S-1; B, primer Tth T16S-3; C, primer HTK-1; D, primer HTK-2. (B) Diagnostic PCR analysis of the chromosomal rrsA locus of HB8 and HG 286. L, size markers; wt, DNA from wild-type HB8; Δ, DNA from the ΔrrsA∷ htk1 mutant HG 286. Predicted sizes of the PCR products in base pairs are indicated below the gel.
This construct was introduced into T. thermophilus HB8 by natural transformation (Koyama et al. 1986), with kanamycin resistance resulting from recombination and allelic replacement at the site of homology, rrsA. The presence of the null allele was confirmed by diagnostic PCR (Fig. 2), as was the absence of the bla gene of pUC18 (data not shown). One isolate, designated HG 286, was kept for further characterization and mutant selections, and the null allele of this strain is hereafter referred to as ΔrrsA∷ htk1.
Thermus spp. have a noncanonical arrangement of rRNA genes, with two 16S rRNA gene copies, rrsA and rrsB, that are unlinked to and transcribed independently of two 23S rRNA-5S rRNA-tRNAGly operons, rrlA-rrfA-glyT and rrlB-rrfB-glyT (Hartmann et al. 1987; Hartmann and Erdmann 1989). The deletion mutant HG 286, therefore, contains a single intact 16S rRNA gene (rrsB), while retaining two 23S rRNA-5S rRNA-tRNAGly operons. The resulting imbalance in rRNA gene copy number was found to produce a 21% increase in doubling time. Nevertheless, repeated subculturing of the deletion mutant indicated that the htk insertion is genetically stable under the conditions examined.
Amino acid substitutions in ribosomal protein S12
Spontaneous streptomycin-resistant mutants were selected by plating on TEM plates containing 100 μg/mL streptomycin. These arose at a frequency of around 10−7, comparable to the frequencies of other spontaneous antibiotic-resistant mutants of T. thermophilus that we have described previously (Gregory et al. 2001a, b; Cameron et al. 2004; Gregory et al. 2005b). We tentatively identified the mutations by PCR amplification and sequencing of rrsB and rpsL. As expected, some of the mutants isolated on streptomycin contained amino acid substitutions in ribosomal protein S12. Among these were the previously described K42R and K87R substitutions conferring streptomycin resistance, as well as R85C conferring streptomycin pseudodependence (Gregory et al. 2001a). There also appeared two that we had not identified in our previous study, the streptomycin-resistant T40I and K53E substitutions. K53E has been found previously by Carter and co-workers using wild-type T. thermophilus HB8 (Carter 2002; A. Carter and V. Ramakrishnan, pers. comm.). As the focus of this study was the examination of base substitutions in the central pseudoknot of 16S rRNA, these S12 amino acid substitutions are not considered further.
Identification of pseudoknot substitutions
The 16S rRNA base substitutions that were identified in these selections are U13C, U20G, C912A, A914G, and A915G, all located in and around the central pseudoknot. Three of these residues, U13, U20, and A915 engage in a base triple with U13 forming a Hoogsteen interaction with A915 (Fig. 1B,D), while stacking under this base triple is a type II “sheared” G–A pair consisting of G21 and A914 (Fig. 1B,E). C912 forms with G885 the Watson–Crick base pair closing helix 27 (Fig. 1B,F). The base identities of U13, U20, A914, and A915 are highly conserved across the entire phylogenetic spectrum, with greater than 97% conservation among all species and 99% conservation among bacteria (comparative RNA database) (Cannone et al. 2002). C912, on the other hand, is conserved among bacterial, chloroplast, and mitochondrial ribosomes (over 97%), while a U is found at this position in archaeal and eukaryotic cytoplasmic ribosomes. Interestingly, C912U is the substitution found in streptomycin-resistant chloroplast mutants (Montandon et al. 1985; Gauthier et al. 1988; Harris et al. 1989), potentially explaining the intrinsic insensitivity of archaeal and eukaryotic cytoplasmic ribosomes to streptomycin. Despite the degree of conservation of these residues, none of the base substitutions produces a greatly debilitating effect on growth rate. The U13C mutant was found to grow at the wild-type rate (52 min), while the U20G and A915G mutants displayed only minimal increases in doubling time (59 min). The most significant growth defects were observed for the C912A and A914G mutations (65 min doubling times). These effects on growth are similar to those produced by ribosomal protein S12 substitutions (Gregory et al. 2001a).
The levels of streptomycin resistance of the pseudoknot mutants are comparable to or slightly lower than those of ribosomal protein S12 mutants of T. thermophilus (Gregory et al. 2001a). Cells from overnight cultures were streaked for single colonies on TEM plates containing 0, 100, 200, 500, or 1000 μg/mL streptomycin and incubated at 72°C. Alternatively, 10 μL of serial dilutions of overnight cultures were spotted onto streptomycin-containing TEM plates. Both methods produced identical results. At 100 μg/mL, wild-type controls HB8 and HG286 showed no growth (wild-type T. thermophilus shows some inhibition at 10 μg/mL), while all mutants were uninhibited at 200 μg/mL. At 500 μg/mL, the U13C mutant was uninhibited, the U20G, A914G and A915G mutants produced colonies of reduced size and the C912A mutant was completely inhibited. All mutants were completely inhibited at 1000 μg/mL.
Since a U23C base substitution in helix 1 of 16S rRNA has been shown to produce a cold-sensitive, subunit assembly defect in E. coli (Dammel and Noller 1993), the growth of wild-type and streptomycin-resistant T. thermophilus mutants was compared at 72°C and 62°C. Cells from overnight cultures grown at 72°C were streaked for single colonies on TEM plates and incubated at the two temperatures. No cold-sensitive phenotypes were observed.
Genetic analysis
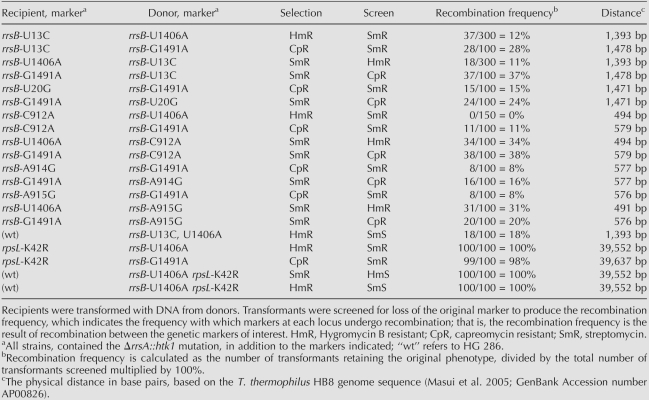
The ability of T. thermophilus to import chromosomal DNA from growth medium and undergo efficient homologous recombination (Koyama et al. 1986) provides a powerful tool for the genetic analysis of mutations. Recombination frequencies in T. thermophilus are sufficiently high that mapping requires markers to be tightly linked, e.g., within the same gene (Hoshino et al. 1994). We obtained such markers from our collection of spontaneous mutants resistant to either of the antibiotics capreomycin or hygromycin B. We had previously characterized such mutants derived from T. thermophilus strain IB-21 (Gregory et al. 2005b) and readily isolated similar mutants of T. thermophilus HB8 for the purpose of this study. These included the hygromycin B-resistance mutation U1406A, and the capreomycin-resistance mutation G1491A. Streptomycin-resistance mutations were genetically mapped by natural transformation relative to these mutations.
The U1406A or G1491A mutations were each transferred from HB8 to HG 286 by transformation to produce strains HG 470 (genotype ΔrrsA∷ htk1 rrsB-U1406A) and HG 473 (ΔrrsA∷ htk1 rrsB-G1491A), respectively. The retention of the ΔrrsA∷ htk1 null allele was confirmed by diagnostic PCR, and the presence of the introduced mutation was confirmed by sequencing rrsB. Streptomycin-resistance mutations were then each mapped in trans by recombination against U1406A or G1491A. For instance, the U13C mutant was transformed with DNA from HG 470, selecting hygromycin B resistance and screening for loss of streptomycin resistance. Acquisition of hygromycin B resistance and retention of streptomycin resistance requires a recombination event occurring between U13 and U1406. The fraction of hygromycin B resistant transformants having lost streptomycin resistance is a reflection of linkage of the two mutations. Reciprocal crosses, in which HG 470 or HG 473 were transformed with DNA from the streptomycin-resistant mutants, were also performed. The results of these analyses are shown in Table 1.
TABLE 1.
Recombination frequencies between mutant alleles of rrsB and rpsLresistant
Crosses in cis were also performed. Strains HG 476 (ΔrrsA∷ htk1 rrsB-U13C, U1406A), HG 478 (ΔrrsA∷ htk1 rrsB-A915G, U1406A), and HG 480 (ΔrrsA∷ htk1 rrsB-A915G, G1491A) were constructed by transformation. DNA from each of these strains was used to transform HG 286, selecting either streptomycin resistance and screening hygromycin B resistance, as in the case of U1406A, or capreomycin resistance as in the case of G1491A, and vice versa. The results from these crosses are also presented in Table 1. Our results resemble those of Hoshino et al. (1994) who found that linkage could be established for tightly linked markers in Thermus spp. by transformation. In general, it can be seen that recombination frequencies are sufficiently high to establish linkage between mutations within the 16S rRNA gene, making genetic mapping by this method straightforward. Curiously, the genetic cross rrsB-C912A versus rrsB-U1406A produced an aberrant recombination frequency. When the rrsB-C912A mutant was used as the recipient and the rrsB-U1406A as the donor allele, no recombinants were recovered. However, the reciprocal cross, rrsB-U1406A versus rrsB-C912A, gave a 34% recombination frequency. While we do not have an explanation for this observation, similar marker effects have been observed in genetic crosses in E. coli (Crawford and Preiss 1972; Stadler and Kariya 1973).
The genome sequences of T. thermophilus HB27 (Henne et al. 2004) and HB8 (Masui et al. 2005) place both 16S rRNA genes and ribosomal protein genes at great enough distances from one another that, given the high recombination frequency, they should behave as genetically unlinked. To establish the absence of linkage between rrsB and rpsL, the streptomycin-resistant rpsL-K42R mutant HG 425 was crossed with the hygromycin B-resistant rrsB-U1406A mutant HG 470 by transformation, selecting hygromycin B resistance and screening for loss of streptomycin resistance (see Table 1). All hygromycin B-resistant transformants screened as streptomycin resistant, indicating that there is no effective genetic linkage between rrsB and rspL. The cis cross was also performed. Strains HG 483 (ΔrrsA∷ htk1 rrsB-U1406A rpsL-K42R) and HG 486 (ΔrrsA∷ htk1 rrsB-G1491A rpsL-K42R) were constructed by transformation and the genotype of each strain was confirmed by sequencing. Crosses with HG 286 also indicated that there is no linkage between rrsB and rpsL. These results demonstrate the utility of a strain having a single genetic locus encoding 16S rRNA and genetic mapping of mutations by transformation and recombination.
Effects of base substitutions on 16S rRNA local structure
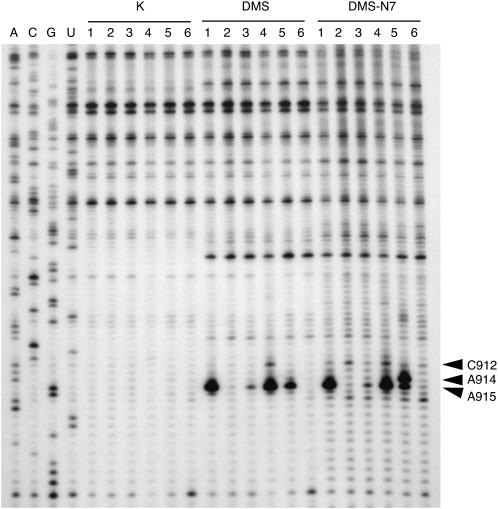
Previous studies with E. coli ribosomes have indicated that base substitutions in the central pseudoknot, including U13A, U13C, A914U, or A914G (Pinard et al. 1993), and C912U or A915G (Leclerc et al. 1991), interfere with streptomycin binding. As streptomycin interacts with 16S rRNA exclusively through backbone contacts (Fig. 1B,D–F), such effects on binding must be a consequence of altered base–base arrangements perturbing backbone trajectory. To investigate the structural effects of these base substitutions, we probed our mutant T. thermophilus 70S ribosomes with the base-specific reagents dimethyl sulfate (DMS), 2-keto-3-ethoxybutyraldehyde (Kethoxal) and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide-metho-p-toluene sulfonate (CMCT). In the U13–U20–A915 base triple configuration, the N1 of A915 is free to react with DMS, and is remarkably hyperreactive to this alkylating agent (Fig. 3). Strong reactivity of this position has previously been observed for E. coli ribosomes (Moazed and Noller 1987).
FIGURE 3.
Structure probing of wild-type and mutant ribosomes in the helix 27 region of 16S rRNA with DMS. Wild-type ribosomes (lane 1); U13C (lane 2); U20G (lane 3); C912A (lane 4); A914G (lane 5); and A915G (lane 6). K, unmodified ribosomes; DMS, modification with dimethylsulfate; DSM–N7, modification with dimethylsulfate followed by reduction with NaBH4 and cleavage with aniline to detect modification at the N7 of Gs. Arrowheads indicate the positions of C912, A914, and A915.
DMS transfers a methyl group via an SN2 displacement mechanism, and reactivity is largely determined by the nucleophilicity of the reacting amine (Streitwieser and Heathcock 1981). Enhanced nucleophilicity of the A915–N1 could be explained by the hydrogen bonding arrangement involving this base. In this base triple the A915–N7 hydrogen bonds with the U13–N3, the A915–N6 participates in two hydrogen bonds with the U13–O4 and the U20–O2, while the U13–O4 in turn participates in a hydrogen bond with the U20–N3. Multiple hydrogen bonding is expected to be cooperative and have the effect of increasing the electronegativity of the A915–N6 amino group (Saenger 1984). Transfer of this enhanced electronegativity to the A915–N1 via resonance could in turn result in its increased nucleophilicity and DMS reactivity. As shown in Figure 3, the effects of base substitutions indicate that the DMS hyper-reactivity of the A915–N1 is indeed established by this particular hydrogen bonding configuration. Loss of the U20–O2 hydrogen bond to A915–N6 with the U20G substitution causes the A915–N1 to react to DMS at a level similar to other accessible adenosines. In contrast, A915 becomes completely refractory to DMS modification in the U13C mutant. This complete loss of DMS reactivity probably results from formation of novel hydrogen bonding interactions involving the A915–N1, the most obvious candidate being a Watson–Crick interaction with U20. The U13C substitution, in addition to abolishing multiple hydrogen bonding interactions, is also likely to create steric clashes involving the exocyclic amino group, favoring displacement of this base toward the major groove. Such a displacement could easily explain the streptomycin-resistance phenotype caused by this base substitution.
The DMS reactivity of A915 is also diminished by the A914G substitution, suggesting that stacking interactions between the U13–U20–A915 base triple and the G21–A914 type II sheared pair contribute to the DMS hyperreactivity of A915. That the integrity of the G21–A914 type II sheared configuration is compromised by the A914G substitution is indicated by the increased reactivity of the G–N7 at 914 to DMS (Fig. 3). This reactivity excludes the possibility of an isosteric sheared G–G pair with a single N2–N7 hydrogen bond in this mutant. Thus, these two bases must engage in a substantially different arrangement, perhaps explaining why the A914G substitution produces the most dramatic effect on growth.
Finally, replacement of C912 with A produces weak reactivity of this position to DMS, suggesting that the terminal base pair of helix 27, rather than forming a type I pseudo-Watson–Crick pair, now forms a type II G–A pair with G885, with the A–N1 at 912 accessible to solvent. No changes in reactivity were observed in helix 1 (data not shown), suggesting that U13 and U20 continue to engage in hydrogen bonding interactions in these mutant ribosomes.
While structure probing with base-specific reagents can provide some indication as to the effects of mutations on ribosome structure, knowledge of the precise conformational rearrangements of these residues will require atomic-resolution X-ray crystallographic analysis. The presence of these mutations in T. thermophilus HB8 increases the likelihood that such information will be obtained in the near future.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and genetic methods
All mutants were derived from the Japanese T. thermophilus strain HB8, ATCC27634 (Oshima and Imahori 1974), obtained from the American Type Culture Collection. T. thermophilus was cultivated aerobically in ATCC medium 1598 (Thermus Enhanced Medium, or TEM) at 72°C. Plates contained 2.8% Difco agar. Spontaneous antibiotic-resistant mutants were isolated by plating ∼109 cells from saturated overnight cultures on TEM plates containing 30 μg/mL kanamycin sulfate, 50 μg/mL hygromycin B, 50 or 100 μg streptomycin sulfate, or 100 or 200 μg/mL capreomycin sulfate. All antibiotics were purchased from Sigma Chemical Corp.
Chromosomal DNA was prepared essentially as described by Marmur (Marmur 1961) and transformation was performed as described by others (Koyama et al. 1986). Single colonies were patched onto TEM plates containing the appropriate antibiotic and incubated at 72°C. PCR amplifications for cloning were performed using Pfu DNA polymerase (Stratagene) or for sequencing using Taq DNA polymerase (Promega). Part of rrsA and rrsB corresponding to positions from −56 to 575 (numbering of the mature 16S rRNA molecule) was amplified using primers Tth 16S-BoxA (5′-GCAGGGGGATCTTGAAAAGGGG-3′) and Tth 16S-D (5′-CTTTACGCCCAGTGAATCCGGG-3′). Part of rrsA and rrsB corresponding to positions 795–1544 was amplified using primers Tth 16S-A (5′-CCCGGGTAGTCCACGCCCTAAACG-3′) and Tth 16S-H (5′-AGAAAGGAGGTGATCCAGCCGCAC-3′). PCR products were sequenced directly using a PCR sequencing kit (USB) or by the University of California, Davis Sequencing service. Oligonucleotide primers were obtained from Operon Technologies and Invitrogen.
To assay the genetic stability of the ΔrrsA∷ htk1 mutation, three independent 10 mL overnight cultures of HG 286 were grown in TEM and subcultured three times by diluting 1:200 into the same medium. Single colonies were obtained at the start and at the finish of the experiment by plating serial dilutions onto TEM plates. Fifty colonies from each culture were screened for loss of kanamycin resistance by patching onto TEM plates plus 30 μg/mL kanamycin sulfate.
Oligonucleotides used for construction of an rrsA deletion mutant
Oligonucleotides used for the PCR amplification of 16S rRNA gene fragments and for amplification of htk were as follows: For the upstream fragment, oligonucleotides Tth P16S-1 (5′-GTGGTCCTCGAGGTACCAGCTCCCCGAG-3′) and Tth P16S-2 (5′-CGAGGGCAGCGGATCCCCTCAATGAAGC-3′) were used. These oligonucleotides introduce Kpn I and Bam HI cloning sites, respectively (restriction enzyme recognition sites underlined). For the downstream fragment, oligonucleotides Tth T16S-1 (5′-GGTGCGGCTGGATCCCCTCCTTTCTAAG-3′) and Tth T16S-2 (5′-CTCTAGAGGATCAAGCTTGAGGACCACC-3′) were used. These oligonucleotides introduce Bam HI and Hind III cloning sites, respectively. Oligonucleotide Tth T16S-3 (5′-CGCGCCGGGTGAAGCTTTGCCCCTTTC-3′) was used for examining the final deletion mutant. The htk gene was amplified using the oligonucleotides HTK-1 (5′-GAATTCGAGCTCGGATCCCGTTGACGGCG-3′) and HTK-2 (5′-GCTTGCATGCGGATCCGTAACCAACATG-3′) which both introduce Bam HI sites. Oligonucleotides used to amplify the bla gene of pUC18 were Bla-1 (5′-GAAGAGTATGAGTATTCAACATTTCC-3′) and Bla-2 (5′-CCAATGCTTAATCAGTGAGGCACC-3′).
Preparation of 70S ribosomes and structure probing with base-specific modifying reagents
70S ribosomes from T. thermophilus HB8 and mutant derivatives were prepared from midlog phase cultures grown as described previously (Cameron et al. 2004). Chemical modification with dimethyl sulfate (DMS), 2-keto-3-ethoxybutyraldehyde (Kethoxal) and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide-metho-p-toluene sulfonate (CMCT) was carried out according to Merryman and Noller (Merryman and Noller 1998), except that DMS was diluted 1:10 or 1:20 prior to use, and modification with Kethoxal was carried out at pH 7.2. DMS modification of the N7 of G residues was monitored by primer extension following treatment with NaBH4 and aniline according to Krol and Carbon (1989). Two independent preparations of ribosomes from each wild-type and mutant strain were examined. Probing of helix 27 was performed using oligonucleotide Tth 16S-B (5′-CGAATTAAACCACATGCTCCACCG-3′) and helix 1 was probed using oligonucleotide Tth 16S-U (5′-GGCCCGCACGACTTGCATGTCTTAGG-3′).
ACKNOWLEDGMENTS
This work was supported by grant GM19756 from the U.S. National Institutes of Health (to A.E.D.). We are grateful to Sarah Delaney and Daniel Jarem for insightful comments. We also are indebted to Riccardo Belardinelli for technical assistance and to Judith Nathanson for her expertise with the preparation of the figures.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1374809.
REFERENCES
- Borges K.M., Bergquist P.L. Genomic restriction map of the extremely thermophilic bacterium Thermus thermophilus HB8. J. Bacteriol. 1993;175:103–110. doi: 10.1128/jb.175.1.103-110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D.M., Thompson J., Gregory S.T., March P.E., Dahlberg A.E. Thiostrepton-resistant mutants of Thermus thermophilus . Nucleic Acids Res. 2004;32:3220–3227. doi: 10.1093/nar/gkh644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone J.J., Subramanian S., Schnare M.N., Collett J.R., D'Souza L.M., Du Y., Feng B., Lin N., Madabusi L.V., Muller K.M., et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr J.F., Gregory S.T., Dahlberg A.E. Severity of the streptomycin resistance and streptomycin dependence phenotypes of ribosomal protein S12 of Thermus thermophilus depends on the identity of highly conserved amino acid residues. J. Bacteriol. 2005;187:3548–3550. doi: 10.1128/JB.187.10.3548-3550.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P. Cambridge University; Cambridge, UK: 2002. “Structural studies of the 30S ribosomal subunit”. Ph.D. thesis. [Google Scholar]
- Carter A.P., Clemons W.M., Brodersen D.E., Morgan-Warren R.J., Wimberly B.T., Ramakrishnan V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Crawford I.P., Preiss J. Distribution of closely linked markers following intragenic recombination in Escherichia coli . J. Mol. Biol. 1972;71:717–733. doi: 10.1016/s0022-2836(72)80034-2. [DOI] [PubMed] [Google Scholar]
- Dammel C.S., Noller H.F. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes & Dev. 1993;7:660–670. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- Davies J., Gilbert W., Gorini L. Streptomycin, suppression and the code. Proc. Natl. Acad. Sci. 1964;51:883–890. doi: 10.1073/pnas.51.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.D., Chen L.L., Tai P.C. Misread protein creates membrane channels: An essential step in the bactericidal action of aminoglycosides. Proc. Natl. Acad. Sci. 1986;83:6164–6168. doi: 10.1073/pnas.83.16.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W.L. The PyMol molecular graphics system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Frattali A.L., Flynn M.K., De Stasio E.A., Dahlberg A.E. Effects of mutagenesis of C912 in the streptomycin binding region of Escherichia coli 16S ribosomal RNA. Biochim. Biophys. Acta. 1990;1050:27–33. doi: 10.1016/0167-4781(90)90136-p. [DOI] [PubMed] [Google Scholar]
- Gauthier A., Turmel M., Lemieux C. Mapping of chloroplast mutations conferring resistance to antibiotics in Chlamydomonas: Evidence for a novel site of streptomycin resistance in the small subunit rRNA. Mol. Gen. Genet. 1988;214:192–197. doi: 10.1007/BF00337710. [DOI] [PubMed] [Google Scholar]
- Gorini L. Streptomycin and misreading of the genetic code. In: Nomura M., et al., editors. Ribosomes. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1974. pp. 791–803. [Google Scholar]
- Gregory S.T., Cate J.H.D., Dahlberg A.E. Streptomycin-resistant and streptomycin-dependent mutants of the extreme thermophile Thermus thermophilus . J. Mol. Biol. 2001a;309:333–338. doi: 10.1006/jmbi.2001.4676. [DOI] [PubMed] [Google Scholar]
- Gregory S.T., Cate J.H.D., Dahlberg A.E. Spontaneous erythromycin resistance mutation in a 23S rRNA gene, rrlA, of the extreme thermophile Thermus thermophilus IB-21. J. Bacteriol. 2001b;183:4382–4385. doi: 10.1128/JB.183.14.4382-4385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S.T., Carr J.F., Dahlberg A.E. A mutation in the decoding center of Thermus thermophilus 16S rRNA suggests a novel mechanism of streptomycin resistance. J. Bacteriol. 2005a;187:2200–2202. doi: 10.1128/JB.187.6.2200-2202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory S.T., Carr J.F., Rodriguez-Correa D., Dahlberg A.E. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus . J. Bacteriol. 2005b;187:4804–4812. doi: 10.1128/JB.187.14.4804-4812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski K.B., Rodnina M.V. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 2004;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- Harris E.H., Burkhart B.D., Gillham N.W., Boynton J.E. Antibiotic resistance mutations in the chloroplast 16S and 23S rRNA genes of Chlamydomonas reinhardtii: Correlation of genetic and physical maps of the chloroplast genome. Genetics. 1989;123:281–292. doi: 10.1093/genetics/123.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R.K., Erdmann V.A. Thermus thermophilus 16S rRNA is transcribed from an isolated transcription unit. J. Bacteriol. 1989;171:2933–2941. doi: 10.1128/jb.171.6.2933-2941.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R.K., Ulbrich N., Erdmann V.A. An unusual rRNA operon constellation: In Thermus thermophilus HB8 the 23S/5S rRNA operon is a separate entity from the 16S rRNA operon. Biochimie. 1987;69:1097–1104. doi: 10.1016/0300-9084(87)90009-5. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Yano T., Kuramitsu S., Kagamiyama H. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett. 2001;506:231–234. doi: 10.1016/s0014-5793(01)02926-x. [DOI] [PubMed] [Google Scholar]
- Henne A., Bruggemann H., Raasch C., Wiezer A., Hartsch T., Liesegang H., Johann A., Lienard T., Gohl O., Martinez-Arias R., et al. The genome sequence of the extreme thermophile Thermus thermophilus . Nat. Biotechnol. 2004;22:547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- Hoshino T., Hidaka Y., Kosuge T., Hasegawa M., Saito T., Nakahara T. Transformation as a tool for genetic fine mapping of closely located mutations in Thermus thermophilus . FEMS Microbiol. Lett. 1994;117:175–179. [Google Scholar]
- Koyama Y., Hoshino T., Tomizuka N., Furukawa K. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 1986;166:338–340. doi: 10.1128/jb.166.1.338-340.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol A., Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- Kurland C.G., Hughes D., Ehrenberg M. Limitations of translational accuracy. In: Neidhardt F.C., et al., editors. Escherichia coli Salmonella typhimurium: Cellular and molecular biology. 2nd ed. American Society for Microbiology; Washington, DC: 1996. pp. 979–1004. [Google Scholar]
- Leclerc D., Melançon P., Brakier-Gingras L. Mutations in the 915 region of Escherichia coli 16S ribosomal RNA reduce the binding of streptomycin to the ribosome. Nucleic Acids Res. 1991;19:3973–3977. doi: 10.1093/nar/19.14.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J. Streptomycin resistance: a genetically recessive mutation. J. Bacteriol. 1951;61:549–550. doi: 10.1128/jb.61.5.549-550.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmur J. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 1961;3:208–218. [Google Scholar]
- Masui R., Kurokawa K., Nakagawa N., Tokunaga F., Koyama Y., Shibata T., Oshima T., Yokoyama S., Yasunaga T., Kuramitsu S. 2005. NCBI. Thermus thermophilus HB8, complete genome [Google Scholar]
- Merryman C., Noller H.F. Footprinting and modification-interference analysis of binding sites on RNA. In: Smith C.J.W., editor. RNA: Protein interactions. A practical approach. IRL Press; Oxford, UK: 1998. pp. 237–253. [Google Scholar]
- Moazed D., Noller H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987;327:389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Montandon P.E., Nicolas P., Schurmann P., Stutz E. Streptomycin-resistance of Euglena gracilis chloroplasts: Identification of a point mutation in the 16S rRNA gene in an invariant position. Nucleic Acids Res. 1985;13:4299–4310. doi: 10.1093/nar/13.12.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle J.M., Murphy F.V., Tarrry M.J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Oshima T., Imahori K. Description of Thermus thermophilus (Yoshida and Oshima), comb. nov., a nonsporulating thermophilic bacterium from a Japanese thermal spa. Int. J. Syst. Bacteriol. 1974;24:102–112. [Google Scholar]
- Pinard R., Payant C., Melançon P., Brakier-Gingras L. The 5′ proximal helix of 16S rRNA is involved in the binding of streptomycin to the ribosome. FASEB J. 1993;7:173–176. doi: 10.1096/fasebj.7.1.7678560. [DOI] [PubMed] [Google Scholar]
- Pinard R., Payant C., Brakier-Gingras L. Mutations at positions 13 and/or 914 in Escherichia coli 16S ribosomal RNA interfere with the initiation of protein synthesis. Biochemistry. 1995;34:9611–9616. doi: 10.1021/bi00029a038. [DOI] [PubMed] [Google Scholar]
- Poot R.A., van den Worm S.H., Pleij C.W., van Duin J. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 1998;26:549–553. doi: 10.1093/nar/26.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T., Noller H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger W. Principles of nucleic acid structure. Springer; New York: 1984. p. 117. [Google Scholar]
- Schlüenzen F., Tocilj A., Zarivach R., Harms J., Gluehmann M., Janell D., Bashan A., Bartels H., Agmon I., Franceschi F., et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Springer B., Kidan Y.G., Prammananan T., Ellrott K., Bottger E.C., Sander P. Mechanisms of streptomycin resistance: Selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 2001;45:2877–2884. doi: 10.1128/AAC.45.10.2877-2884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler D., Kariya B. Marker effects in the genetic transduction of tryptophan mutants of E. coli . Genetics. 1973;75:423–439. doi: 10.1093/genetics/75.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streitwieser A., Jr, Heathcock C.H. Introduction to organic chemistry. Macmillan; New York: 1981. [Google Scholar]
- Vila-Sanjurjo A., Lu Y., Aragonez J.L., Starkweather R.E., Sasikumar M., O'Connor M. Modulation of 16S rRNA function by ribosomal protein S12. Biochim. Biophys. Acta. 2007;1769:462–471. doi: 10.1016/j.bbaexp.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Brodersen D.E., Clemons W.M., Jr, Morgan-Warren R.J., Carter A.P., Vonrhein C., Hartsch T., Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Yeh K.C., To K.Y., Sun S.W., Wu M.C., Lin T.Y., Chen C.C. Point mutations in the chloroplast 16s rRNA gene confer streptomycin resistance in Nicotiana plumbaginifolia . Curr. Genet. 1994;26:132–135. doi: 10.1007/BF00313800. [DOI] [PubMed] [Google Scholar]