Abstract
The Cfr methyltransferase confers combined resistance to five different classes of antibiotics that bind to the peptidyl transferase center of bacterial ribosomes. The Cfr-mediated modification has previously been shown to occur on nucleotide A2503 of 23S rRNA and has a mass corresponding to an additional methyl group, but its specific identity and position remained to be elucidated. A novel tandem mass spectrometry approach has been developed to further characterize the Cfr-catalyzed modification. Comparison of nucleoside fragmentation patterns of A2503 from Escherichia coli cfr+ and cfr− strains with those of a chemically synthesized nucleoside standard shows that Cfr catalyzes formation of 8-methyladenosine. In addition, analysis of RNA derived from E. coli strains lacking the m2A2503 methyltransferase reveals that Cfr also has the ability to catalyze methylation at position 2 to form 2,8-dimethyladenosine. The mutation of single conserved cysteine residues in the radical SAM motif CxxxCxxC of Cfr abolishes its activity, lending support to the notion that the Cfr modification reaction occurs via a radical-based mechanism. Antibiotic susceptibility data confirm that the antibiotic resistance conferred by Cfr is provided by methylation at the 8 position and is independent of methylation at the 2 position of A2503. This investigation is, to our knowledge, the first instance where the 8-methyladenosine modification has been described in natural RNA molecules.
Keywords: Cfr, methyltransferase, antibiotic resistance, 8-methyladenosine, mass spectrometry
INTRODUCTION
The peptidyl transferase center (PTC) on the large ribosomal subunit is mainly composed of the central loop of domain V of 23S rRNA and contains binding sites for various antibiotics of clinical and veterinary importance. It is the site of peptide elongation, and therefore an efficient place for drugs to block protein synthesis. Consequently, the PTC is the target for alterations that prevent drug binding and thereby cause antibiotic resistance. Although the resistance mechanisms established so far in this region are mostly RNA mutations, a newly discovered methylation at the PTC mediated by the Cfr methyltransferase (Kehrenberg et al. 2005) confers combined resistance to five different classes of antibiotics that bind to the PTC (Long et al. 2006). The phenotype was named PhLOPSA for resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics (Long et al. 2006). The cfr gene was originally discovered on a multiresistance plasmid isolated during a surveillance study of florfenicol resistance in Staphylococcus spp. isolates of animal origin (Schwarz et al. 2000) and later shown to encode a methyltransferase that methylates nucleotide A2503 of Escherichia coli 23S rRNA (Kehrenberg et al. 2005).
In E. coli, there is a natural m2A methylation at A2503 (Kowalak et al. 1995) mediated by the RlmN methyltransferase (Toh et al. 2008). Although the lack of the natural methylation at A2503 confers a slight increase in susceptibility to tiamulin, hygromycin A, sparsomycin, and linezolid (Toh et al. 2008), this methylation is considered to be a housekeeping modification rather than a bona fide antibiotic resistance determinant. Cfr adds an additional methyl group at A2503 and also, at least in the case of E. coli, reduces the natural 2′-O-ribose methylation at position C2498 (Kehrenberg et al. 2005). Cfr and RlmN contain regions of homology, including a cysteine-rich motif that is characteristic of radical SAM enzymes (Sofia et al. 2001; Toh et al. 2008), a superfamily whose members catalyze a diverse set of reactions including unusual methylations, isomerizations, sulfur insertions, and anaerobic oxidations. Based on the homology between Cfr and RlmN, it has recently been proposed that Cfr causes a previously unobserved methylation of position 8 of A2503 (m8A2503), or alternatively converts m2A to a likewise novel 2-ethyladenosine (Toh et al. 2008).
The aim of this study was to identify the adenosine modification catalyzed by Cfr at position 2503 of 23S rRNA. As A2503 is the substrate of both the Cfr and RlmN methyltransferase, the current study used four E. coli strains that either lacked or contained one or both of these methyltransferase genes to enable identification of the product(s) of Cfr action on A2503. The complete characterization of a novel modification is far from trivial, and typically requires a combination of initial structural analysis by mass spectrometry followed by chemical synthesis of candidate modified nucleosides. To properly characterize the products of Cfr modification, we developed an online nanoliquid chromatography electrospray ionization tandem mass spectrometry (nano-LC-ESI-MSn) approach on an Agilent 6340 ion trap mass spectrometer. Mass spectrometry has successfully been employed in the structural characterization of nucleobases and nucleosides (for reviews, see Crain 1990a; Banoub et al. 2005). Mass spectrometric studies of RNA nucleosides involve collision-induced dissociation (CID) in multiple tandem MS steps (MSn), which typically commences with neutral loss of ribose followed by a concerted ring opening and elimination of ammonia from the protonated nucleobase. Subsequent fragmentation of the deaminated base produces fragments of various masses dependent on individual structure of the base in question (Nelson and McCloskey 1992, 1994; Dudley et al. 2005; Jensen et al. 2007). The similarity in MS fragmentation behavior among the different samples in this study made it necessary to perform up to five tandem MS steps (MS5) in order to structurally characterize the novel modifications. Chromatography on a graphitized carbon column made it possible to separate the differently modified, but isobaric adenosine nucleosides.
In this work, mass spectrometry is used to demonstrate that methylation of position 8 is the main product of Cfr action upon reaction with A2503, and that Cfr also has a less pronounced capability to methylate position 2 of A2503 to produce 2,8-dimethyladenosine. Mutagenesis experiments are used to corroborate that Cfr is a radical SAM methyltransferase by showing that mutations of the cysteines in the presumed radical SAM motif CxxxCxxC abolish Cfr activity. Moreover, it is confirmed that antibiotic resistance is caused by methylation at position 8 of A2503 23S RNA and is independent of the m2A modification.
RESULTS
Analysis of a 23S RNA subfragment containing A2503 by reverse-phase HPLC
As both Cfr and RlmN modify A2503 in 23S rRNA (Fig. 1C), ribosomes were isolated from four strains of E. coli with and without the methyltransferase genes (Table 1). Nucleosides from hydrolysis of a subfragment containing positions 2483–2527 of 23S rRNA (Fig. 1B) were characterized using reverse phase liquid chromatography with UV detection (RP-HLPC/UV) by gradient separation on a standard C18 stationary phase at neutral pH (Fig. 2). Nucleosides originating from both the isolated rRNA fragment and the annealed DNA oligonucleotide used during sample purification were successfully resolved in the chromatogram. The majority of the nucleosides could be identified by comparison of retention times of commercially available standards analyzed under identical chromatographic conditions (data not shown). The observed elution order of the nucleosides is similar to the elution order observed by Pomerantz and McCloskey (1990).
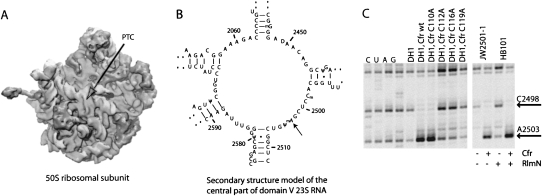
FIGURE 1.
(A) The position of the PTC in the cleft of the 50S ribosomal subunit. (B) The secondary structure of the central loop of domain V of 23S rRNA flanking the methylated nucleotide A2503 that is positioned in the PTC. (C) Primer extension analysis of reverse transcriptase stops on 23S RNA from E. coli caused by modifications at A2503 and C2498. The region shown is limited to the nucleotides at and just proceeding A2503. Lanes marked C, U, A, and G refer to dideoxysequencing reactions. Other labels refer to the strains used for ribosome isolation and the absence or presence of methyltransferases. The slightly stronger bands in two lanes (the C112A and C115A mutants) are due to loading heterogeneities. The presence or absence of the Cfr and RlmN methyltransferases in the four strains used for MS analysis is indicated to the right, below the gel.
TABLE 1.
Minimal inhibitory concentrations (MIC) for the strains investigated in the presence or absence of the Cfr and RlmN methyltransferases
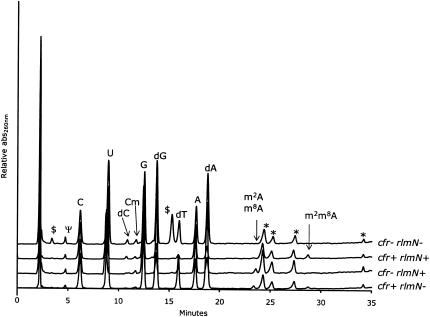
FIGURE 2.
HPLC-UV chromatograms of the four samples. The guanosine signal has the same height in all chromatograms, wherefore the relative intensities of other signals are directly comparable. Peaks marked with an asterisk are sample preparation artifacts present in an enzyme-only hydrolysis, and the peaks marked with $ are unknown contaminants of nonnucleoside nature. The peaks corresponding to the expected modified adenosines are marked for visualization purposes.
A series of small peaks appears in the elution window between 23 and 29 min, depending on the presence of the A2503-targeting methyltransferases RlmN and Cfr (Fig. 2). In the wild-type (wt) sample (cfr−/rlmN+) the peak at ∼23.5 min can be identified as m2A based on its relative retention time (Pomerantz and McCloskey 1990). The addition of the cfr gene (cfr+/rlmN+) results in the disappearance of the m2A signal and the appearance of a peak eluting around 28.8 min that likely corresponds to the hypermethylated A2503. When only the cfr gene is present (cfr+/rlmN−), a signal similar to the one from m2A appears at ∼23.5 min. Based on the relative retention time this peak may be interpreted as a monomethylated A2503. The similar retention times of the peaks in the cfr+/rlmN− and the cfr−/rlmN+ samples suggests that they correspond to nucleosides of a closely related chemical structure such as m2A and a hypothesized m8A. A weak signal at 28.8 min is also observed in the cfr+/rlmN− sample, indicating that Cfr alone has the ability to hypermodify A2503. The cfr−/rlmN− sample gives rise to the expected absence of chromatographic characteristics of modified A2503 nucleosides. As the chromatograms alone are only indicative of possible structures, further analyses were necessary.
Analysis by graphite column LC-ESI-MSn
It is apparent from Figure 2 that a major problem in using C18 reverse-phase chromatography to characterize the Cfr A2503 product is its coelution with m2A formed by RlmN. We therefore developed a nano-LC-ESI-MSn method using a porous graphitized carbon (PGC) two-column setup. Nano-LC/MS applications generally use a two-column setup with a short enrichment column placed in front of the analytical column to reduce sample-loading time and to desalt and focus analytes at the top of the analytical column prior to gradient separation and MS analysis. The use of C18-type stationary phases for enrichment is not likely to succeed due to the hydrophilic nature of nucleosides. Nucleosides such as pseudouridine and cytidine that elute isocratically under aqueous conditions on C18 stationary phases are not retained on the enrichment column and will thus be lost during loading of the sample. PGC provides different retention and separation properties compared with C18-type stationary phases, and has been used to separate highly polar compounds, geometric isomers, and glucosides (Polyakova and Row 2005; Pereira 2008). The rRNA nucleoside samples from the four strains were reanalyzed by nano-LC-ESI-MSn on a PGC stationary phase and separated using a formic acid/acetonitrile gradient.
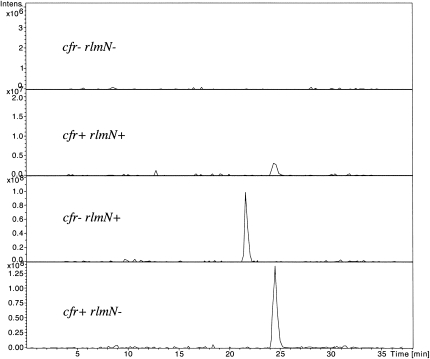
The isobaric m2A and m8A nucleosides have an expected pseudomolecular ion [M+H]+ at m/z 282 using positive electrospray ionization. The extracted ion chromatograms (XIC) at m/z 282 clearly display two fully resolved and intense peaks at 21.8 and 24.5 min in the cfr−/rlmN+ and the cfr+/rlmN− samples, respectively (Fig. 3). A weak signal at m/z 282 is also observed at 24.5 min in the cfr+/rlmN+sample and as expected no peak at m/z 282 is observed in the cfr−/rlmN− background. Thus, the altered stationary phase chemistry provided by PGC is able to fully resolve the Cfr A2503 product from m2A down to the baseline (Fig. 3), whereas the C18 chromatography afforded essentially no separation (Fig. 2).
FIGURE 3.
Extracted ion chromatogram of monomethylated adenosine species (m/z 282). Peaks were resolved using a PGC column on an Agilent Chip-LC electrospray ion trap tandem mass spectrometer.
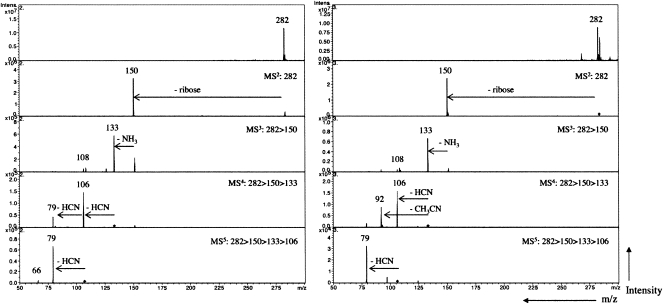
We have recently demonstrated higher-order tandem mass spectrometry on an ion trap instrument as a valuable tool in characterizing isobaric modified nucleosides (Jensen et al. 2007), and therefore this technique was implemented to characterize the Cfr products. The tandem mass spectra resulting from collision-induced activation at m/z 282 from cfr+/rlmN− (unknown methylation of A2503) and cfr−/rlmN+ (m2A2503), respectively, are presented in Figure 4. For both compounds, initial fragmentation of the intact nucleoside yields a single fragment ion at m/z 150 corresponding to the characteristic neutral loss of ribose (132 Da). The dominant fragmentation of nucleoside molecular ions in electrospray ionization is the CID of the N–C glycosidic bond that leads to the replacement of the sugar moiety with a hydrogen atom (Wilson and McCloskey 1975; Qian et al. 2007). Following isolation and fragmentation of the nucleobase at m/z 150, a second neutral loss of 17 Da is observed corresponding to the loss of NH3 and a minor peak at m/z 108 corresponding to the loss of cyanamide (NH2CN). Neutral loss of ammonia is also a characteristic of CID of nucleobases (Nelson and McCloskey 1992; Jensen et al. 2007; Qian et al. 2007); hence, MS3 was insufficient to distinguish the two isobaric adenosine derivatives.
FIGURE 4.
Tandem collision induced dissociation mass spectra (MS5) of m/z 282 from cfr+/rlmN− (left panel) and cfr−/rlmN+ (right panel).
Further fragmentation revealed different fragment ion pattern for the two adenosine derivatives. In the cfr−/rlmN+ m2A2503, fragmentation of m/z 133 in MS4 yields two intense peaks at m/z 106 and m/z 92 (Fig. 4, right panel), indicating neutral loss of HCN and acetronitrile (CH3CN). This is a marked difference from the analogous analysis of the cfr+/rlmN− sample, where sequential HCN losses generate peaks at m/z 106 and m/z 79 (Fig. 4, left panel). The loss of HCN and CH3CN from 2-methyl adenine has been explained by an initial opening of the pyrimidine ring at N1–C2 as a result of protonation at N1 of the adenine substructure (Nelson and McCloskey 1992; Tureček and Chen 2005). Thus, the observed losses of HCN or CH3CN following ring opening at N1–C2 revealed the methyl substitution at C2 of A2503 as expected in the cfr−/rlmN+ sample. Subsequent fragmentation of the dominant ion m/z 106 yields a single fragment ion at m/z 79 indicating a second loss of HCN. In the cfr+/rlmN− (Fig. 4, left panel), MS4 fragmentation of the deammoniated nucleobase at m/z 133 yields two peaks at m/z 106 and m/z 79 corresponding to two sequential expulsions of HCN (−27 Da). Sequential loss of HCN from adenines may be explained by the opening of the pyrimidine ring at N1–C2 as mentioned above. The absence of CH3CN-loss at this stage shows that Cfr does not methylate C2 of A2503 (cf. spectrum 4 of Fig. 4, right panel). Isolation and fragmentation of m/z 106 gives an intense peak at m/z 79 that substantiates the sequential loss of HCN and a minor peak at m/z of 66 that corresponds to the loss of acetonitrile (CH3CN) from m/z 106.
In summary, the adenosine methylations induced by Cfr and RlmN are different as deduced from MS5 (Fig. 4), the PGC chromatographic retention time of the Cfr A2503 product is distinct from both m1A, m2A, and m6A (data not shown) and Cfr is homologous to RlmN that methylates an aromatic carbon atom. Taken together, these observations suggest that Cfr may catalyze formation of m8A2503. In order to test this possibility, especially as m8A has not previously been identified in natural RNA, a pure m8A nucleoside was synthesized according to Van Aerschot et al. (1993) and analyzed by PGC ion trap MS5. The retention time of the synthetic m8A was indistinguishable from the Cfr-induced product of A2503, and the MS5 series of both species were identical (for tandem MS data on synthetic m8A and a graphical representation of the proposed fragmentation patterns, see Supplemental Figs. S1, S2, respectively). Hence, we conclude that the monomethylated product of Cfr in the absence of RlmN is m8A2503.
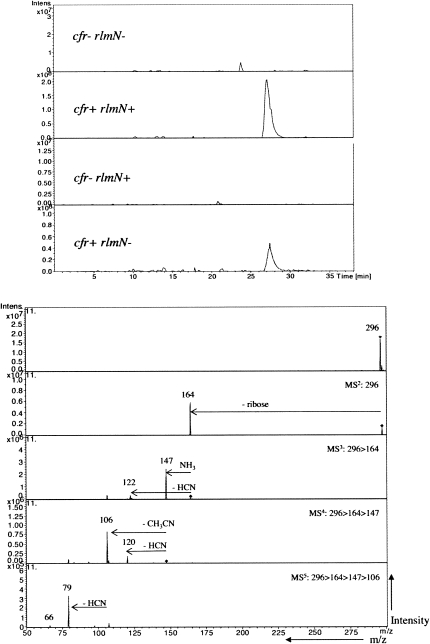
Another compound of interest is the unknown peak eluting at 27.8 min in both the cfr+/rlmN+ and cfr+/rlmN− samples (Fig. 2). Based on the MS5 data for the monomethylated A2503, we suggest that the presence of both enzymes together produces an m2m8A2503. In the cfr+/rlmN+ sample, the XIC of m/z 296 (the mass of protonated, dimethylated adenosine) displays an intense peak at 27.7 min (Fig. 5, top panel). As observed for the monomethylated adenosines, the most intense peaks in the MS2 and MS3 spectra correspond to the dominant loss of ribose (m/z 164) and ammonia (m/z 147), respectively (Fig. 5, bottom panel). Minor losses of NH2CN (42 Da) as well as ammonia and acetonitrile (17 Da + 41 Da) from m/z 164 are also observed. Assuming that fragmentation of m/z 147 proceeds through an opening of the pyrimidine ring at N1–C2, loss of acetonitrile (41 Da) to m/z 106 indicates a methylation at C2 as described for m2A above. Subsequent fragmentation of m/z 106 yields fragment ions at m/z 79 and a minor m/z 66 as observed for m8A. Taken together, this fragmentation genealogy indicates that the peak at m/z 296 is from 2,8-dimethyladenosine present in the cfr+/rlmN+ sample. A similar PGC LC-MS analysis of the m/z 296 from the cfr+/rlmN− sample (Fig. 5, top panel and data not shown) revealed identical behavior with respect to chromatographic retention time and MSn fragmentation, leading us to the conclusion that Cfr has an obscured ability to methylate position 2 of A2503 after the primary methylation at position 8 is complete.
FIGURE 5.
Extracted ion chromatogram (top panel) and tandem collision induced dissociation mass spectra (MS5, lower panel) of m/z 296 from the cfr+/rlmN+ sample.
Classification of the Cfr methyltransferase and its in vivo effect
Based on sequence homology, it has been proposed that Cfr is a radical SAM enzyme (Kehrenberg et al. 2005; Toh et al. 2008). Although the MS data presented above show that Cfr methylates an aromatic carbon atom, it does not address whether the reaction has a radical-based mechanism. To test whether Cfr is likely to be a true radical SAM enzyme, the CxxxCxxC motif in Cfr, which is a characteristic of radical SAM enzymes (Sofia et al. 2001), was mutagenised. Each of the cysteines in the motif was replaced by alanine, and the mutagenized Cfr expressed in E. coli. The effect of each mutation was investigated by primer extension analysis of 23S rRNA isolated from mutant E. coli strains (Fig. 1C) and via antibiotic susceptibility testing of these strains (Table 1). The primer extension analysis (Fig. 1C) clearly shows a strong reduction in band intensity for position A2503 in 23S rRNA from cells with Cfr mutated in the radical SAM motif (at amino acid position 115, 119, and 122) compared with cells with wild-type Cfr. The intensities of the bands arising from the primer extension stop at 23S rRNA position A2503 in the strains with the above cysteine mutations correspond to that observed for 23S rRNA isolated from a strain lacking Cfr, indicating that these mutated Cfr proteins are inactive. The mutated cysteines in the radical SAM motif are thus essential for Cfr activity and highly indicative of a radical SAM methyltransferase mechanism. A control mutation at cysteine 113 showed no significant reduction in Cfr activity according to primer extension analysis. The primer extension analysis of 23S RNA from the four strains used for MS analysis is also presented in Figure 1C. The results are consistent with the expected modification pattern in that there is no visible stop at A2503 when neither of the two methyltransferases are present, a strong stop band at A2503 when Cfr is present, a weak band at A2503 when RlmN is present, and the near absence of primer extension stop at C2498 when Cfr is present.
Measurement of the minimal inhibitory concentrations (MIC) of antibiotics for strains expressing the different Cfr mutants is an independent assay of Cfr activity. As mentioned in the introduction, Cfr confers a PhLOPSa phenotype with resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. The florfenciol and valnemulin MIC values after induction of the cfr gene are significantly reduced with the C112A, C116A, and C119A mutations in the CxxxCxxC motif relative to wt Cfr and the C110A control mutant (Table 1); the corresponding MICs for the four strains used for MS analysis are also presented in Table 1. Comparisons of the obtained MICs are complicated by the different E. coli background strains and by the fact that the RlmN knock out strain JW2501-1 is less sensitive to the antibiotics than HB101. Nevertheless, it is clear that Cfr is an antibiotic resistance determinant and provides decreased sensitivity regardless of the methylation status at position 2 of A2503. This is also in accordance with the small effect of RlmN on antibiotic sensitivity reported previously (Toh et al. 2008).
DISCUSSION
During the last decade, it has been shown that Cfr confers resistance to a series of antibiotics and that Cfr methylates A2503 in E. coli 23S rRNA. However, the precise location of the methylation in the nucleotide could not be deduced. The present work demonstrated that Cfr methylates position 8 of A2503, a hitherto unknown methylation in natural RNA molecules, and that Cfr possesses a masked m2A activity.
The full characterization of the Cfr-mediated modification required the development of a novel LC-MSn setup, where up to MS5 was necessary for unambiguous determination of the methylated A2503 derivatives. The characterization of both unmodified and modified nucleosides is desirable in many biological and pharmaceutical studies, but the task is analytically challenging due to the high polarity and structural similarity of the analytes. In this study, the two isobaric adenosine modifications m2A and m8A were successfully resolved using a PGC stationary phase, thereby enabling structural characterization based on their MS5 fragmentation patterns. The retention mechanisms of PGC stationary phase are different from those observed on C18 type stationary phases and provide markedly greater retention and selectivity for polar compounds (reviewed in Pereira 2008). The overall retention on PGC is determined by the balance of two factors: a hydrophobic interaction as for C18 type stationary phases and charged-induced interactions of a polar analyte with the polarizable surface of the graphite particles (Lepont et al. 2001; Pereira 2008). In accordance with our data, the use of a PGC stationary phase coupled with MS detection to resolve mixtures of nucleosides and their mono-, di-, and triphosphates surpassed a variety of C18 type stationary phases specifically designed to retain polar compounds (Xing et al. 2004).
Tandem mass spectrometry has previously successfully been employed in the structural characterization of nucleobases and nucleosides. Structural characterization on the ion-trap type instrument used here is achieved by isolating the ions of interest and submitting them to sequential fragmentation by CID in MSn. The molecular structure of the parent ion is then reconstructed based on the MSn fragmentation pattern. However, fragment ions produced may vary significantly depending on the type of instrument used for structural characterization. For example, Nelson and McCloskey (1992) observed loss of NH3 (17 Da), NH2CN (42 Da), and CH3CN (41 Da) from m/z 150 in the CID spectrum of 2-methyladenine using high-energy collisions in a Fast Atom Bombardment instrument. The dominant fragment ion of 2-methyladenine in our study is m/z 133, whereas other fragment ions are observed at very low intensity in the fragment ion spectrum of 2-methyladenine (Fig. 4, right panel). The difference may be explained by the ion trap instrument's preference for the lowest energy fragmentation pathways. The online LC/MS5 identification of m8A and comparison with a synthetic standard underscores the usefulness of an ion trap instrument for characterization of an unknown compound without the use of stable isotope labeling or NMR approaches. The method is simple to implement and provides the needed chromatographic selectivity to resolve isobaric modifications in a single run.
The straightforward explanation for the antibiotic resistance conferred by the methylation of position 8 on A2503 is a steric interaction between the added methyl group and the antibiotic. A mechanism, in which C8 methylation directly hinders drug binding, is supported by the recent X-ray structure of linezolid bound to the Deinococcus radiodurans 50S ribosomal subunit (Wilson et al. 2008). Here, the C8 position of A2503 is abutting the drug in such a manner that a methyl group at this position would point into the drug binding site and interfere directly with drug binding. A very similar positioning of A2503 is also observed in the Haloarcula marismortui 50S-linezolid complex (Ippolito et al. 2008). Moreover, a recently refined model of the Deinococcus radiodurans 50S subunit without bound antibiotic shows the same orientation of the A2503 base (Harms et al. 2008).
The orientation of A2503 in the above structures is different from previously published X-ray crystallographic data on the Deinococcus radiodurans 50S ribosomal subunit complexed with chloramphenicol, clindamycin, or tiamulin (Schlunzen et al. 2001; Schlunzen et al. 2004), where the base is oriented away from the bound antibiotics. Thus, an earlier conclusion about the Cfr-based modification having an indirect effect on the drug binding pocket (Kehrenberg et al. 2005) must be revised according to the new structural models. We suggest that the methylation at the C8 position has a direct effect on drug binding by placing a methyl group in the drug binding pocket. However, the mechanism behind the reduced C2498 ribose methylation upon A2503 C8 methylation and the manner in which the latter affects the RNA structure to inhibit RNase T1 cleavage at G2502 remain unclear. These effects are both observed independently of the C2 methylation of A2503 (data not shown), and we speculate that the rRNA may adopt a more rigid local structure upon synthesis of m8A2503.
The MIC data demonstrate that methylation of position 8 is the dominant antibiotic resistance determinant, but indigenous modification at position 2 also contributes to low-level resistance. This has been demonstrated by Toh and coworkers who showed that the absence of rlmN results in a modest increase in sensitivity toward selected peptidyl transferase-targeting antibiotics (Toh et al. 2008). The same phenomenon was observed when U2504 is not converted to pseudouridine and when U2552 is not 2′-O-methylated (Toh and Mankin 2008). We have indications of a similar effect in an E. coli strain lacking the ribose methylation on C2498 (data not shown). We have previously suggested that one function of post-transcriptional modifications is to modulate intermolecular RNA–RNA contacts, because most rRNA modifications are found in such regions. It is, however, notable that A2503 and U2504 do not participate in intermolecular RNA–RNA interactions, substantiating a “housekeeping” antibiotic resistance resulting from indigenous modifications at these two nucleotides.
The cfr gene has been found on transposons and in different geographical locations, suggesting that it can be spread within the microbial community (Kehrenberg and Schwarz 2006; Kehrenberg et al. 2007). It is alarming that one methyltransferase confers resistance to five different classes of clinically relevant antimicrobial agents. In addition, this mechanism of resistance functions in both Gram-positive and Gram-negative bacteria (Long et al. 2006). Although the mutagenesis data presented in this work strongly suggests that Cfr is an enzyme belonging to the Radical SAM family, its origin remains unresolved. The cfr gene has recently been reported in Staphylococcus spp. isolates from humans (Toh et al. 2007; Mendes et al. 2008), underlining its dissemination potential. The identification of the specific modification catalyzed by Cfr is an important step forward in the development of approaches to inhibit its undesired activity in pathogenic bacteria.
MATERIALS AND METHODS
Materials for nucleoside analysis
Commercially available nucleoside standards were purchased from Sigma Aldrich. Ultrapure water (18.2 MΩ) was produced using an Elga Purelab Ultra Water System. LC/MS grade formic acid was purchased from Merck, whereas other LC/MS-grade solvents were purchased from Sigma Aldrich. Ammonium acetate was purchased from J.T. Baker.
8-Methyladenosine was synthesized at the Nucleic Acid Center, University of Southern Denmark, by palladium-catalyzed cross-coupling of a tetraalkyltin with a halogenated purine nucleoside according to Van Aerschot et al. (1993). The product was verified by positive ion ESI/MS and 1H and 13C NMR spectroscopy (data not shown).
Sample preparation
E. coli strains harboring all possible combinations of the rRNA methyltransferase-encoding genes cfr and rlmN were grown and ribosomes isolated according to Kehrenberg et al. (2005). DH1 and HB101 are standard laboratory strains while JW2501-1 is a Keiko Collection strain obtained from the E. coli Genetic Resource Center. Isolation of a 48 nucleotide rRNA subfragment (CGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGC) encompassing positions 2480–2527 of 23S rRNA by specific hybridization, PAGE purification, and single-strand digestion with mung bean nuclease was performed as described by Andersen et al. (2004). The isolated RNA subunit was hydrolyzed to nucleosides using nuclease P1, phosphodiesterase I, and alkaline phosphatase according to Crain (1990b).
HPLC/UV-analysis
Reverse-phase chromatography was performed on a GE HealthCare Ettan LC using a Phenomenex Luna C18(2) column (2 × 250 mm, 5 μm particles, 100 Å pores). Nucleosides were separated using a linear gradient of (A) 40 mM ammoniumacetate, adjusted to pH 6 with glacial acetic acid, and (B) 40% acetonitrile at a flowrate of 250 μL/min at 40°C using UV signal at 260 nm for detection. The gradient was 0% B isocratic at 2 min followed by a linear increase to 25% B at 27 min and 60% B at 37 min. Prior to each analysis, the column was preconditioned with 100% A for 15 min.
Nano Chip ESI-ITMS
Mass analysis was performed using an Agilent Chip Cube nano-LC/ESI Ion-Trap system. Online enrichment and analysis was performed using an Agilent 1100 HPLC system consisting of a loading pump operating at 4 μL/min and an analytical pump operating at 300 nL/min. The automated chip handler, the “chip-cube,” is mounted directly in front of an Agilent XCT Ultra 6340 ion trap with a microvalve for flow-path switching, providing ultra low dead volume. The chip consists of an integrated 9 mm, 160 nL PGC 5 μm particles enrichment column, a 150 mm × 75μm PGC 5 μm particles analytical column and a nanospray emitter. Samples were loaded onto the enrichment column in 0.1% formic acid (FA). Gradient separation was achieved using (A) 0.1% FA and (B) 90% acetonitrile, 0.1% FA. Gradient was initially at 10% B, increase to 20% B at 10 min, 50% B at 20 min, 90% B at 25 min, and held for 3 min; then the system returned to 10% B and was allowed to reequilibrate for 5 min prior to the next injection.
Spectra were recorded in positive ion mode by scanning in the range m/z 220–400 with a scan rate of 8000 μ/sec, an ion current count set at 400,000, and maximum accumulation time of 200 msec. The following MS settings, optimized by infusion of 8-methyladenosine at 10 pmol/μL, were used for all experiments: drying gas, 4 L/min; dry gas temperature, 325°C; skimmer, 40 V; capillary exit, 112.2; lens 1, −50 V; and lens 2, −60 V; and the voltage applied to the chip emitter was set at 1850 V, drawing a current of 50–60 nA. To obtain CID fragment spectra in the low m/z range, the fragmentation amplitude was manually adjusted to 0.4 V for MS and 0.6 V for MSn to secure a potential well deep enough to avoid ejection of fragment ions from the ion trap.
Construction of plasmids encoding Cfr mutants
Plasmids encoding Cfr mutants with single cysteine to alanine mutations at Cys113, Cys115, Cys119, and Cys122 were constructed in two steps using overlap extension PCR. The plasmid pCfrHisN, containing the wild-type cfr gene cloned into the NdeI and HindIII sites of plasmid pLJ102 (Johansen et al. 2006), was used as a template. In the first step, two overlapping fragments were made where each was amplified with an outer primer complementary to sequences flanking the 5′ or 3′ end of the cfr gene and a mutagenic primer in the opposite orientation containing the appropriate mutations. In the second step, the two outer primers were used to amplify fragments containing full-length mutant cfr genes, where the two overlapping fragments from the first step were used as the template. The fragments were cloned into the NdeI and HindIII sites of plasmid pLJ102 to form plasmids pCfrHisN110A, pCfrHisN112A, pCfrHisN116A, and pCfrHisN119A. These plasmids and pCfrHisN were used to transform E. coli strain DH1. Full details of primers and procedures will be published elsewhere.
Primer extension analysis
Ribosomes were extracted with phenol and ribosomal RNA was precipitated with ethanol, resuspended in water, and monitored by primer extension analysis with AMV reverse transcriptase (Finnzymes, Espoo, Finland). The Cy5-labeled deoxyoligonucleotide primer (5′-GAACAGCCATACCCTTG-3′), complementary to nucleotides 2540–2556 of E. coli 23S rRNA, was used. The cDNA extension products were separated on 6% polyacrylamide sequencing gels. The positions of the stops were visualized by fluorescence scan and identified by referencing to dideoxynucleotide sequencing reactions on 23S rRNA that were electrophoresed in parallel.
Drug susceptibility testing
Drug susceptibility testing was done in a microtiter plate format by measuring optical density values at 450 nm with a Victor 3 spectrophotometer (Perkin-Elmer). LB medium was inoculated with single colonies and incubated overnight. The cultures were diluted to OD450 = 0.01 and 100 μL diluted culture was mixed with 100 μL of antibiotic solutions in a series with twofold concentration steps. Strains marked in Table 1 were induced by adding 1 mM IPTG.
The tested concentration ranges were: florfenicol 1–128 μg/mL and valnemulin 16–256 μg/mL. The MIC was defined as the drug concentration at which the growth of the cultures was completely inhibited after 48 h incubation at 37°C.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
Elzbieta Purta and Stephen Douthwaite are thanked for providing an E. coli gene knockout strain for the 23S rRNA C2498 methyl transferase prior to publication and Tatjana Kristensen is thanked for excellent technical assistance. Daniel N. Wilson is acknowledged for sharing the coordinates of the D. radiodurans 50S-linezolid X-ray structure prior to their release. A.M.B.G. was supported by the Danish Biotechnology Instrument Center. Additional financial support was from the Danish Natural Science Research Council, the Danish Medical Research Council, and the Danish National Research Foundation.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1371409.
REFERENCES
- Andersen T.E., Porse B.T., Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banoub J.H., Newton R.P., Esmans E., Ewing D.F., Mackenzie G. Recent developments in mass spectrometry for the characterization of nucleosides, nucleotides, oligonucleotides, and nucleic acids. Chem. Rev. 2005;105:1869–1915. doi: 10.1021/cr030040w. [DOI] [PubMed] [Google Scholar]
- Crain P.F. Mass-spectrometric techniques in nucleic-acid research. Mass Spectrom. Rev. 1990a;9:505–554. [Google Scholar]
- Crain P.F. Preparation and enzymatic-hydrolysis of DNA and RNA for mass-spectrometry. Methods Enzymol. 1990b;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- Dudley E., Tuytten R., Bond A., Lemiere F., Brenton A.G., Esmans E.L., Newton R.P. Study of the mass spectrometric fragmentation of pseudouridine: Comparison of fragmentation data obtained by matrix-assisted laser desorption/ionisation postsource decay, electrospray ion trap multistage mass spectrometry, and by a method utilising electrospray quadrupole time-of-flight tandem mass spectrometry and in-source fragmentation. Rapid Commun. Mass Spectrom. 2005;19:3075–3085. doi: 10.1002/rcm.2151. [DOI] [PubMed] [Google Scholar]
- Harms J.M., Wilson D.N., Schluenzen F., Connell S.R., Stachelhaus T., Zaborowska Z., Spahn C.M., Fucini P. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol. Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Ippolito J.A., Kanyo Z.F., Wang D., Franceschi F.J., Moore P.B., Steitz T.A., Duffy E.M. Crystal structure of the oxazolidinone antibiotic linezolid bound to the 50S ribosomal subunit. J. Med. Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- Jensen S.S., Ariza X., Nielsen P., Vilarrasa J., Kirpekar F. Collision-induced dissociation of cytidine and its derivatives. J. Mass Spectrom. 2007;42:49–57. doi: 10.1002/jms.1136. [DOI] [PubMed] [Google Scholar]
- Johansen S.K., Maus C.E., Plikaytis B.B., Douthwaite S. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol. Cell. 2006;23:173–182. doi: 10.1016/j.molcel.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Schwarz S. Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob. Agents Chemother. 2006;50:1156–1163. doi: 10.1128/AAC.50.4.1156-1163.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C., Schwarz S., Jacobsen L., Hansen L.H., Vester B. A new mechanism for chloramphenicol, florfenicol and clindamycin resistance: Methylation of 23S ribosomal RNA at A2503. Mol. Microbiol. 2005;57:1064–1073. doi: 10.1111/j.1365-2958.2005.04754.x. [DOI] [PubMed] [Google Scholar]
- Kehrenberg C., Aarestrup F.M., Schwarz S. IS21-558 insertion sequences are involved in the mobility of the multiresistance gene cfr . Antimicrob. Agents Chemother. 2007;51:483–487. doi: 10.1128/AAC.01340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak J.A., Bruenger E., McCloskey J.A. Post-transcriptional modification of the central loop of domain-V in Escherichia-coli 23-S ribosomal-RNA. J. Biol. Chem. 1995;270:17758–17764. doi: 10.1074/jbc.270.30.17758. [DOI] [PubMed] [Google Scholar]
- Lepont C., Gunatillaka A.D., Poole C.F. Retention characteristics of porous graphitic carbon in reversed-phase liquid chromatography with methanol–water mobile phases. Analyst (Lond.) 2001;126:1318–1325. doi: 10.1039/b102719k. [DOI] [PubMed] [Google Scholar]
- Long K.S., Poehlsgaard J., Kehrenberg C., Schwarz S., Vester B. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptograrnin A antibiotics. Antimicrob. Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R.E., Deshpande L.M., Castanheira M., DiPersio J., Saubolle M.A., Jones R.N. First report of cfr-mediated resistance to linezolid in human staphylococcal clinical isolates recovered in the United States. Antimicrob. Agents Chemother. 2008;52:2244–2246. doi: 10.1128/AAC.00231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.C., McCloskey J.A. Collision-induced dissociation of adenine. J. Am. Chem. Soc. 1992;114:3661–3668. [Google Scholar]
- Nelson C.C., McCloskey J.A. Collision-induced dissociation of uracil and its derivatives. J. Am. Soc. Mass Spectrom. 1994;5:339–349. doi: 10.1016/1044-0305(94)85049-6. [DOI] [PubMed] [Google Scholar]
- Pereira L. Porous graphitic carbon as a stationary phase in HPLC: Theory and applications. J. Liquid Chromatogr. Relat. Technol. 2008;31:1687–1731. [Google Scholar]
- Polyakova Y., Row K.H. HPLC of some polar compounds on a porous graphitized carbon Hypercarb (TM) column. J. Liquid Chromatogr. Relat. Technol. 2005;28:3157–3168. [Google Scholar]
- Pomerantz S.C., McCloskey J.A. Analysis of RNA hydrolyzates by liquid-chromatography mass-spectrometry. Methods Enzymol. 1990;193:796–824. doi: 10.1016/0076-6879(90)93452-q. [DOI] [PubMed] [Google Scholar]
- Qian M., Yang S., Wu H., Majumdar P., Leigh N., Glaser R. Ammonia elimination from protonated nucleobases and related synthetic substrates. J. Am. Soc. Mass Spectrom. 2007;18:2040–2057. doi: 10.1016/j.jasms.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlunzen F., Pyetan E., Fucini P., Yonath A., Harms J.M. Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol. Microbiol. 2004;54:1287–1294. doi: 10.1111/j.1365-2958.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- Schlunzen F., Zarivach R., Harms J., Bashan A., Tocilj A., Albrecht R., Yonath A., Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- Schwarz S., Werckenthin C., Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri . Antimicrob. Agents Chemother. 2000;44:2530–2533. doi: 10.1128/aac.44.9.2530-2533.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia H.J., Chen G., Hetzler B.G., Reyes-Spindola J.F., Miller N.E. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: Functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S.M., Mankin A.S. An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors. J. Mol. Biol. 2008;380:593–597. doi: 10.1016/j.jmb.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S.M., Xiong L.Q., Arias C.A., Villegas M.V., Lolans K., Quinn J., Mankin A.S. Acquisition of a natural resistance gene renders a clinical strain of methicillin-resistant Staphylococcus aureus resistant to the synthetic antibiotic linezolid. Mol. Microbiol. 2007;64:1506–1514. doi: 10.1111/j.1365-2958.2007.05744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S.M., Xiong L.Q., Bae T., Mankin A.S. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tureček F., Chen X.H. Protonated adenine: Tautomers, solvated clusters, and dissociation mechanisms. J. Am. Soc. Mass Spectrom. 2005;16:1713–1726. doi: 10.1016/j.jasms.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Van Aerschot A.A., Mamos P., Weyns N.J., Ikeda S., De Clercq E., Herdewijn P.A. Antiviral activity of C-alkylated purine nucleosides obtained by cross-coupling with tetraalkyltin reagents. J. Med. Chem. 1993;36:2938–2942. doi: 10.1021/jm00072a013. [DOI] [PubMed] [Google Scholar]
- Wilson D.N., Schluenzen F., Harms J.M., Starosta A.L., Connell S.R., Fucini P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc. Natl. Acad. Sci. 2008;105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M.S., McCloskey J.A. Chemical ionization mass-spectrometry of nucleosides. Mechanisms of ion formation and estimations of proton affinity. J. Am. Chem. Soc. 1975;97:3436–3444. doi: 10.1021/ja00845a026. [DOI] [PubMed] [Google Scholar]
- Xing J.S., Apedo A., Tymiak A., Zhao N. Liquid chromatographic analysis of nucleosides and their mono-, di-, and triphosphates using porous graphitic carbon stationary phase coupled with electrospray mass spectrometry. Rapid Commun. Mass Spectrom. 2004;18:1599–1606. doi: 10.1002/rcm.1524. [DOI] [PubMed] [Google Scholar]