Abstract
mRNA localization is a widespread mode of delivering proteins to their site of function. The embryonic axes in Drosophila are determined in the oocyte, through Dynein-dependent transport of gurken/TGF-α mRNA, containing a small localization signal that assigns its destination. A signal with a similar secondary structure, but lacking significant sequence similarity, is present in the I factor retrotransposon mRNA, also transported by Dynein. It is currently unclear whether other mRNAs exist that are localized to the same site using similar signals. Moreover, searches for other genes containing similar elements have not been possible due to a lack of suitable bioinformatics methods for searches of secondary structure elements and the difficulty of experimentally testing all the possible candidates. We have developed a bioinformatics approach for searching across the genome for small RNA elements that are similar to the secondary structures of particular localization signals. We have uncovered 48 candidates, of which we were able to test 22 for their localization potential using injection assays for Dynein mediated RNA localization. We found that G2 and Jockey transposons each contain a gurken/I factor-like RNA stem–loop required for Dynein-dependent localization to the anterior and dorso–anterior corner of the oocyte. We conclude that I factor, G2, and Jockey are members of a “family” of transposable elements sharing a gurken-like mRNA localization signal and Dynein-dependent mechanism of transport. The bioinformatics pipeline we have developed will have broader utility in fields where small RNA signals play important roles.
Keywords: transposable elements, intracellular RNA localization, bioinformatics, Drosophila, RNA secondary structure, Dynein
INTRODUCTION
mRNA localization is a commonly used mechanism for delivering proteins to their cytoplasmic site of function (St Johnston 2005; Muller et al. 2007). Examples are known from a wide range of eukaryotic systems. cis-Acting RNA signals of diverse length, sequence, and structure are recognized by trans-acting protein factors that in most cases facilitate the association of the RNA cargo with a molecular motor that transports it to its correct intracellular destination. Well-characterized examples range from Ash1 mRNA in Saccharomyces cerevisiae (Chartrand et al. 2002) to Vg1 mRNA localization in Xenopus (Mowry and Melton 1992; Deshler et al. 1997) and actin mRNA in chicken fibroblast cells (Kislauskis et al. 1994).
In Drosophila, mRNA localization and translational regulation play an essential role in determining the polarity of the oocyte and embryo. Localized mRNAs include oskar (osk) and nanos (nos) at the posterior, and bicoid (bcd) at the anterior, as well as gurken (grk) RNA, which encodes a TGFα signal, targeted to the dorso–anterior corner of the oocyte. Grk protein is required to establish both the anterior–posterior and dorso–ventral axes of the oocyte and embryo, by providing positional information to overlying follicle cells. grk RNA is synthesized in nurse cells, transported to the oocyte along microtubules (MTs) (Caceres and Nilson 2005; Clark et al. 2007) and concentrated at the posterior, where it is translated. It is then transported to the anterior face of the oocyte, and then dorsally to form a cap between the nucleus and the adjacent follicle cells where it is again translated (MacDougall et al. 2003). The movement of grk RNA along MTs is mediated by Dynein and requires the grk localization signal (GLS), a small stem–loop within the coding region that is recognised by a number of proteins that together are presumed to specify the destination (Van De Bor et al. 2005).
The I factor, a non-LTR retrotransposon, encodes a transcript that is localized within the oocyte in a similar manner to grk mRNA. The I factor RNA contains a cis-acting RNA localization signal, the I factor localization signal (ILS), that is related to the GLS in secondary structure and competes for shared components of the localization machinery (Seleme et al. 2005; Van De Bor et al. 2005). Like grk, the I factor is transcribed in nurse cells (Seleme et al. 1999) and is transported into the oocyte, first to the posterior pole and then to the dorso–anterior corner, where it forms a cap overlying the nucleus.
Non-LTR retrotransposons are transcribed into a full-length RNA transposition intermediate that moves to the cytoplasm to be translated and then enters the nucleus, together with at least the reverse transcriptase and nuclease encoded by the second open reading frame. Once in the nucleus, non-LTR RNA is reverse transcribed into DNA that is integrated at a new site by a process called target primed reverse transcription (Cost et al. 2002). Transposition of the I factor takes place in the female germ-line and we have suggested that its RNA transposition intermediate is localized at the dorso–anterior corner of the oocyte to facilitate nuclear entry and transposition (Van De Bor et al. 2005). All non-LTR retrotransposons are believed to undergo target primed reverse transcription, raising the possibility that the transposition intermediates of other non-LTR elements in Drosophila could contain grk-like localization elements.
cis-Acting signals required for localization of other RNAs within the Drosophila oocyte have been mapped with varying degrees of precision. These include bcd, fs(1)K10 (K10), orb, and nos (Macdonald and Struhl 1988; Kim-Ha et al. 1993; Macdonald et al. 1993; Serano and Cohen 1995; Gavis et al. 1996). With the exception of the K10 and orb localization signals, that are very similar in sequence and structure (Cohen et al. 2005), no obvious similarities in either primary sequence or secondary structure have been found (Davis and Ish-Horowicz 1991; Francis-Lang et al. 1996; Simmonds et al. 2001; Bullock et al. 2003).
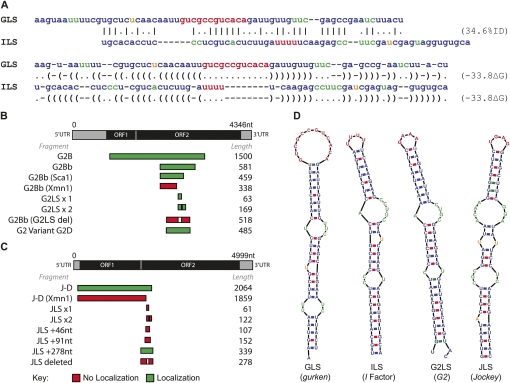
Although there is only 35% sequence identity between the GLS and ILS (Van De Bor et al. 2005), their secondary structures show striking similarities, as predicted by Mfold (Zuker 2000). This similarity suggests that for these localization signals, secondary structure may be of key importance for the interaction of trans-acting proteins with the mRNA cargos. Detection of localization signals that are functionally related to the GLS and ILS will therefore require the detection of sequences with the potential to form similar secondary structures.
Here, we describe a bioinformatics pipeline, which we have used to screen all the transposable elements identified in the genome sequence of D. melanogaster (release 5.1) for potential localization signals similar in secondary structure to the GLS and ILS. We have identified two elements, G2 (Di Nocera 1988) and Jockey (Mizrokhi et al. 1988; Priimagi et al. 1988) that contain localization sequences that can direct the correct localization of RNA injected into stage 8/9 oocytes. We propose that such secondary structure motifs may play a role in the transposition of several non-LTR retrotransposons in Drosophila by allowing their RNA transposition intermediates to move to the nucleus of cells in which they transpose.
RESULTS AND DISCUSSION
RNA2DSearch
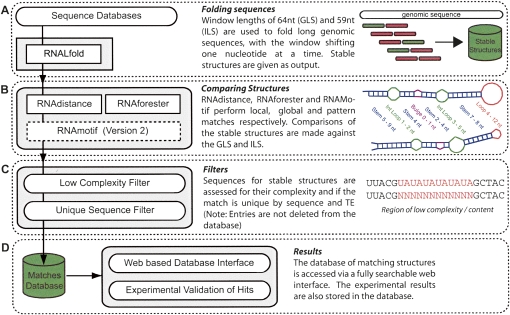
To assess whether the GLS and ILS share primary sequence similarity, we carried out a pairwise sequence alignment (Needleman and Wunsch 1970), using nucleotide substitution matrices. We found only limited sequence similarity between the two localization sequences (Fig. 1A). Moreover, BLAST (Altschul et al. 1990) searches in the Drosophila sequence databases using the GLS failed to find the ILS and vice versa. In contrast, secondary structure predictions of the GLS and ILS using Mfold (Zuker 2000), highlight obvious similarities at the secondary structural level (Fig. 1D). We also performed searches using RSEARCH (Klein and Eddy 2003), a stochastic context free grammar (SCFG) approach, which incorporates secondary structure information in the searches. While RSEARCH is a more powerful method than BLAST, it still fails to retrieve the ILS using the GLS as bait in the search and vice versa. This is because SCFGs are designed to find homologous RNAs and the GLS and ILS are not sufficiently similar in sequence and structure to be considered homologs; however, the secondary structure similarity leads us to believe there is a conserved motif required for binding the proteins of the localization machinery (Fig. 1A). This suggests the possibility of searching the genome for sequences that could be transcribed into RNA with a secondary structure motif similar to a consensus derived from the dorso–anterior localization signal of grk and the I factor. To achieve this goal we have developed a bioinformatics pipeline that we named RNA2DSearch (Fig. 2; see Materials and Methods), able to search for RNA secondary structure motifs on a genomic-wide scale and to compare them to a reference structure using several existing RNA bioinformatics resources.
FIGURE 1.
Comparison of the GLS and ILS. (A) Sequence and 2D secondary similarity. The GLS and ILS sequences show a low sequence identity of 35% when aligned with the Needleman–Wunsch global pairwise alignment algorithm with nucleotide substitution matrices (Needleman and Wunsch 1970). A Dynalign suboptimal structure prediction and alignment shows similarity in the secondary structures (Mathews 2005). Experimental validation of the predicted RNA localization signals: gene structures and mapping of regions necessary and sufficient for localization. (B) G2 structure showing position of the G2LS and the sequences tested experimentally. (C) Jockey structure showing the JLS position and the sequences tested experimentally. The green indicates localizing fragments, and red non-localizing. Comparison of the localizing hits found by RNA2DSearch to the GLS and ILS. (D) The predicted secondary structures, using Mfold (Zuker 2000), show the GLS and ILS share similar structures. The G2LS and JLS show a high degree of secondary structure similarity to the GLS and ILS. (A,D) Sequences are colored by secondary structure (red:loop; green:internal loop; orange:bulge).
FIGURE 2.
Flow diagram summarizing the RNA2DSearch pipeline. (A) Transposable element sequences are folded and collate using RNALfold. (B) Secondary structures of the stable structures are compared against known localizing signals GLS and ILS using RNAdistance (Hofacker 2003), RNAforester (Hochsmann et al. 2003) and RNAMotif (Macke et al. 2001) (Version 2). (C) Sequence analysis steps such as measuring sequence complexity with Dust and ensuring the matches are non-redundant by sequence and transposable element. (D) Structures similar to the GLS and ILS are collated in a relational database and linked to a custom web based interface for searching and annotation by the user.
Identification of two additional non-LTRs RNA localization signals
As the localization signal within I factor RNA may play a role in transpositon we wanted to determine whether other transposable elements (TEs) contain related localization signals. Release 5.1 of the D. melanogaster genome sequence contains 6003 copies of 155 unique transposable elements, containing over 7.5 × 106 nucleotides (www.flybase.org). Each TE family is likely to include both active and inactive members, and these cannot be distinguished a priori, making it impractical to test RNA from individual members of each family for their localization ability. In order to identify likely candidates to test we screened the entire set of TE sequences using RNA2DSearch. The extent of the transcribed region of most transposable elements has yet to be determined, although transcripts of non-LTR retrotransposons cover the complete sequence of the element. All the hits that we tested come from the transcribed region of the transposable element concerned, and correspond to the sequence of the transcribed strand.
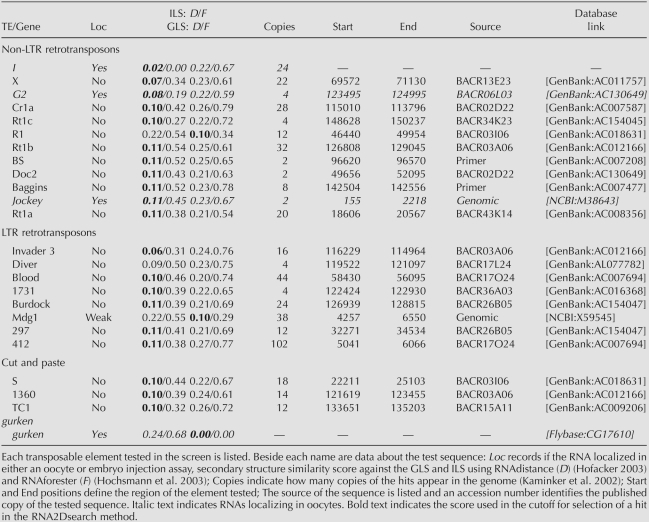
Predicted stable stem–loop structures were ranked according to their similarity to the ILS and GLS as indicated by the “full” structure similarity score assigned by RNAdistance (Table 1; Hofacker 2003). Elements with an RNAdistance score of 11 or less, considering only those with the lowest score in the case of elements with two or more candidates. We identified 48 elements matching these criteria, and of these we tested the 22 for which we could obtain a BAC clone containing genomic DNA of the particular element concerned. Testing the other elements was not practical, as PCR strategies cannot readily amplify the unique copies of a transposable element from a genomic sequence containing highly related individual TE sequences. These comprised 11 non-LTR retrotransposons, eight LTR retrotransposons, and three transposons (cut and paste elements). We transcribed fluorescently labeled RNA from each using DNA containing the predicted stem–loops, and injected these into stage 8/9 oocytes. Two RNAs, one from the element G2 (TE19713) and the other from Jockey (TE20045) localized with a pattern comparable to that of I factor and grk RNAs (Fig. 3A–D). We have named these stem–loop structures as G2LS, for G2 localization signal and JLS for Jockey localization signal.
TABLE 1.
Summary of the transposable elements tested from the RNA2DSearch predictions
FIGURE 3.
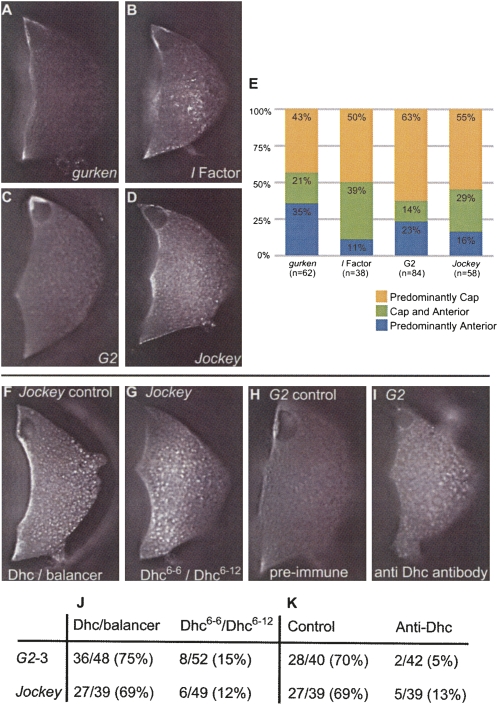
A comparison of the localization patterns of RNA containing the GLS and ILS to the Jockey and G2 hits found with RNA2DSearch. The localization patterns of labeled RNAs injected into stage 8/9 oocytes. Pictures were taken 40 min post injection. (A) grk RNA, full length transcript. (B) I factor RNA containing the ILS within a 1.7kb transcript. (C) G2 RNA: G2B (Fig. 1B) containing G2LS. In (A–C) the localized RNA can be seen forming a cap at the nucleus with some injected RNA remaining at the anterior cortex. (D) Jockey RNA J-D (Fig. 1C), containing the JLS. This RNA mainly localizes to the anterior cortex but can form a weak cap. (E) The proportion of oocytes showing a particular localization pattern after injection of RNA with grk, I factor, G2 or Jockey localization motifs (usually within large fragments of 1.5–2.2 kb) is indicated with n at the base of each bar. G2 and Jockey localization signals require Dynein for their localization. (F) Control, Jockey RNA injected into a Dhc / balancer oocyte. (G) Jockey RNA injected into trans-heterozygous hypomorphic Dhc mutant oocytes (Dhc 6-6/Dhc 6-12). (H) Control, G2 RNA injected into an oocyte preinjected with a preimmune serum, showing no affect. (I) G2 RNA injected into an oocyte that had been preinjected with an antibody raised against Dhc. (J–K) Fraction and percentage of RNA localization events over the total number of injected oocytes.
We tested whether the G2LS and JLS are responsible for the ability of G2 and Jockey RNAs to localize by injecting a deletion series of RNAs flanking the putative localization signals (Fig. 1B,C). All RNAs containing the putative G2 stem–loop localized and this was abolished if the stem–loop sequence was deleted. Furthermore, G2LSx1, an RNA containing the G2 stem–loop alone also localized (Fig. 1B). We conclude that the G2LS is both necessary and sufficient for localization. Interestingly, an RNA (G2LSx2) that contains two tandem copies of the G2LS localizes more efficiently than RNA with a single copy (Fig. 1B). We obtained comparable results for Jockey fragments with and without the JLS demonstrating that the motif is also necessary for localization. However, the JLS alone is not sufficient for localization (Fig. 1C; see Materials and Methods).
The G2LS and JLS are transported within the oocyte by Dynein
We have previously shown that movement of RNA containing either the GLS or ILS to the dorso–anterior corner of the oocyte requires the major minus end directed microtubule (MT) motor, cytoplasmic Dynein (Van De Bor et al. 2005). To test whether RNAs containing the G2LS or JLS localize by a similar mechanism, we used hypomorphic alleles of Dhc as well as an inhibitory anti-Dhc monoclonal antibody (Sharp et al. 2000) that we have previously shown disrupts the localization of RNA containing the GLS or ILS (Van De Bor et al. 2005). Injection of anti-Dhc antibody into oocytes 10–15 min before injection of fluorescently labeled RNA containing either the G2LS or the JLS significantly reduced the movement of these RNAs to the dorso–anterior corner (Fig. 3I,K) indicating that Dynein is required for their localization. Control injections of similar concentrations of a rabbit pre-immune serum (Fig. 3F,K), or a control mouse ascites fluid (Sigma) (data not shown), did not inhibit localization (see Materials and Methods). We also found that the efficiency of localization of injected G2LS or JLS containing RNAs was reduced in oocytes of females trans-heterozygous for two hypomorphic Dynein heavy chain mutants (Dhc6-6/Dhc6-12) (Fig. 3G,J). Injections of the same RNAs into control oocytes from females heterozygous for either of the mutations (Dhc6-6/+ or /Dhc6-12/+) localized at similar efficiencies to wild-type egg chambers (Fig. 3J). We conclude that G2 and Jockey localize within the oocyte by a similar Dynein-dependent mechanism to grk and I factor RNAs and require related localization machinery.
In total, we have identified three non-LTR retrotransposons (ILS, G2LS, and JLS) that contain a sequence that can direct the localization of RNA to the same site as grk mRNA within the oocyte. As part of their life cycle, non-LTR retrotransposons reverse transcribe a full length RNA at a genomic site. This is primed by a 3′OH at a single strand break in chromosomal DNA. We propose that the RNA localization signals that we have identified, allow the non-LTR transposons concerned to target their RNA to the host nucleus during transposition. A similar RNA nuclear localization signal would not be required for either LTR retrotransposons or transposons as the former reverse transcribe their RNA in the cytoplasm using a tRNA primer, while the latter move without an RNA intermediate. Indeed the potential localization signals that we identified in LTR retrotransposons or transposons did not direct localization of RNA in stage 8/9 oocytes (Table 1).
Having defined a total of four related signals (GLS, ILS, G2LS, and JLS) for localization to the dorso–anterior corner of the oocyte we are well placed to further define primary and secondary structure features that give them their ability to localization. This can be done using the G2LS and JLS to refine the search parameters of RNA2DSearch and this should allow us to discover other non-LTR transposable elements with similar modes of mRNA localization. We should also be able to more easily identify those features that are essential for their activity. Furthermore, the bioinformatics pipeline we have developed is highly flexible and at the same time rapid, thus allowing multiple searches of genomic databases for any relatively small RNA structural motif.
MATERIALS AND METHODS
Transposable element sequence databases
Sequences annotated as being transposable elements from FlyBase (Release 5.1; www.flybase.org) were used. Annotations are from Kaminker et al. (2002); supplementary materials were also utilized.
Folding sequences into stable secondary structures
A discussion of the bioinformatics approaches applicable to RNA motif searching can be found in Hamilton and Davis (2007). Locally stable structures of a defined length were identified by folding the input sequence, which could be an entire genome, in overlapping windows set at the defined length, progressing along the sequence, one nucleotide at a time using RNALfold (Hofacker et al. 2004). In this analysis window lengths of 64 and 59 nucleotides were selected, as these are the lengths of the GLS and ILS, respectively.
Comparing secondary structures to the reference structures
Each structure was compared to the reference structures, in this case the GLS and ILS. Separate comparisons were made using RNAdistance (Hofacker 2003) and RNAforester (Hochsmann et al. 2003). RNAdistance performs global structure comparisons, and in contrast, RNAforester was used to compare structures on a local scale, and find similarities between substructures. Both RNAdistance and RNAforester provide scores for the secondary structure similarities (scaled to be in the range 0–1), the lower the score, the more similar the structures being compared. RNAdistance scores of below 0.11, against either the GLS or ILS, were used as a cutoff for being considered as a hit. This threshold was chosen as it reflects the point at which the frequency of hits greatly increases. An additional step using RNAMotif (Macke et al. 2001) was included in a modification of the RNA2DSearch method. RNAMotif does not make direct comparisons to a reference structure, but instead uses descriptions files, based on the predicted secondary structures of the reference sequences (and primary sequences). The descriptions used incorporate all the main features common to both the GLS and ILS, accommodating small differences between the two structures (see Fig. 2B). The RNAMotif step was still able to predict the G2 and JLS, but produced less false positive hits.
Excluding low complexity sequences
The program Dust (Morgulis et al. 2006) was used to identify low complexity regions within sequences selected as having the potential to form secondary structures similar to the reference structure, the percentage of low complexity in each sequence was calculated so that these can be filtered out if desired (see Fig. 2C). A strict filter for removal of low complexity was used.
Availability of programs and supporting data
The code for RNA2DSearch is open source and freely available from the RNA2DSearch website (www.rna2dsearch.com). The code is provided as a set of Perl scripts and modules. In addition there must be working installations of RNAVienna, Mfold, and RNAMotif for RNA2DSearch to run. We have also provided the search results in raw text files for downloading, RNAMotif description files and full links to the Flybase TE database used.
Experimental testing of RNA sequences
The DNA templates used for in vitro transcription of labeled RNA were obtained in one of three ways. (1) Amplification from specific BAC clones containing the specific element insertion of interest (C.H.O.R.I. BAC Resource). (2) By direct amplification from the genome (Jockey and Mdg1). (3) Reannealing of appropriate oligonucleotides to create the motif (BS and Baggins) and subcloning this DNA into pGEM-T (Promega) for in vitro transcription. (See Table 1 for details). Amplification steps (1 and 2) incorporated a T7 primer for direct transcription from the appropriate strand (GTAATACGACTCACTATAGGGAGA). High fidelity polymerases Phusion (Finnzymes) or Platinum pfx (Invitrogen) were used for PCR amplification. Primers were designed to amplify between 800 and 1600 bases surrounding the hits of interest. The control grk RNA was full length as described in Figure 3 of Neuman-Silberberg and Schupbach (1993). The I factor control RNA was a Kpn1 to Cla1 fragment of the I factor cloned into pBluescript and digested with BsmI prior to transcription.
Some potential localization motifs were tested in a less stringent assay, the ability to move to the apical side of nuclei at blastoderm, another Dynein-dependent process (Bullock and Ish-Horowicz 2001; Wilkie and Davis 2001). The stem–loop from the LTR retrotransposon mdg1 localized weakly in this assay but failed to localize in the oocyte. We define a transcript to have not localized when it forming a diffuse signal throughout the ooplasm.
Fly strains
Flies were maintained on standard yeast–cornmeal–agar medium at 23°C. Localization of RNA was tested in embryos or oocytes from the wild-type Oregon R strain, Dhc 6-6 / Dhc 6-12 trans-heterozygotes (a gift from T. Hayes, University of Minnesota, Minneapolis, MN).
Imaging.
Imaging and injection were performed on a widefield DeltaVision microscope (Applied Precision, Olympus lX70, and Roper Coolsnap HQ) using an Olympus 20x/0.75NA objective lens and a Burleigh micromanipulator from Scientifica.
Synthesis of fluorescently labeled, capped RNA
RNA was transcribed in vitro using T7 or Sp6 RNA polymerase (Wilkie and Davis 2001) labeled with cy3-UTP (Perkin Elmer). RNA was purified over miniquick RNA columns (Roche Applied Science), precipitated and resuspended in DEPC treated water to a minimum concentration of 200 ng/μL for injection.
Injection of RNA and antibodies
RNA was injected into either living Drosophila melanogaster blastoderm embryos or oocytes. Eggs were collected at 23°C and dechorionated using 50% bleach solution. Partially dehydrated eggs were injected at blastoderm (stages 4–5) in halocarbon oil 95. Oocytes were dissected in halocarbon oil 95 and separated into individual ovarioles directly onto coverslips as described in (Parton and Davis 2006). RNA or antibody was injected using sterile Femtotips (Eppendorf). The anti-Dhc antibody was kindly supplied by D. Sharp (Albert Einstein College of Medicine, New York) injected at 11 mg/mL. Either control mouse ascites fluid (Sigma M8273) or a rabbit preimmune serum were used as controls at equivalent concentrations to anti-Dhc. Antibody was preinjected into oocytes 10–15 min prior to the injection of RNA; all injection steps were completed within 45 min of oocyte dissection.
ACKNOWLEDGMENTS
R.S.H. was supported by a Medical Research Council Career Establishment Grant and an eDIKT grant from the Science and Technologies Facilities Council (STFC) (to I.D.). E.H. and C.J. are supported by a BBSRC project grant (to D.F. and I.D.). G.V. was supported by a studentship from the Greek Government. V.V.D.B. was supported by an HFSP postdoctoral fellowship. I.D. was supported by a Wellcome Trust Senior Research Fellowship (067413). We thank Ian Moss who provided data on G2 variants as part of his Honours Project and Jim Brodie for help testing the F element.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1264109.
REFERENCES
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- Bullock S.L., Zicha D., Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22:2484–2494. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres L., Nilson L.A. Production of gurken in the nurse cells is sufficient for axis determination in the Drosophila oocyte. Development. 2005;132:2345–2353. doi: 10.1242/dev.01820. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X.H., Huttelmaier S., Donato D., Singer R.H. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- Clark A., Meignin C., Davis I. A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development. 2007;134:1955–1965. doi: 10.1242/dev.02832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R.S., Zhang S., Dollar G.L. The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element. RNA. 2005;11:1017–1029. doi: 10.1261/rna.7218905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I., Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- Deshler J.O., Highett M.I., Schnapp B.J. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276:1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- Di Nocera P.P. Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res. 1988;16:4041–4052. doi: 10.1093/nar/16.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis-Lang H., Davis I., Ish-Horowicz D. Asymmetric localization of Drosophila pair-rule transcripts from displaced nuclei: Evidence for directional nuclear export. EMBO J. 1996;15:640–649. [PMC free article] [PubMed] [Google Scholar]
- Gavis E.R., Lunsford L., Bergsten S.E., Lehmann R. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development. 1996;122:2791–2800. doi: 10.1242/dev.122.9.2791. [DOI] [PubMed] [Google Scholar]
- Hamilton R.S., Davis I. RNA localization signals: Deciphering the message with bioinformatics. Semin. Cell Dev. Biol. 2007;18:178–185. doi: 10.1016/j.semcdb.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochsmann M., Toller T., Giegerich R., Kurtz S. Local similarity in RNA secondary structures. Proc. IEEE Comput. Soc. Bioinform. Conf. 2003;2:159–168. [PubMed] [Google Scholar]
- Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., Priwitzer B., Stadler P.F. Prediction of locally stable RNA secondary structures for genome-wide surveys. Bioinformatics. 2004;20:186–190. doi: 10.1093/bioinformatics/btg388. [DOI] [PubMed] [Google Scholar]
- Kaminker J.S., Bergman C.M., Kronmiller B., Carlson J., Svirskas R., Patel S., Frise E., Wheeler D.A., Lewis S.E., Rubin G.M., et al. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 2002;3:0084.1–0084.20. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J., Webster P.J., Smith J.L., Macdonald P.M. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- Kislauskis E.H., Zhu X., Singer R.H. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R.J., Eddy S.R. RSEARCH: Finding homologs of single structured RNA sequences. BMC Bioinformatics. 2003;4:44. doi: 10.1186/1471-2105-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P.M., Kerr K., Smith J.L., Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118:1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- Macdonald P.M., Struhl G. cis-Acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- MacDougall N., Clark A., MacDougall E., Davis I. Drosophila gurken (TGFα) mRNA localizes as particles that move within the oocyte in two Dynein-dependent steps. Dev. Cell. 2003;4:307–319. doi: 10.1016/s1534-5807(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Macke T.J., Ecker D.J., Gutell R.R., Gautheret D., Case D.A., Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H. Predicting a set of minimal free energy RNA secondary structures common to two sequences. Bioinformatics. 2005;21:2246–2253. doi: 10.1093/bioinformatics/bti349. [DOI] [PubMed] [Google Scholar]
- Mizrokhi L.J., Georgieva S.G., Ilyin Y.V. Jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988;54:685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Morgulis A., Gertz E.M., Schaffer A.A., Agarwala R. WindowMasker: Window-based masker for sequenced genomes. Bioinformatics. 2006;22:134–141. doi: 10.1093/bioinformatics/bti774. [DOI] [PubMed] [Google Scholar]
- Mowry K.L., Melton D.A. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Muller M., Heuck A., Niessing D. Directional mRNA transport in eukaryotes: Lessons from yeast. Cell. Mol. Life Sci. 2007;64:171–180. doi: 10.1007/s00018-006-6286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg F.S., Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFα-like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- Parton R., Davis I. Lifting the fog: Image restoration by deconvolution. In: Celis J.E., editor. Cell biology. Academic; New York: 2006. pp. 187–200. [Google Scholar]
- Priimagi A.F., Mizrokhi L.J., Ilyin Y.V. The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologous to some retroviral proteins. Gene. 1988;70:253–262. doi: 10.1016/0378-1119(88)90197-7. [DOI] [PubMed] [Google Scholar]
- Seleme M.C., Busseau I., Malinsky S., Bucheton A., Teninges D. High-frequency retrotransposition of a marked I factor in Drosophila melanogaster correlates with a dynamic expression pattern of the ORF1 protein in the cytoplasm of oocytes. Genetics. 1999;151:761–771. doi: 10.1093/genetics/151.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleme M.C., Disson O., Robin S., Brun C., Teninges D., Bucheton A. In vivo RNA localization of I factor, a non-LTR retrotransposon, requires a cis-acting signal in ORF2 and ORF1 protein. Nucleic Acids Res. 2005;33:776–785. doi: 10.1093/nar/gki221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serano T.L., Cohen R.S. A small predicted stem–loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Rogers G.C., Scholey J.M. Cytoplasmic Dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000;2:922–930. doi: 10.1038/35046574. [DOI] [PubMed] [Google Scholar]
- Simmonds A.J., dosSantos G., Livne-Bar I., Krause H.M. Apical localization of wingless transcripts is required for wingless signaling. Cell. 2001;105:197–207. doi: 10.1016/s0092-8674(01)00311-7. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: The intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Van De Bor V., Hartswood E., Jones C., Finnegan D., Davis I. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev. Cell. 2005;9:51–62. doi: 10.1016/j.devcel.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Wilkie G.S., Davis I. Drosophila wingless and pair-rule transcripts localize apically by Dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- Zuker M. Calculating nucleic acid secondary structure. Curr. Opin. Struct. Biol. 2000;10:303–310. doi: 10.1016/s0959-440x(00)00088-9. [DOI] [PubMed] [Google Scholar]