Abstract
siRNAs mediate sequence-specific gene silencing in cultured mammalian cells but also silence unintended transcripts. Many siRNA off-target transcripts match the guide-strand “seed region,” similar to the way microRNAs match their target sites. The extent to which this seed-matched, microRNA-like, off-target silencing affects the specificity of therapeutic siRNAs in vivo is currently unknown. Here, we compare microRNA-like off-target regulations in mouse liver in vivo with those seen in cell culture for a series of therapeutic candidate siRNAs targeting Apolipoprotein B (APOB). Each siRNA triggered regulation of consistent microRNA-like off-target transcripts in mouse livers and in cultured mouse liver tumor cells. In contrast, there was only random overlap between microRNA-like off-target transcripts from cultured human and mouse liver tumor cells. Therefore, siRNA therapeutics may trigger microRNA-like silencing of many unintended targets in vivo, and the potential toxicities caused by these off-target gene regulations cannot be accurately assessed in rodent models.
Keywords: APOB, RNAi, off-target, therapeutic
INTRODUCTION
The first demonstration of RNAi in mammalian cells in 2001 (Elbashir et al. 2001a) was soon followed by suggestions that this technology could be used for development of a new class of human therapeutics (Agami 2002). In contrast to small-molecule or biological therapeutics that target properties of a gene product (for example, enzymatic activity or cell surface localization), RNAi therapeutics are expected to interfere with the transcripts of disease-causing genes on the basis of nucleotide sequence. Thus, RNAi therapeutics have the potential to address all transcribed genes without regard to gene product properties. Progress in developing RNAi-based therapeutics has been rapid. Therapeutic targets have been silenced by small interfering RNAs (siRNAs) in rodents (Soutschek et al. 2004) and in primates (Zimmermann et al. 2006; John et al. 2007), and candidate therapeutics are now in advanced clinical testing (de Fougerolles et al. 2007). To date, some candidate siRNA therapeutics have shown good efficacy with little evidence of toxicity (de Fougerolles et al. 2007), but other candidates have shown nonspecific off-target effects (Kleinman et al. 2008).
Early studies suggested that siRNA-based target silencing was exquisitely specific, as single nucleotide mismatches with an intended target greatly reduced silencing efficacy (Tuschl et al. 1999; Elbashir et al. 2001b; Hutvagner and Zamore 2002). However, subsequent microarray studies showed that siRNAs also silence many unintended target transcripts (Jackson et al. 2003, 2006b; Bilanges and Stokoe 2005; Birmingham et al. 2006). Unintended transcripts may be silenced with kinetics and concentration-dependence indistinguishable from the intended target, although generally with lesser magnitude (Jackson et al. 2003). Off-target silencing is now widely accepted as a complication of in vitro studies using siRNAs (Echeverri et al. 2006) and can trigger unintended phenotypes (Lin et al. 2005; Bartz et al. 2006; Jackson et al. 2006a). Most of these unintended targets share sequence complementarity in their 3′UTR regions with residues 1–8 (or contain seed hexamer strings matching nucleotides 1–6, 2–7, or 3–8) of the siRNA guide strand (Jackson et al. 2003, 2006b; Birmingham et al. 2006). This seed-matched, off-target silencing by siRNAs is reminiscent of target regulation by microRNAs (Lim et al. 2005), and for convenience, we will hereafter refer to it as “microRNA-like” silencing.
For siRNAs to be used as therapeutic molecules, it is especially important that they effectively silence transcripts of the intended target gene while showing minimal activity against transcripts of other, unintended genes. It remains unclear whether or not microRNA-like silencing of unintended targets poses a complication for siRNA-based therapeutics. To address this possibility, we have taken advantage of recently developed techniques for silencing of therapeutic targets in vivo following systemic injection of siRNAs encapsulated in lipid nanoparticles (LNPs) (Morrissey et al. 2005b). Here, we present a comparison of microRNA-like off-target silencing by siRNAs in cultured human and mouse cells with that seen in the livers of mice treated with LNPs loaded with siRNAs. For these experiments, we have used APOB as a model target to examine the specificity of therapeutic siRNAs. As the carrier protein in low-density lipoprotein (LDL), APOB is central to maintenance of blood cholesterol levels. Thus, APOB is an attractive target for therapeutic siRNAs, but is considered a “nondruggable” protein. siRNAs targeting APOB were effective in vivo at target silencing and at reducing plasma cholesterol levels in mice (Soutschek et al. 2004) and nonhuman primates (Zimmermann et al. 2006). We have used APOB siRNAs encapsulated in LNPs to test microRNA-like off-target silencing in mice. We have also compared microRNA-like off-target profiles in cultured mouse and human cells.
RESULTS
Selection of siRNAs that silence human and mouse APOB
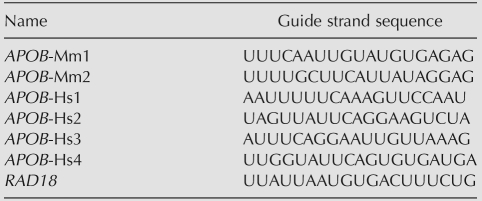
We selected a total of six siRNAs targeting APOB, including four siRNAs targeting both human and mouse transcripts, and two siRNAs targeting only the mouse transcript (Table 1). Candidate siRNAs were selected primarily for sequence conservation between man and mouse, with consideration for established siRNA design criteria (Khvorova et al. 2003). Candidate siRNAs were synthesized in chemically modified form (Morrissey et al. 2005b) and screened empirically to identify those having greatest efficacy (i.e., ability to silence APOB). As shown in Figure 1, all selected APOB siRNAs effectively silenced the target gene in both human liver-derived HUH7 and PLC/PRF/5 cells. All siRNAs also triggered robust silencing of Apob mRNA (90%–95%) in mouse liver-derived Hepa1-6 cells 24 h post-transfection and in mouse livers 3 d post-injection (data not shown).
TABLE 1.
Sequences of siRNAs used in this study
FIGURE 1.
Efficacy and specificity of APOB siRNAs. (A) The percentage of APOB mRNA remaining after transfection of APOB siRNAs, relative to cells treated with transfection reagent in the absence of RNA duplex, at 6, 12, 24, and 48 h in two human liver-derived cell lines, HUH7 and PLC/PRF/5. (B) A heat map of expression profiles of HUH7 and PLC/PRF/5 liver-derived human cell lines 6, 12, 24 (HUH7 only), and 48 h after transfection of six APOB siRNAs and the positive control RAD18 siRNA. Transcripts regulated with P-value < 0.001 in two or more experiments are shown; the position of APOB is marked.
We used microarray profiling to examine the specificity of APOB siRNAs. Human liver tumor-derived cell lines HUH7 and PLC/PRF/5 were transfected with one of the six APOB siRNAs or a positive control siRNA targeting RAD18. As shown in Figure 1B, each of the siRNAs triggered a distinct off-target signature. Profiling of additional human cell lines (HeLa, HCT116, and human mammary epithelial cells) gave similar results (data not shown). To distinguish direct (primary) and indirect (secondary) transcript regulations by the siRNAs, we assessed time-dependent transcriptional changes triggered by each of the candidate APOB siRNAs. We showed previously that transcripts down-regulated early after transfection were characterized by 3′UTR regions matching the siRNA seed sequences (Jackson et al. 2003, 2006b). We designated transcripts down-regulated at 12 h post-transfection and confirmed at an additional time point as “early off-targets.”
To assess the degree of microRNA-like off-target regulation, we examined APOB early off-targets for siRNA seed hexamer strings in their 3′UTRs. We compared the frequencies of these strings with their frequencies in the “universe” of all 3′UTRs of transcripts represented on the microarray. For all 4096 possible hexamer words, we calculated the fold-enrichment, the P-value of enrichment, and the rank order of enrichment (by P-value) in 3′UTRs of APOB early off-targets. Early off-targets triggered by the four mouse–human-homologous APOB siRNAs, one of the mouse-specific siRNAs, and the positive control siRNA were all highly enriched for transcripts whose 3′UTRs contained hexamer words matching siRNA guide strand seed regions. For example, in the PLC/PRF/5 profiles, 47%–80% of early off-targets had 3′UTRs matching the central seed hexamer of the guide strand (bases 2–7) of each siRNA (enrichment, two- to fivefold; P-values, 5e-27 to1e-62; and rank, 1 to 4 out of 4096 possible hexamers). Similar numbers were obtained for HUH7 cells (data not shown). Thus, as expected, these siRNAs demonstrated microRNA-like off-target activity in vitro. In contrast, one of the mouse-specific siRNAs (APOB-Mm1) triggered down-regulation of very few transcripts, and no significant enrichment for any seed hexamer matches could be detected in the early off-targets. This example shows that it is possible to empirically identify siRNAs that are highly specific in their silencing properties.
Mouse in vitro and in vivo off-target regulations show strong overlap
We next sought to compare gene expression changes triggered by siRNAs in cell culture with those in livers of mice treated with LNP-encapsulated siRNAs. We selected three mouse–human-homologous APOB siRNAs and transfected them into mouse Hepa1-6 cells. In parallel experiments, we encapsulated the siRNAs and injected them into mice. We then compared gene expression profiles from cultured cells with those from livers of treated mice. Many of the regulations observed in mouse livers treated with APOB siRNA-loaded LNPs were siRNA-independent responses to APOB silencing and/or LNP delivery (data not shown). In particular, many of the gene expression changes detected in livers at 24–48 h were LNP-dependent. By 72 h, these had mitigated enough to allow for more accurate interpretation of siRNA-dependent effects. Silencing of APOB by the siRNAs examined here was still very strong at 72 h, making it likely that microRNA-like off-target silencing also remained robust at this time. To identify siRNA sequence-specific transcriptional changes common between cultured cells and mouse livers, we used ANOVA analysis to identify transcripts whose mean regulation varied between siRNAs.
We used two approaches for this analysis. We identified siRNA-specific in vivo signatures and compared them to early off-targets identified in vitro. Each transcript was tested by ANOVA for regulation by each APOB siRNA relative to all other siRNAs within the in vivo profiles. The in vivo selected siRNA-specific transcripts overlapped significantly with early off-targets selected independently in vitro (15%–58% overlap, P-values, 1e-6 to to 6e-49) (data not shown).
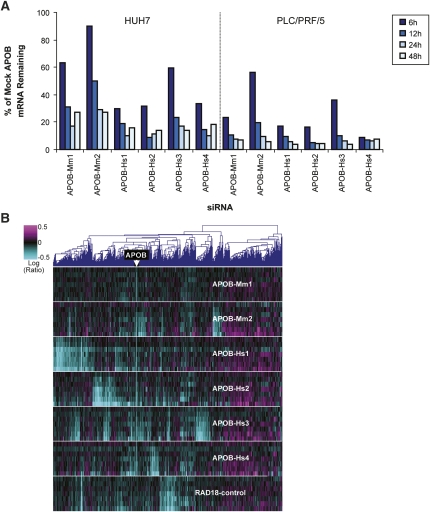
We also selected transcripts whose regulation differed between siRNAs in the combined in vivo and in vitro profiles. A heat map representation of the regulation of these transcripts showed strong overlap between patterns from mouse cells and mouse liver (Fig. 2A). In contrast, there was little overlap between the patterns of different siRNAs. The overlap between ANOVA-selected transcripts down-regulated in vivo and in vitro was quantified for two siRNAs in Figure 2B; the third siRNA showed similar results (data not shown). On the average, 60% of siRNA early off-targets down-regulated in Hepa1-6 cells were also down-regulated in a siRNA-specific manner in mouse livers.
FIGURE 2.
Sequence-dependent off-target regulations are consistent in vitro and in vivo. (A) Heat map representation of hexamer-containing transcripts regulated in mouse cell lines and mouse livers in vivo. Shown are expression profiles of mouse livers 72 h after treatment with a single 3-mg/kg dose of each of three APOB siRNA LNPs, three mice per siRNA; and of Hepa1-6 mouse liver-derived tissue culture cells 6, 12, 24, and 48 h after transfection of three APOB siRNAs. The heat map is restricted to genes selected by one-way ANOVA for siRNA-specific regulation (1458 transcripts representing 1288 mouse genes whose mean regulation differed between siRNAs with ANOVA P-value < 0.001). (Yellow boxes) Blocks of transcripts regulated both in cell lines and liver by the indicated siRNA. The position of Apob is marked. (B) The degree of overlap of mouse in vivo and in vitro signatures of APOB-Hs1 and APOB-Hs4 within ANOVA-selected transcripts is shown graphically as Venn diagrams. The down-regulated gene sets are the union of the down-regulated genes in three individual experiments shown in A.
Overlapping in vivo and in vitro selected siRNA-specific transcripts were enriched for 3′UTR seed hexamer matches (enrichment, 1.9-fold to 2.3-fold; P-values, 6e-5 to 5e-7; and rank, 1 to 5). Signatures common to in vitro and in vivo profiles for APOB-Hs1, APOB-Hs2, and APOB-Hs3 were also significantly enriched for transcripts whose 3′UTRs contained matches to the siRNA central seed hexamers (enrichment, 2.0-fold to 3.3-fold; P-values, 8e-10 to 3e-15; and rank, 1 to 3). Thus, significant overlap in siRNA-specific, microRNA-like off-target regulation in vivo and in vitro was clearly observable despite differences in delivery, transcript expression, and tissue complexity between tissue culture cells and mouse livers.
Human and mouse off-target regulations are distinct
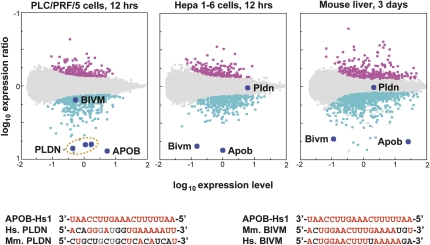
We next addressed the specificity of siRNA therapeutics in mouse and human cells. We compared the identities of transcripts regulated in mouse and human liver-derived cell lines for each siRNA, initially focusing on APOB-Hs1, which showed strong and specific off-target regulation (Fig. 3). In both human and mouse cells, APOB-Hs1 silenced a number of off-target transcripts to a similar extent as the intended target (∼10-fold). However, the identity of the strongly silenced targets was species specific. One of the best-silenced off-target transcripts in human cells was Pallidin (PLDN), a lysosomal storage disease gene (Fig. 3, left panel). FASTA alignment of APOB-Hs1 and human PLDN demonstrates 9 nucleotides of contiguous complementarity, including the entire seed sequence. However, APOB-Hs1 does not down-regulate Pldn in mouse cells in vitro (Fig. 3, center panel) or in mouse liver in vivo (Fig. 3, right panel). The APOB-Hs1 alignment site in human PLDN is not conserved in the mouse sequence. APOB-Hs1 triggers strong regulation of a different off-target transcript, Bivm (basic immunoglobulin-like variable motif-containing, a putative housekeeping gene), in Hepa1-6 cells (Fig. 3, center panel) and in mouse liver (Fig. 3, right panel). BIVM was only weakly down-regulated in human cells (Fig. 3, left panel). Mouse Bivm shows a long, central alignment of 14 contiguous bases to APOB-Hs1, but this site is disrupted by a mismatch in the human gene. These findings show that the same siRNA can trigger strong regulation at the RNA level of different off-targets in human and mouse.
FIGURE 3.
APOB-Hs1 strongly regulates off-target genes. (Left panel) An expression profile of PLC/PRF/5 cells at 12 h post-transfection of APOB-Hs1. (Center panel) An expression profile of Hepa1-6 cells 12 h post-transfection of APOB-Hs1. (Right panel) An expression profile of mouse liver 72 h post-delivery of APOB-Hs1. PLDN, BIVM, and APOB transcripts are labeled. (Bottom panel) The best FASTA alignments of the APOB-Hs1 guide strand with mouse and human (left) PLDN and (right) BIVM transcripts. (Red) Perfect matches; (gray) G-U pairs; (black) mismatches.
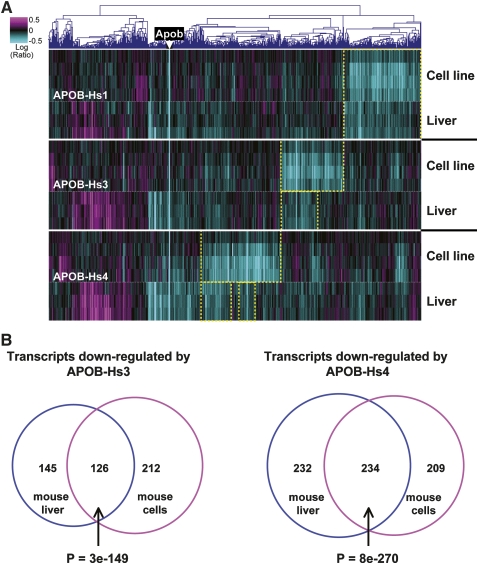
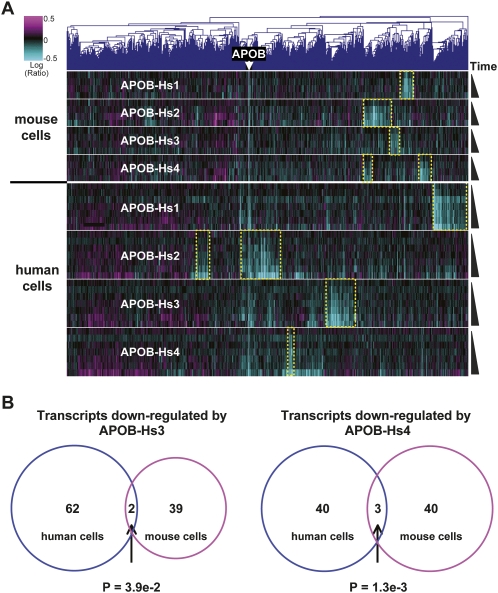
We also employed a more global approach to examining the specificity of siRNA therapeutics in mouse and human cell lines. As shown in Figure 4A, a heat map analysis showed little overlap between off-target transcripts triggered by the different siRNAs in cultured human and mouse liver cancer cells. The overlap between human and mouse early off-targets was quantified for two siRNAs in Figure 4B; the third siRNA showed similar results (data not shown). Among the genes that overlap between human and mouse cell lines in Figure 4B, none are common between the two siRNAs, and neither set shows significant biological annotation. Thus, the off-target transcript profiles were largely distinct in humans and mouse cells. Despite the differences in transcript identities, 3′UTRs of siRNA off-targets in both species were enriched for seed hexamer matches. In mouse Hepa1-6 cell expression profiles, 43%–77% of early off-targets had 3′UTRs matching central seed hexamers (enrichment, twofold to fourfold; P-values, 3e-13 to 1e-63; and rank, 1 to 2). These values were nearly identical to the values showing enrichment for seed hexamer matches in human PLC/PRF/5 cells (Fig. 1B). Thus, both human and mouse cells showed similar enrichment for seed matches, but human off-targets were unlikely to be down-regulated in mouse cells, and vice versa.
FIGURE 4.
Poor overlap between sequence-dependent regulations in human and mouse. (A) A heat map of expression profiles of HUH7 and PLC/PRF/5 liver-derived human cells and Hepa1-6 liver-derived mouse cells 6, 12, 24, and 48 h after transfection of APOB siRNAs. Transcripts regulated with P-value < 0.01 in two or more experiments are shown; the position of APOB is marked. (Yellow boxes) Blocks of transcripts regulated in siRNA-specific fashion in human or mouse cells. (B) Transcripts down-regulated 12 h and confirmed 6 or 48 h after transfection of APOB-Hs1 or APOB-Hs4 in PLC/PRF/5 cells were compared to transcripts down-regulated in Hepa1-6 cells 12 h post-transfection and confirmed at 6 or 48 h. Transcripts with coefficients of variation <50% were excluded from the down-regulated gene sets as uninformative. Analysis was restricted to a set of 13,733 mouse/human ortholog pairs with defined 3′UTRs, identified by reciprocal best BLAST matches between species, and common to the human and mouse microarrays. Please note that the number of transcripts analyzed in the figure is different from what is shown in Figure 2 because of different analysis procedures.
To understand why the siRNA transcript profiles differed across species, we examined the hexamer composition of the 3′UTRs of orthologous human and mouse transcripts. Human orthologs of mouse early off-targets and mouse orthologs of human early off-targets showed reduced content of siRNA-complementary 3′UTR hexamers relative to the early off-targets. While seed hexamer enrichment was significantly greater among early off-targets than enrichment for randomized versions of the seed hexamers (P < 7e-4, Kolmogorov–Smirnov goodness-of-fit hypothesis test), among orthologs, seed hexamer enrichment was equivalent to enrichment for randomized hexamers (P > 0.03). Thus, cross-species orthologs of early off-targets showed only chance enrichment for siRNA seed hexamers. This suggested that cross-species differences in 3′UTR composition might affect off-target activity between species. Indeed, we found that although overall hexamer content of the human and mouse transcriptomes was highly correlated (r > 0.9), only 36% of randomly selected hexamers in human 3′UTRs were found in the orthologous mouse 3′UTR (data not shown).
To confirm the effect of conservation of 3′UTR sequences on siRNA targeting, we projected early off-target genes onto pairs of human and mouse orthologs as classified by the number of distinct hexamer seed matches in both species. Classification demonstrated a random relationship between human and mouse seed match content (Supplemental Fig. 1, left panel); there was no preference for ortholog pairs to have equivalent seed content. Transcripts with three distinct hexamer seeds were most likely to be targeted within a species (Supplemental Fig. 1, center and right panels), in agreement with previous studies on microRNA targeting (Grimson et al. 2007). However, as the number of transcripts with multiple distinct seed matches in both species was only a small fraction of the total, most early off-targets in each species were found in transcripts whose orthologs had fewer distinct seed matches. These results suggest that species-specific 3′UTR composition contributes to species specificity of siRNA off-target activity.
DISCUSSION
Expression profiling experiments with cultured cells have shown that siRNAs trigger regulation of unintended transcripts with sequence complementarity to the siRNA seed region. Here we have demonstrated that in vivo siRNA treatments can recapitulate many of the same sequence-dependent early off-target regulations seen in vitro. The degree of overlap between signatures generated in vitro and in vivo was remarkable, given the many experimental differences, including siRNA delivery methods, cell environments, biological context, basal transcript expression levels, and tissue compositions. Transcripts regulated in common between livers and cultured cells were enriched for those with 3′UTR sequences complementary to the siRNA seed region. This microRNA-like silencing of unintended transcripts having seed region complementarity is therefore a fundamental property of most siRNAs in vitro and in vivo. Since microRNA-like off-target effects can lead to unwanted phenotypes in vitro, the same may also hold true in vivo. Although the limited data available have not shown off-target effects of siRNAs in vivo, it will be important to be vigilant for these as more data become available.
Transcripts regulated by microRNA-like seed-dependent silencing are largely different between cultured mouse and human liver cancer cells. Off-target signatures triggered by each siRNA in human cells were enriched for transcripts whose 3′UTRs matched siRNA seed hexamers, but the mouse orthologs of those transcripts were not likewise enriched. Conversely, off-target signatures generated in mouse cells were enriched for transcripts whose 3′UTRs contained seed hexamer matches, but the human orthologs of those transcripts contained only chance levels of these hexamer motifs. Thus, the same siRNA sequence regulates different sets of seed-matching transcripts in human and mouse, but with selectivity determined by context (e.g., species).
Unlike the microRNA-like silencing by siRNAs that we have studied here, target regulation by bona fide microRNAs is believed to be markedly conserved across species. Many microRNAs are highly conserved between humans and mice (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Paddison et al. 2002; Lim et al. 2003). Many microRNA target sites also show sequence conservation in 3′UTR regions (Lewis et al. 2003). Therefore, in contrast to what we have demonstrated for siRNA off-targets, significant overlap is expected between microRNA targets in human and mouse.
There are several possible explanations for the likely differences in cross-species conservation between siRNA off-targets and microRNA targets. One possibility is that there are as yet undiscovered differences between target sites for siRNA off-targets and microRNA targets. For microRNAs, seed hexamer (or heptamer, or octamer) matches are important for target recognition, but these seed matches are not always sufficient for repression. Other characteristics may help specify targeting, including nucleotide composition near the site and position in the 3′UTR (Grimson et al. 2007). While characteristics of siRNA off-target sites are less well defined than those of microRNA target sites, a model for predicting experimental microRNA target sites was likewise able to predict siRNA off-target sites (Grimson et al. 2007). This suggests that microRNA target sites and siRNA off-target sites are similar in their targeting determinants. Another possibility is that there are many more nonconserved microRNA targets than currently recognized. While many microRNA target sites are clearly conserved, the relative numbers of conserved versus nonconserved, species-specific sites is unclear. Genome-scale experiments similar to those done here but using microRNA agonists and/or antagonists will be necessary to elucidate the balance between different types of microRNA target sites.
However, the most likely explanation for the likely difference in cross-species conservation in siRNA off-targets and microRNA targets is that, in contrast to microRNA target sites, there has been no evolutionary selection for siRNA off-target sites. Despite extreme conservation of specific regulatory sites in 3′UTRs (Xie et al. 2005), these regions do not show overall strong sequence conservation (Siepel et al. 2005). Therefore, a siRNA that does not match conserved 3′UTR motifs would likely match a spectrum of off-target sites that were not well conserved. Differences in 3′UTR sequence composition between mouse and human transcripts would therefore be expected to lead to differences in the identity of off-target transcripts in each species.
An important consequence of the lack of species conservation of siRNA off-targets is that experiments in mice or mouse cells do not accurately predict off-target regulations in humans. Thus, it is not possible to know from studies in rodents most of the siRNA off-targets that will be regulated in humans. Since these off-target regulations can lead to undesired phenotypes, possible toxicities in humans may escape detection in rodents. In fact, our results suggest that human cell lines are more appropriate than live mouse studies for predicting sequence-dependent off-target activity of human therapeutic siRNAs. With the controversy over the testing of first-time-in-human therapeutics (Nada and Somberg 2007), it seems prudent to take efforts to reduce and/or understand microRNA off-target regulations by potential therapeutic siRNAs through the use of human cell line expression profiling.
MATERIALS AND METHODS
siRNA transfection and delivery in vivo
The sequences of siRNAs used in this study are shown in Table 1. All siRNAs were chemically modified as previously described (Morrissey et al. 2005a,b). Hepa1-6 mouse hepatoma cell line and HUH7 and PLC/PRF/5 human hepatoma cell lines (obtained from ATCC) were transfected in 6-well plates using Lipofectamine RNAiMAX (Invitrogen) and siRNA duplexes at a final concentration of 10 nM. For in vitro analysis, RNA was extracted at 6, 12, 24, and 48 h post-transfection. For in vivo studies, mouse livers were harvested 3 d following a single administration of APOB siRNA (3 mg/kg) formulated in LNPs (Morrissey et al. 2005a,b). LNPs were made using the cationic lipid CLinDMA (2-{4-[(3β)-cholest-5-en-3-yloxy]-butoxy}-N,N-dimethyl-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-1-amine), cholesterol, and PEG-DMG (monomethoxy(polyethyleneglycol)-1,2-dimyristoylglycerol) in 50.3:44.3:5.4 molar ratio. siRNAs were incorporated in the LNPs with high encapsulation efficiency by mixing siRNA in buffer into an ethanolic solution of the lipid mixture, followed by a stepwise diafiltration process. Cholesterol was purchased from Northern Lipids. CLinDMA and PEG-DMG were synthesized by Sirna Therapeutics. The encapsulation efficiency was determined using a SYBR Gold fluorescence assay. The particle size and Zeta potential measurements were performed using a Brookhaven ZetaPal particle sizer. The mean siRNA encapsulation efficiency was 85% ± 5%. The mean particle size was 135 ± 10 nm with a polydispersity of 0.10 ± 0.02. The LNPs had a positive surface charge density of 20 ± 5 mV.
RNA analysis
Total RNA was purified using an RNeasy kit (QIAGEN). Silencing of APOB was measured using TaqMan Gene Expression Assays (Applied Biosystems). Genome-scale expression analysis was performed using custom microarrays (Affymetrix) containing oligonucleotide probes corresponding to ∼22,000 human genes or 28,000 mouse genes. RNA from siRNA-transfected cultured cells was compared to RNA from cells treated with transfection reagent in the absence of RNA duplex. RNA from siRNA-treated mouse livers was compared to RNA from livers of mice treated with PBS. Three mice were analyzed for each treatment or control group. Each siRNA treatment was ratioed to a virtual pool of the PBS treatments. Microarray analysis was performed as described (Irizarry et al. 2003). Data were analyzed using Rosetta Resolver software. Microarray data are available in GEO (http://www.ncbi.nlm.nih.gov/geo/info/linking.html).
Analysis methods
Regulated transcripts were identified in microarray gene expression signatures using a P-value cut-off (P < 0.01). Transcripts with a coefficient of variation <50% across all experiments in a single cell line were excluded as uninformative. No cuts were placed on fold-change in expression. One-way ANOVA analysis was performed using Rosetta Resolver and MATLAB software. Regulated transcripts were tested for enrichment of hexamer word frequencies relative to a background set (i.e., the set of genes represented on the microarray) using the hypergeometric distribution. The P-value for enrichment in a set of 3′UTRs of each of 4096 possible hexamers was calculated and subjected to a Bonferroni correction. A set of 13,733 mouse/human ortholog pairs with defined 3′UTRs was identified by reciprocal best BLAST matches between species and used for cross-species comparisons.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Chandra Vargeese, Keith Bowman, Weimin Wang, and Ye Zhang for supplying the LNP preparation techniques; the Rosetta Gene Expression Laboratory for performing the microarray hybridizations; the Sirna Therapeutics staff for technical assistance; the Rosetta Inpharmatics staff for technical assistance and help with the analysis; and Oscar Puig, Eugen Buehler, and John Thompson for helpful discussions.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/10.1261/rna.1326809.
REFERENCES
- Agami R. RNAi and related mechanisms and their potential use for therapy. Curr. Opin. Chem. Biol. 2002;6:829–834. doi: 10.1016/s1367-5931(02)00378-2. [DOI] [PubMed] [Google Scholar]
- Bartz S.R., Zhang Z., Burchard J., Imakura M., Martin M., Palmieri A., Needham R., Guo J., Gordon M., Chung N., et al. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol. Cell. Biol. 2006;26:9377–9386. doi: 10.1128/MCB.01229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilanges B., Stokoe D. Direct comparison of the specificity of gene silencing using antisense oligonucleotides and RNAi. Biochem. J. 2005;388:573–583. doi: 10.1042/BJ20041956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A., Anderson E.M., Reynolds A., Ilsley-Tyree D., Leake D., Fedorov Y., Baskerville S., Maksimova E., Robinson K., Karpilow J., et al. 3′UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- de Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J. Interfering with disease: A progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri C.J., Beachy P.A., Baum B., Boutros M., Buchholz F., Chanda S.K., Downward J., Ellenberg J., Fraser A.G., Hacohen N., et al. Minimizing the risk of reporting false positives in large-scale RNAi screens. Nat. Methods. 2006;3:777–779. doi: 10.1038/nmeth1006-777. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Harboth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001b;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G., Zamore P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Leake D., Reynolds A., Schelter J., Guo J., Johnson J.M., Lim L., Karpilow J., Nichols K., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006a;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Burchard J., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006b;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M., Constien R., Akinc A., Goldberg M., Moon Y.A., Spranger M., Hadwiger P., Soutschek J., Vornlocher H.P., Manoharan M., et al. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007;449:745–747. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kleinman M.E., Yamada K., Takeda A., Chandrasekaran V., Nozaki M., Baffi J.Z., Albuquerque R.J., Yamasaki S., Itaya M., Pan Y., et al. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans . Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin S., Ruan S., Anderson M.G., McDowell J.A., Kroeger P.E., Fesik S.W., Shen Y. siRNA-mediated off-target gene silencing triggered by a 7nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey D.V., Blanchard K., Shaw L., Jensen K., Lockridge J.A., Dickinson B., McSwiggen J.A., Vargeese C., Bowman K., Shaffer C.S., et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005a;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V., et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005b;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Nada A., Somberg J. First-in-man (FIM) clinical trials post-TeGenero: A review of the impact of the TeGenero trial on the design, conduct, and ethics of FIM trials. Am. J. Ther. 2007;14:594–604. doi: 10.1097/MJT.0b013e31813737dd. [DOI] [PubMed] [Google Scholar]
- Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutschek J., Akinc A., Bramlage B., Charisse K., Constien R., Donoghue M., Elbashir S., Geick A., Hadwiger P., Harborth J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Tuschl T., Zamore P.D., Lehmann R., Bartel D.P., Sharp P.A. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann T.S., Lee A.C., Akinc A., Bramlage B., Bumcrot D., Fedoruk M.N., Harborth J., Heyes J.A., Jeffs L.B., John M., et al. RNAi-mediated gene silencing in nonhuman primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]