Abstract
MicroRNAs (miRNAs) are small noncoding ribonucleotides that bind mRNAs and function mainly as translational repressors in mammals. MicroRNAs have been implicated to play a role in many diseases, including diabetes. Several reports indicate an important function for miRNAs in insulin production as well as insulin secretion. We have recently carried out a screen in the pancreatic β-cell line MIN6 to identify miRNAs with altered abundance in response to changes in glucose concentrations. This screen resulted in identification of 61 glucose-regulated miRNAs from a total of 108 miRNAs detectable in MIN6 cells. Many of the identified miRNAs, including miR-124a, miR-107, and miR-30d were up-regulated in the presence of high glucose. Only a few of the miRNAs, including miR-296, miR-484, and miR-690 were significantly down-regulated by high glucose treatment. Interestingly, we found that overexpression of miR-30d, one of the miRNAs up-regulated by glucose, increased insulin gene expression, while inhibition of miR-30d abolished glucose-stimulated insulin gene transcription. Overexpression or inhibition of miR-30d did not have any effect on insulin secretion. These data suggest that the putative target genes of miR-30d may be negative regulators of insulin gene expression.
Keywords: miRNA, glucose, pancreas, MIN6, diabetes, miR-30d
INTRODUCTION
A major group of endogenous small noncoding ribonucleotides called microRNAs (miRNAs), isolated from plants and mammals, have been shown to play an important role in regulation of gene expression at the post-transcriptional level (Bartel 2004; Tang 2005; Nilsen 2007). In mammals, miRNAs regulate the stability and/or translation efficiency of their target gene transcripts by partially binding to the 3′ UTR. The human genome is predicted to have close to 1000 potential miRNAs (John et al. 2004), and 30% of human transcripts are predicted to be regulated by miRNAs (Lewis et al. 2005; Xie et al. 2005). Although thousands of miRNAs have been identified from various organisms and stored in the miRBase (Griffiths-Jones et al. 2008), the identification of the physiological target genes for most of the mammalian miRNAs has been very challenging. This is due to the fact that in mammals each miRNA can regulate potentially more than 200 different transcripts, and the same target gene can be regulated by many miRNAs.
The levels of several miRNAs are altered in various diseases such as cancer (Fabbri et al. 2007; Skaftnesmo et al. 2007; Molnar et al. 2008), neurodegeneration (Fiore et al. 2008; Nelson et al. 2008), and cardiovascular disease (Ikeda et al. 2007; Scalbert and Bril 2008). Recent reports indicate that miRNAs are also important in the establishment of diabetes and may regulate insulin production and insulin secretion from the pancreatic β cells (Cuellar and McManus 2005; Joglekar et al. 2007; Poy et al. 2007). A conditional deletion of Dicer in pancreatic progenitor cells suggests that Dicer is essential for β-cell development (Lynn et al. 2007). Furthermore, 107 different miRNAs have been identified to be expressed in the developing mouse pancreas (Lynn et al. 2007). Several miRNAs including miR-375 and miR-124a, which are highly expressed in pancreatic β cells, have been implicated to negatively regulate insulin exocytosis (Poy et al. 2004; Krek et al. 2005; Lovis et al. 2008). In addition, both miR-375 and miR-124a appear to have important roles in pancreas development (Baroukh et al. 2007; Kloosterman et al. 2007; Poy et al. 2007).
Many genes in pancreatic β cells are up- or down-regulated in response to changes in blood glucose levels, and thereby regulate the optimal production and secretion of insulin (Ohneda et al. 2000; Poitout et al. 2006; Baroukh et al. 2007). Insulin secretion, as well as insulin gene transcription, insulin mRNA stability, and insulin mRNA translation and insulin processing have been demonstrated to be regulated by glucose in pancreatic β cells. Since glucose is an important nutrient signal for β cells, we tested whether glucose also regulates the expression of miRNAs in pancreatic β cells. We report here the identification of several glucose-regulated miRNAs from the pancreatic β-cell line MIN6, which may play an important role in glucose regulation of β-cell function. One of the glucose-induced miRNAs, miR-30d, was further investigated for its role in insulin gene transcription and insulin secretion.
RESULTS
Identification of glucose-regulated miRNAs from the pancreatic β-cell line MIN6
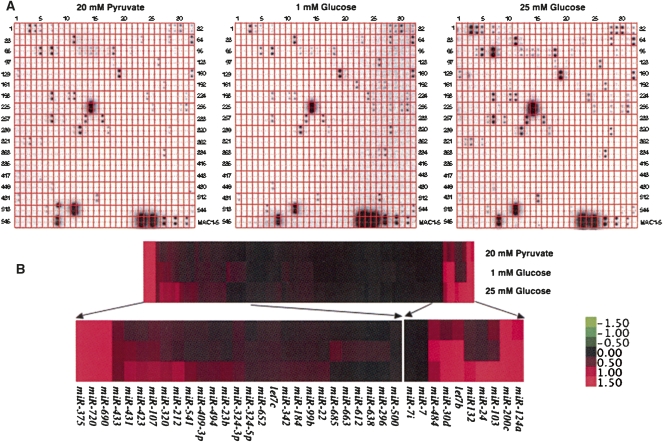
In order to identify glucose-regulated miRNAs in pancreatic β cells, we have incubated the mouse insulinoma cell line MIN6 with 20 mM pyruvate (no glucose control), 1 or 25 mM glucose for 16 h, and prepared small RNA samples. After labeling, the isolated small RNAs were hybridized to an array platform containing a collection of mouse/human miRNAs as described previously (Fig. 1A; Supplemental Table 1; Tang et al. 2007) . Using this simple array platform, we have identified 108 different miRNAs expressed in the MIN6 cells, many of which were regulated in response to high glucose (Fig. 1A). Hierarchical clustering of the glucose-regulated miRNAs using the Cluster 3.0 software (de Hoon et al. 2004) indicated that about 50 of the identified miRNAs were significantly up-regulated by high glucose (Fig. 1B; Supplemental Table 1). On the other hand, expression of several miRNAs, including miR-296, miR-484, miR-612, miR-638, miR-663, and miR-690 were significantly down-regulated after treatment with high glucose.
FIGURE 1.
Identification of glucose-regulated miRNAs from the pancreatic β-cell line MIN6 by miRNA array analysis. (A) Total RNA isolated from MIN6 cells treated with 20 mM pyruvate, 1 or 25 mM glucose for 16 h was separated on a 15% PAGE gel. After isolation of small RNAs from the gel, they were radiolabeled and hybridized to the array membrane. (B) Hierarchical clustering of the identified miRNAs based on their expression profiles using Gene Cluster 3.0 (average linkage and Euclidean distance as similarity measure). The data obtained for each miRNA were normalized to external and internal controls and median centered prior to clustering. The expression levels ranged from +1.5 log10 to −1.5 log10.
Validation of expression of the glucose-regulated miRNAs in MIN6 cells
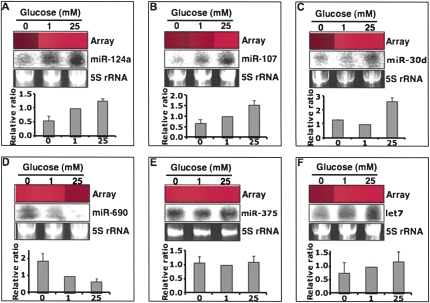
To confirm the expression of the identified miRNAs, the glucose-regulated expression for six of the miRNAs (miRNA-124a, miRNA-107, miRNA-30d, miRNA-690, miRNA-375, and miRNA-let7) was analyzed in MIN6 cells treated with 20 mM pyruvate, 1 or 25 mM glucose, by Northern blotting (Fig. 2). As observed in the original screen, expression of miRNA-124a, miRNA-107, and miRNA-30d was up-regulated by treatment of MIN6 cells with 25 mM glucose compared with cells incubated with 1 mM glucose or 20 mM pyruvate (Fig. 2A–C). Furthermore, we could also confirm the down-regulation of miRNA-690 expression by high glucose (Fig. 2D). Expression of miRNA-375, which has been reported to be one of most abundant miRNAs in pancreatic β cells, was similar under all the three conditions (Fig. 2E). Analysis of the expression of the normally ubiquitously expressed miRNA let-7 revealed a slight increase in expression on 1 or 25 mM glucose compared with 20 mM pyruvate (Fig. 2F). In summary, while the expression of miR-124a, miR-107, and miR-30d are up-regulated by high glucose, the levels of miR-690 appear to be down-regulated after treatment with 25 mM glucose.
FIGURE 2.
Northern blot validation of the glucose-regulated miRNAs from the array data. Total RNA (100 μg per sample) from MIN6 cells treated with 20 mM pyruvate (no glucose control), 1 or 25 mM glucose was separated on a 15% PAGE gel under denaturing conditions, blotted, and hybridized with 32P-labeled miRNA probes. 5S rRNA stained with ethidium bromide was used as a loading control and miRNA levels were compared with 1 mM glucose, which was set as onefold. The expression patterns of miR-124a (A), miR-107 (B), miR-30d (C), miR-690 (D), miR-375 (E), and let7 (F) were consistent with the array data. The array bar for each miRNA was exported from the clustering tree. Red bar indicates increased levels, while a black bar means no signal was detected in the array.
Overexpression of miR-30d enhances glucose-regulated insulin gene transcription, but not insulin secretion
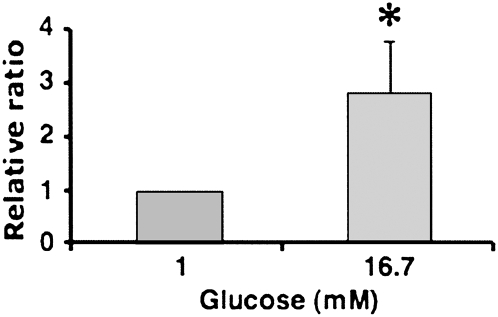
To investigate the role of the glucose-induced miRNAs in pancreatic β-cell function, we selected the glucose-induced miRNA miR-30d for further analysis. We first tested whether the levels of miR-30d were increased by high glucose in primary mouse pancreatic islets. Pancreatic islets contain five different types of cells, of which 60%–80% are insulin-producing β cells. We could confirm the regulation of miR-30d levels by glucose also in isolated mouse islets using real-time RT-PCR (Fig. 3). Exposure of pancreatic islets to high glucose (16.7 mM) for 16 h resulted in an almost threefold increase of miR-30d levels, which was comparable to the increase observed in MIN6 cells incubated with 25 mM glucose. This confirms that the levels of miR-30d are elevated by high glucose in both insulinoma cells and isolated primary mouse islets.
FIGURE 3.
Glucose-induced miR-30d expression in isolated mouse islets. Mouse pancreatic islets were isolated, purified, and incubated with 1 or 16.7 mM glucose in RPMI for 16 h. After this incubation period, RNA was extracted and used for real-time PCR to measure miR-30d levels. The level of miR-30d expression was normalized to the β-cell-specific transcription factor NeuroD1 mRNA levels. The presented data are the average of three independent experiments ± SD; (*) P < 0.05.
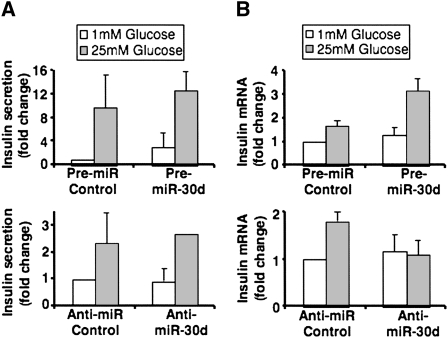
To further investigate the role of miR-30d in pancreatic β-cell function, we have overexpressed or inhibited miR-30d in the MIN6 pancreatic β-cell line and measured insulin production and secretion. Overexpression of miR-30d by transfection of pre-miR-30d into the MIN6-cell line had no significant effect on insulin secretion on 1 or 25 mM glucose compared with the negative control (Fig. 4A, top). Conversely, inhibition of miR-30d using anti-miR-30d oligonucleotides in the same cell line also did not alter insulin secretion (Fig. 4A). Next, we tested whether miR-30d plays a role in regulation of insulin gene transcription by quantifying insulin I mRNA levels by real-time RT-PCR in MIN6 cells, where miR-30d was overexpressed or inhibited. As indicated in Figure 4B (top), overexpression of miR-30d resulted in increased expression of the mouse insulin I gene by high glucose. Consistent with this finding, inhibition of miR-30d completely abolished the glucose-induced expression of the insulin I gene compared with the negative control, which displayed a twofold induction in insulin gene expression by high glucose (Fig. 4B, bottom). These data suggest an important role for miR-30d in the regulation of insulin gene transcription by glucose.
FIGURE 4.
MiR-30d affects insulin gene transcription, but not insulin secretion in MIN6 cells. MIN6 cells were transfected with miR-30d precursor or miR-30d inhibitor and with corresponding negative controls. After 48 h, the cells were incubated with 1 or 25 mM glucose and insulin secretion (A) and insulin I mRNA levels (B) were quantified using mouse insulin ELISA and real-time PCR, respectively. The presented data are the average of three independent experiments ± SD.
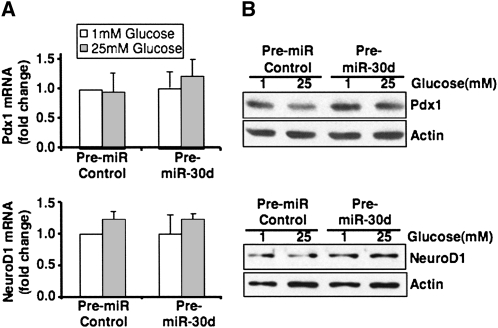
miR-30d-induced insulin gene transcription does not involve the β-cell-specific transcriptional activators Pdx1 and NeuroD1
Glucose regulation of insulin gene transcription is mediated by β-cell-specific transcription factors, including Pdx1 and NeuroD1. Of these two transcription factors, NeuroD1 contains a predicted binding site for miR-30d at the 3′ UTR, and this binding site is conserved between the human and mouse 3′ UTR region of NeuroD1. We tested whether miR-30d regulates the levels of NeuroD1 and found that overexpression or inhibition of miR-30d did not change NeuroD1 mRNA or protein levels significantly (Fig. 5, bottom). There was also no change in the levels of Pdx-1 by overexpression of miR-30d (Fig. 5, top). These data suggest that induction of insulin gene expression by miR-30d is due to inhibition of an unknown insulin gene repressor.
FIGURE 5.
Overexpression of miR-30d has no effect on Pdx1 and NeuroD1 mRNA and protein levels. MIN6 cells were transfected with miR-30d precursor or control precursor miRNA. After 48 h, cells were incubated with 1 or 25 mM glucose in DMEM for 16 h. (A) Total RNA was extracted and used for real-time PCR to measure Pdx1 and NeuroD1 mRNA levels. The data shown are the average of three independent experiments ± SD. (B) Western blot analysis of Pdx-1 and NeuroD1 protein levels in 1 or 25 mM glucose incubated MIN6 cells after transfection with the miR-30d precursor or control precursor.
DISCUSSION
We report here the identification of miRNAs from the pancreatic β-cell line MIN6 that are glucose regulated using our recently established miRNA array platform (Tang et al. 2007). We have identified 108 different miRNAs that were expressed in MIN6 cells, many of which are up-regulated by high glucose, such as miR-30d, miR-124a, and miR-107. Only a few miRNAs, including miR-690, were down-regulated by exposure to 25 mM glucose. The levels of miR-375, which appear to be the most abundant miRNA in pancreatic β cells, were not altered by glucose. Several of the identified miRNAs were also induced by glucose in isolated primary mouse islets (Fig. 3; data not shown).
It is not surprising that miRNA levels in pancreatic β cells are regulated by nutrients such as glucose, considering that β cells are in general very responsive to changes in blood glucose levels (Ohneda et al. 2000; Poitout et al. 2006). Insulin gene transcription, insulin mRNA stability, insulin mRNA translation, preproinsulin processing, and insulin secretion are very tightly controlled by changes in blood glucose levels (Itoh and Okamoto 1980; Ohneda et al. 2000; Wicksteed et al. 2001; Schuit et al. 2002; Poitout et al. 2006). Thus, some of the identified glucose-regulated miRNAs may play an important role in insulin production and insulin secretion. In fact, further analysis of miR-30d function by overexpression and inhibition studies in MIN6 cells suggests that miR-30d plays an important role in glucose induction of insulin gene expression. Overexpression of miR-30d increased insulin gene expression, while inhibition of miR-30d abolished glucose stimulation of insulin expression. These data suggest that miR-30d is important for down-regulation of an unidentified transcriptional repressor(s) of the insulin gene. Histone deacetylases have been shown to negatively regulate gene expression. However, overexpression or inhibition of miR-30d in MIN6 cells did not have any effect on HDAC1 (class I HDAC) or HDAC7 (class II HDAC) protein levels (data not shown).
The function and the target genes of the glucose-regulated miRNAs found in this study are mostly unknown, except for miR-124a, which appears to have an important role in pancreatic islet development (Baroukh et al. 2007). One of the miR-124a target genes that have been validated in pancreatic β cells is the transcription factor FoxA2, which regulates the expression of important β-cell-specific genes such as Pdx-1, Kir6.2, and Sur-1 (Baroukh et al. 2007). There are also reports implicating miR-124a in the regulation of insulin secretion. Using a computational prediction program (PicTar), it has been suggested that miR124a, coordinately with miR-375 and let-7b, regulates the levels of myotrophin and thereby inhibits insulin secretion (Krek et al. 2005). In a recent study, it has been shown that overexpression of miR-124a in the MIN6-cell line decreases glucose-stimulated insulin secretion by directly affecting the levels of Rab27A (Lovis et al. 2008).
A previous study reported the isolation of 107 different miRNAs from the developing mouse pancreas at the embryonic stage E14.5 (Lynn et al. 2007). Several of the glucose-regulated miRNAs we have identified have also been isolated in this screen (Lynn et al. 2007). Although several miRNAs have been identified to be expressed in the developing pancreas and in adult pancreatic β cells under various conditions, the exact role of these miRNAs in β-cell function are unknown. Further characterization of the miRNAs expressed in pancreatic β cells with respect to their function and β-cell-specific target genes, may shed light on their role in the establishment of diabetes.
MATERIALS AND METHODS
Cell culture and transfections
Mouse insulinoma 6 (MIN6) cells were cultured in Dulbecco's modified Eagle's media (DMEM) containing 25 mM glucose, 15% (v/v) fetal bovine serum, 1% penicillin/streptomycin, 2 mM glutamine, and 100 μM β-mercaptoethanol in humidified 5% CO2, 95% air at 37°C, and used between passage 22 to 30 (Miyazaki et al. 1990). For glucose regulation experiments, MIN6 cells were grown overnight in DMEM with 20 mM pyruvate and then washed with 1× phosphate-buffered saline and transferred to low (1 mM) or high (25 mM) glucose containing medium for the indicated times.
MicroRNA array and data analysis
Total RNA was isolated from MIN6 cells using Trizol reagent (Invitrogen). Fifty micrograms of total RNA from each sample were separated on a 15% denaturing PAGE and small RNAs (14–28 nt) were recovered from the gel and used for the array as described previously (Tang et al. 2007). Briefly, the small RNAs were dephosphorylated with Antarctic phosphatase (New England Biolabs) and then radiolabeled with γ-32P-ATP and PNK. The radiolabeled small RNAs were hybridized to the mouse miRNA array membrane and subsequently detected using a PhosphorImager (Typhoon 9400, Amersham). The obtained data were normalized to internal controls and clustered using Cluster 3.0 (de Hoon et al. 2004). The clustering data were visualized using Java TreeView (Saldanha 2004).
Northern blot validation
Northern blot analysis was performed as previously described (Tang et al. 2003; Tang 2005). Briefly, 50 μg of total RNA isolated from MIN6 cells were separated on a denaturing 15% urea–PAGE gel, electro-transferred to Hybond-N+ membrane (Amersham Biosciences), blotted, and probed using DNA oligonucleotides labeled with γ- P32-ATP. After washing, Northern blots were placed on PhosphorImager screens, and subsequently scanned using the Typhoon Scanner. The radioactive signals were quantified using the ImageQuant software package.
Real-time RT-PCR
To quantify the levels of mature miR-30d in mouse islets, the mirVana qRT-PCR miRNA detection kit (Applied Biosystem) was used in conjunction with real-time RT-PCR with TaqMan MicroRNA Assays (Applied Biosystem, # 4373059), according to the manufacturer's instructions. mir-30d levels were normalized to NeuroD1 mRNA levels in mouse islets. Mouse insulin I, Pdx-1, and NeuroD1 mRNA levels were quantified using real-time RT-PCR with the TaqMan real-time RT-PCR kit (Applied Biosystem). The primers for mouse insulin I were CCTGTTGGTGCACTTCCTAC (forward), TGCAGTAGTTCTCCAGCTGC, and CCCGCCGTGAAGTGGAGGACCC (FAM-labeled probe). The primers for β-actin as internal control for MIN6 cells were CGTGGGCCGCCCTAGGCAACC (forward), TTGGCCTTAGGGTTCAGGGGGG, and TGCCACAGGATTCCATACCCAAGAAGG (HEX-labeled probe). For quantification of Pdx1 and NeuroD1 mRNA levels, standard TaqMan Gene Expression Assays from Applied Biosystem were used. Real-time PCR was performed on an Mx4000 instrument (Stratagene).
Overexpression and inhibition of miR-30d
miR-30d RNA precursor (pre-miR-30d) and negative control miRNA precursor, anti-miR-30d oligonucleotides (oligos), as well as anti-miRNA negative control oligos, were purchased from Applied Biosystem. Oligos were transfected into MIN6 cells as described previously (Andrali et al. 2007). Briefly, 5 μg of miRNA precursor or anti-miRNA oligos were electroporated into MIN6 cells using the Amaxa Nucleofector system (Amaxa, Inc.) according to the manufacturer's instructions. Two days after transfection, cells were treated with low (1 mM) or high (25 mM) glucose without serum for 16 h; then, cell lysates or RNA were prepared and subjected to analysis by Western blotting or real-time RT-PCR, respectively.
Western blot analysis
Cell extracts from MIN6 cells were prepared in lysis buffer (10 mM Tris-Cl, pH 8.0, 140 mM NaCl, 5 mM MgCl2 0.2 mM EDTA, 0.5% Nonidet P-40, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors) (Miyazaki et al. 1990; Andrali et al. 2007). The antibodies used in this study are: Pdx-1 (Upstate, cat.# 07-696), NeuroD1 (Santa Cruz, sc-1086), and β-actin (Sigma).
Insulin secretion assay
Cells were incubated in DMEM containing 1 mM glucose for 16 h and transferred to 1x KRB buffer (119 mM NaCl, 4.74 mM KCl, 2.54 mM CaCl2, 1.19 mM MgSO4, 1.19 mM KH2PO4, 25 mM NaHCO3, 10 mM HEPES [pH 7.4], 0.1 g of bovine serum albumin) for 2 h. The cells were then treated with 1.0 mL of KRB buffer containing 1 mM or 25 mM glucose for 1 h. Secreted insulin levels in the medium were examined using the insulin immunosorbent assay kit (ALPCO) and normalized to total protein content.
Isolation of mouse pancreatic islets
Pancreatic islets were isolated from C57/BL6 mice. The experimental procedures were performed according to the protocol approved by the Animal Care and Use Committee at the University of Kentucky. The isolation and collagenase digestion of the pancreata were performed as previously described (Shewade et al. 1999). Briefly, after isolation, the pancreata were cut into small pieces and subjected to collagenase digestion in DMEM medium supplemented with 1 mg/mL collagenase type V (Sigma), 2 mg/mL STI (Sigma), and 2% BSA fraction V (Sigma). The collagenase digestion was stopped by adding chilled DMEM with 10% fetal calf serum and centrifuged at 1000 rpm for 10 min. After 48 h incubation with 10% fetal calf serum in RPMI-1640, isolated islets were handpicked and transferred to low (1 mM) or high (16.7 mM) glucose in DMEM for 16 h. Total RNA was isolated from islets using the Trizol reagent as described above.
Statistical analysis
Data are expressed as mean ± SD. Statistical analysis was carried out using the two-tailed, unpaired Student's t-test. A level of P < 0.05 was considered statistically significant.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Tiffany Nolan for excellent technical assistance. X.T. is supported by a K01 DK078648-01 from NIDDK. L.M. is supported by a Postdoctoral Fellowship from the American Heart Association, Great Rivers Affiliate. G.T. is supported by the KTRDC, USDA-NRI (2006-35301-17115 and 2006-35100-17433), NSF (MCB-0718029 with Sub award No. S-00000260), and an award from the Kentucky Science and Technology Corporation under Contract No. KSTC-144-401-08-029. S.Ö. is supported by grants R01DK067581 from NIDDK, P20RR020171 from the National Center for Research Resources, and 1-05-CD-15 from the American Diabetes Association.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1211209.
REFERENCES
- Andrali S.S., Qian Q., Ozcan S. Glucose mediates the translocation of NeuroD1 by O-linked glycosylation. J. Biol. Chem. 2007;282:15589–15596. doi: 10.1074/jbc.M701762200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroukh N., Ravier M.A., Loder M.K., Hill E.V., Bounacer A., Scharfmann R., Rutter G.A., Van Obberghen E. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic β-cell lines. J. Biol. Chem. 2007;282:19575–19588. doi: 10.1074/jbc.M611841200. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Cuellar T.L., McManus M.T. MicroRNAs and endocrine biology. J. Endocrinol. 2005;187:327–332. doi: 10.1677/joe.1.06426. [DOI] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Fabbri M., Ivan M., Cimmino A., Negrini M., Calin G.A. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin. Biol. Ther. 2007;7:1009–1019. doi: 10.1517/14712598.7.7.1009. [DOI] [PubMed] [Google Scholar]
- Fiore R., Siegel G., Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim. Biophys. Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Kong S.W., Lu J., Bisping E., Zhang H., Allen P.D., Golub T.R., Pieske B., Pu W.T. Altered microRNA expression in human heart disease. Physiol. Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- Itoh N., Okamoto H. Translational control of proinsulin synthesis by glucose. Nature. 1980;283:100–102. doi: 10.1038/283100a0. [DOI] [PubMed] [Google Scholar]
- Joglekar M.V., Parekh V.S., Hardikar A.A. New pancreas from old: Microregulators of pancreas regeneration. Trends Endocrinol. Metab. 2007;18:393–400. doi: 10.1016/j.tem.2007.10.001. [DOI] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Lagendijk A.K., Ketting R.F., Moulton J.D., Plasterk R.H. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lovis P., Gattesco S., Regazzi R. Regulation of the expression of components of the exocytotic machinery of insulin-secreting cells by microRNAs. Biol. Chem. 2008;389:305–312. doi: 10.1515/BC.2008.026. [DOI] [PubMed] [Google Scholar]
- Lynn F.C., Skewes-Cox P., Kosaka Y., McManus M.T., Harfe B.D., German M.S. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y., Oka Y., Yamamura K. Establishment of a pancreatic β-cell line that retains glucose-inducible insulin secretion: Special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- Molnar V., Tamasi V., Bakos B., Wiener Z., Falus A. Changes in miRNA expression in solid tumors: An miRNA profiling in melanomas. Semin. Cancer Biol. 2008;18:111–122. doi: 10.1016/j.semcancer.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Nelson P.T., Wang W.X., Rajeev B.W. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23:243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Ohneda K., Ee H., German M. Regulation of insulin gene transcription. Semin. Cell Dev. Biol. 2000;11:227–233. doi: 10.1006/scdb.2000.0171. [DOI] [PubMed] [Google Scholar]
- Poitout V., Hagman D., Stein R., Artner I., Robertson R.P., Harmon J.S. Regulation of the insulin gene by glucose and fatty acids. J. Nutr. 2006;136:873–876. doi: 10.1093/jn/136.4.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Poy M.N., Spranger M., Stoffel M. microRNAs and the regulation of glucose and lipid metabolism. Diabetes Obes. Metab. 2007;9(Suppl 2):67–73. doi: 10.1111/j.1463-1326.2007.00775.x. [DOI] [PubMed] [Google Scholar]
- Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Scalbert E., Bril A. Implication of microRNAs in the cardiovascular system. Curr. Opin. Pharmacol. 2008;8:181–188. doi: 10.1016/j.coph.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Schuit F., Flamez D., De Vos A., Pipeleers D. Glucose-regulated gene expression maintaining the glucose-responsive state of β-cells. Diabetes. 2002;51(Suppl 3):S326–S332. doi: 10.2337/diabetes.51.2007.s326. [DOI] [PubMed] [Google Scholar]
- Shewade Y.M., Umrani M., Bhonde R.R. Large-scale isolation of islets by tissue culture of adult mouse pancreas. Transplant. Proc. 1999;31:1721–1723. doi: 10.1016/s0041-1345(99)00077-9. [DOI] [PubMed] [Google Scholar]
- Skaftnesmo K.O., Prestegarden L., Micklem D.R., Lorens J.B. MicroRNAs in tumorigenesis. Curr. Pharm. Biotechnol. 2007;8:320–325. doi: 10.2174/138920107783018390. [DOI] [PubMed] [Google Scholar]
- Tang G. siRNA and miRNA: An insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. A biochemical framework for RNA silencing in plants. Genes & Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Gal J., Zhuang X., Wang W., Zhu H., Tang G. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicksteed B., Herbert T.P., Alarcon C., Lingohr M.K., Moss L.G., Rhodes C.J. Cooperativity between the preproinsulin mRNA untranslated regions is necessary for glucose-stimulated translation. J. Biol. Chem. 2001;276:22553–22558. doi: 10.1074/jbc.M011214200. [DOI] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]