Abstract
BACKGROUND
Aberrant promoter methylation of selective tumor suppressor genes has been detected in squamous intraepithelial lesions (SIL) and invasive cervical cancer. Identification of methylation profiles of genes that can distinguish high-grade SIL (HSIL) from low-grade SIL (LSIL), and cytologically negative for intraepithelial lesion or malignancy (NILM) residual liquid-based Papanicolaou (Pap) tests may be potentially useful as an ancillary test for cervical cancer screening.
METHODS
Using real-time quantitative methylation-specific polymerase chain reaction (PCR) (QMSP), the authors analyzed the frequency and relative level of promoter methylation for DAPK1, IGSF4, SPARC, and TFPI2 in biopsy-confirmed HSIL and LSIL, and NILM residual liquid-based Pap tests. The percentage of methylation (%M) for each gene was calculated using the reference gene, ACTB. The cumulative methylation score for each sample, defined as the sum of %M of all 4 genes, was used to analyze the genes in combination.
RESULTS
For each gene analyzed the frequency and relative level of methylation were increased in HSIL compared with combined NILM/LSIL samples. The cumulative methylation scores were significantly higher in HSIL samples (P < .0001). Area under the receiver operating characteristic (ROC) curve (AUC) demonstrated that methylation of each gene could distinguish HSIL from NILM/LSIL samples (AUC range, 0.6–0.67; P ≤ .0028). The combination of 4 genes showed improved test performance (AUC = 0.76; P <.0001). There was no significant difference in cumulative methylation in HSIL cases with histologic outcomes of cervical intraepithelial neoplasia grade 2 (CIN2) versus CIN3. There was no association between the methylation of any gene and the presence of human papillomavirus.
CONCLUSIONS
The methylation profile of multiple genes in combination can better distinguish HSIL from combined NILM/LSIL samples. Although aberrant DNA methylation has the potential to function as a molecular biomarker of HSIL in liquid-based Pap tests, additional genes that are selectively methylated in HSIL are needed to improve the clinical performance.
Keywords: methylation, HSIL, quantitative MSP, biomarker
The goal of cervical cancer screening programs that utilize the Papanicolaou (Pap) test is to identify and treat women with precancerous high-grade squamous intraepithelial lesions (HSIL) and invasive carcinoma. Based on the results of studies that use the 2001 Bethesda System for cervical cytology reporting, each Pap test interpretation is associated with a defined risk of cervical intraepithelial neoplasia grade 2 (CIN2) or more severe lesion (CIN2+) on histologic follow-up.1,2 Findings from the atypical squamous cells of undetermined significance (ASC-US)/low-grade squamous intraepithelial lesion (LSIL) Triage Study (ALTS), a randomized, multicenter clinical trial with 2-year follow-up, showed that the risk of underlying CIN2+ associated with abnormal cytologic findings on Pap tests interpreted as ASC-US, LSIL, atypical squamous cells cannot exclude HSIL (ASC-H), and HSIL are 17%, 25%, 50%, and 63%, respectively.2
Based on the etiologic role of oncogenic/high-risk human papillomavirus (HR-HPV) in cervical carcinogenesis, HR-HPV DNA testing has become a useful adjunct to the Pap test in some populations and clinical settings and has been incorporated into management algorithms for women with abnormal cytologic findings.3 In women of reproductive age and older women, HR-HPV DNA testing is useful in triaging those with ASC-US to colposcopy.3,4 The use of HR-HPV DNA testing in women with LSIL differs depending on patient age.3 In women of reproductive age and adolescent women with LSIL, the high prevalence of HR-HPV precludes its clinical utility in triaging them to colposcopy.3,5 However, because both the prevalence of HR-HPV and high-grade CIN (CIN2/3) decreases with age, HR-HPV DNA testing can be used in the initial management of postmenopausal women with LSIL to triage to colposcopy.3,6,7 In women with ASC-H, immediate referral to colposcopy is recommended given the increased prevalence of both HR-HPV and CIN2+.2,3 In light of the finding that many of the cervical lesions associated with mildly abnormal cytologic findings will spontaneously regress,8 the identification of other objective biomarkers that could help predict which women have underlying CIN2+ or are at increased risk of progression to CIN2+ could help to eliminate unnecessary colposcopic procedures and have a significant impact on the management of women with abnormal Pap tests.
Aberrant DNA methylation of tumor suppressor genes is a frequent event in most human tumors and may occur early in neoplastic progression.9 Previous studies have identified several candidate tumor suppressor genes that are frequently methylated in invasive cervical carcinoma, such as CDH1, CDH13, CDKN2A, DAPK1, HIC1, IGSF4, RARB, and TWIST1.10–14 More recently, a genome-wide screening study that utilized global demethylation and expression microarray analysis of cervical cancer cell lines identified additional novel genes, including SPARC, TFPI2, SFRP1, MT1G, RRAD, and NMES1, that were methylated in a relatively high percentage of exfoliated cervical samples with invasive carcinoma but not in normal samples.15 The methylation profiles of several genes have also been examined in precursor squamous intraepithelial lesions in Pap test samples, with most genes demonstrating more frequent methylation in HSIL compared with LSIL.14,16,17 These findings suggest that detection of methylated tumor suppressor genes could potentially serve as a diagnostic and/or predictive molecular biomarker for CIN2+.
The aims of the study were to 1) analyze the methylation profile of 4 genes using real-time quantitative methylation-specific polymerase chain reaction (PCR) (QMSP) to determine if each gene, and the genes in combination, could distinguish HSIL from LSIL and negative liquid-based Pap tests; 2) determine if the relative level of methylation was associated with histologic outcome, particularly in the distinction between CIN2 and CIN3; and 3) determine if there was an association between methylation profiles and HPV status and/or type.
MATERIALS AND METHODS
Clinical Samples and Cell Culture
Randomly selected biopsy-confirmed HSIL (n = 39) and LSIL (n = 30), and cytologically negative for intraepithelial lesion or malignancy (NILM; n = 30) residual SurePath liquid-based Pap test samples were identified from the pathology archives of the Johns Hopkins Hospital after obtaining approval by the Johns Hopkins Medicine Institutional Review Board. Blinded review of the cytologic (by K.S.G.) and histologic (by B.M.R.) slides were performed to confirm the diagnoses. The mean patient age was 34 ± 12 standard deviation (SD) years for HSIL, 28 ± 10 SD years for LSIL, and 41 ± 10 SD years for NILM. Human cervical cancer cell lines (SiHa and C33A) were kindly provided by Dr. Chien-Fu Hung (Johns Hopkins University, Baltimore, Md) and originally purchased from American Type Culture Collection (Manassas, Va). The SiHa and C33A cells were maintained in DMEM (Invitrogen, Carlsbad, Calif ) supplemented with 10% fetal bovine serum and penicillin/streptomycin (50 U/mL) at 37°C in a 5% CO2 incubator.
DNA Preparation
Genomic DNA from residual liquid-based Pap tests and cervical cancer cell lines was isolated using the PUREGENE DNA Purification Kit (Gentra Systems, Minneapolis, Minn) according to the manufacturer’s instructions. SiHa genomic DNA was used as the positive methylated control (MC). C33A genomic DNA was used as the negative unmethylated control (UC) for DAPK1, IGSF4, and TFPI2; pooled female white blood cell genomic DNA (Novagen, Madison, Wis) was used as the negative UC for SPARC. Approximately 1 µg of genomic DNA was bisulfite-treated using the EZ DNA Methylation-GOLD Kit (ZYMO Research, Orange, Calif) according to the manufacturer’s instructions. Bisulfite-modified DNA was quantitated by UV spectrophotometer analysis and stored at −20°C until use.
Real-Time QMSP
Primers were purchased from Sigma-Proligo (Woodlands, Tex) and fluorogenic probes were purchased from Integrated DNA Technologies (Coralville, Iowa). The methylation-dependent primers and fluorogenic probe for tumor suppressor genes were designed to specifically recognize bisulfite-modified DNA. The sequences of the methylation-dependent primers and probes were as follows: 1) DAPK1, 5′-AGGGGA TTCGGTAATTCGTAG+C-3′ (forward primer), 5′-CC GAAAACTAACCGAAACGAC+G-3′ (reverse primer), and 6FAM-5′-TCGGCGTTTGGGAGGGATTTGCGTT-3′- BHQ1 (probe); 2) IGSF4, 5′-GGCGTTGTGATTGGTTT GTT+C-3′ (forward primer), 5′-CACCTACCTCAAAC TAACGAC+G-3′ (reverse primer), and 6FAM-5′- T+CGTT+CGGGTTT+CGGAGGT-3′-BHQ1 (probe); 3) SPARC, 5′-TTTCGCGGTTTTTTAGATTGTT+C-3′ (forward primer), 5′-AACGACGTAAACGAAAATATC+G-3′ (reverse primer), and 6FAM-5′-AC+GACAAACAAAAC+GC+GCTCTC-3′-BHQ1 (probe);4)TFPI2, 5′-TTTCGTATAAAGCGGGTATT+C-3′ (forward primer), 5′- ACGACCCGCTAAACAAAAC+G-3′ (reverse primer), and 6FAM-5′-C+GAAAAAAC+GCCTAAC+GAAAAAAAA-3′-BHQ1 (probe). Bases preceded by a plus sign and underlined represent locked nucleic acid residues that have been substituted at sites critical for discrimination of methylated and unmethylated DNA to enhance specificity for methylated alleles.18 Methylation independent primers and probe for the internal reference gene, ACTB, were 5′-TGGTGATGGAGGAGGTTTAGTAAGT- 3′ (forward primer), 5′-AACCAATAAAACCTACTCCTCCCTTAA- 3′ (reverse primer), and 6FAM-5′-ACCACCACCCAACACACAATAACAAACACA-3′-BHQ1 (probe).
QMSP was performed in 96-well plates using the Mx3000P Real-Time PCR System (Stratagene, La Jolla, Calif ). Reactions contained 1× iQ Supermix (BioRad, Hercules, Calif), 30 nM ROX reference dye (Stratagene), 300 nM forward and reverse primers, 100 nM probe, and 100 ng of bisulfite-modified genomic DNA template in a final volume of 25 µL. The realtime PCR conditions were 95°C for 5 minutes, then 45 cycles of 95°C for 15 seconds and 60–65°C (depending on the primer set) for 1 minute. Fluorescence data were collected during the annealing/extension step for determination of the cycle threshold (Ct). Each primer pair was run in a separate well and at least duplicate reactions were performed. To ensure the specificity of reactions, each plate contained wells with water only (no template control, NTC), UC DNA, and MC DNA. Serial dilutions of MC DNA template used to generate standard curves from Ct values demonstrated that the linear range of the assay extended from 100 ng to 16 pg of DNA (about 5 genome equivalents). To determine the relative level of methylation of the gene of interest present in each sample, the percentage of methylation (%M) in the sample was calculated as the ratio of the average DNA quantity of the methylated gene of interest to the average DNA quantity of the internal reference gene ACTB multiplied by 100.
HPV DNA Detection and Typing
HPV detection and genotyping was performed using the PCR-based Roche Linear Array HPV genotyping test (Roche Molecular Systems, Branchburg, NJ) as previously described.19 Thirty-seven HPV genotypes were tested, including 19 oncogenic types (HPV-16, -18, -26, 31, -33, -35, -39, -45, -51, -52, -56, -58, -59, -66, -68, -69, -70, -73, and -82) and 18 nononcogenic types (HPV-6, -11, -40, -42, -53, -54, -55, -61, -62, -64, -67, -71, -72, -81, -83, -84, -IS39, and -CP6108). The designation of HPV types as oncogenic or nononcogenic was based on previous epidemiologic classification.20
Statistical Analysis
Categorical data were compared using the Fisher exact test and continuous data were compared using the unpaired t-test (with Welch correction for unequal variances) or the Mann-Whitney test. Receiver operating characteristic (ROC) curves were generated using the percentage methylation (%M) in HSIL and combined NILM/LSIL samples for each gene. The cumulative methylation scores for each sample, defined as the sum of %M of all 4 genes, were used to generate the ROC curve for the 4 genes in combination. The area under the ROC curve (AUC) was used as a measure of test performance to determine if gene methylation could distinguish between HSIL and combined NILM/LSIL samples.21 Statistical analysis was performed using Analyze-It v.2 Software for Excel (Leeds, UK). P-values <.05 were considered statistically significant.
RESULTS
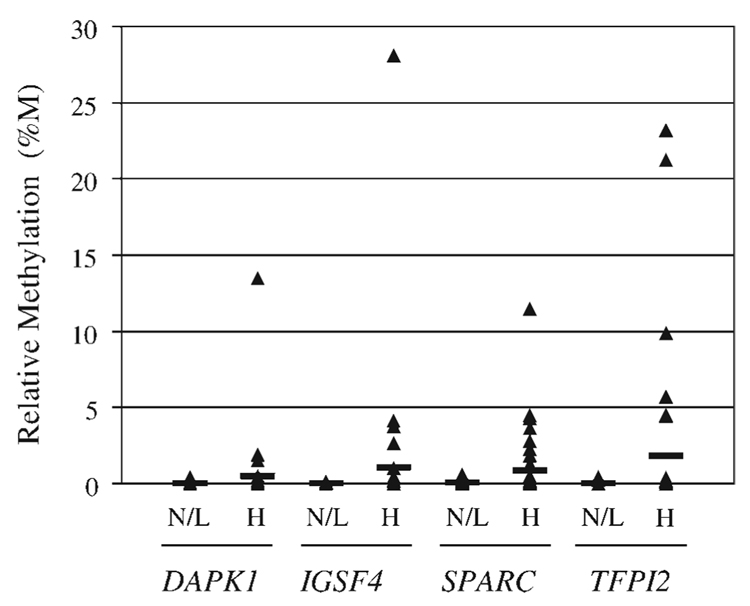
The methylation profile for each of 4 genes (DAPK1, IGSF4, SPARC, and TFPI2) was determined in a cohort of residual liquid-based Pap tests from biopsy-confirmed HSIL and LSIL samples and cytologically negative (NILM) samples using quantitative methylation-specific PCR (QMSP). The QMSP assay had an analytical sensitivity of 16 pg of DNA (approximately 5 genome equivalents) and a linear range of detection from 100 ng to 16 pg of DNA for each gene (data not shown). Qualitative assessment for the presence or absence of methylation at each gene locus demonstrated that methylation was detected more frequently in HSIL than in NILM or LSIL Pap tests (Table 1). Given that both the frequency and relative level of methylation for each of the genes tested were not significantly different between NILM and LSIL samples, comparisons were made between HSIL and combined NILM/LSIL samples to determine if there were differences in methylation between these 2 clinically relevant groups. The relative level of methylation (%M) for each gene in HSIL samples compared with combined NILM/LSIL samples is shown in Figure 1. The range of %M for DAPK1 was 0% to 13.47% (mean, 0.501%) in HSIL and 0% to 0.43% (mean, 0.008%) in combined NILM/LSIL (P 5.1659). The range of %M for IGSF4 was 0% to 28.09% (mean, 1.056%) in HSIL and 0% to 0.09% (mean, 0.004%) in combined NILM/LSIL (P =.1563). The range of %M for SPARC was 0% to 11.44% (mean, 0.868%) in HSIL and 0% to 0.57% (mean, 0.037%) in combined NILM/LSIL (P =.0194). The range of %M for TFPI2 was 0% to 23.18% (mean, 1.784%) in HSIL and 0% to 0.43% (mean, 0.011%) in combined NILM/LSIL (P = .0404). Given the distribution of %M among samples, the nonparametric Mann-Whitney test was used to analyze the differences between median %M in HSIL samples compared with combined NILM/LSIL samples and demonstrated significant differences for all 4 genes (DAPK1, P =.0008; IGSF4, P =.0018; SPARC, P =.0003; and TFPI2, P =.0020).
TABLE 1.
Frequency of Methylation of Gene Loci
| Methylation frequency | |||||
|---|---|---|---|---|---|
| Pap test samples |
DAPK1 No. (%) |
IGSF4 No. (%) |
SPARC No. (%) |
TFPI2 No. (%) |
Any gene No. (%) |
| NILM, n=30 | 1 (3.3) | 3 (10) | 7 (23.3) | 0 (0) | 8 (26.7) |
| LSIL, n=30 | 1 (3.3) | 0 (0) | 3 (10) | 2 (6.7) | 4 (13.3) |
| HSIL, n=39 | 10 (25.6) | 10 (25.6) | 19 (48.7) | 9 (23.1) | 25 (64.1) |
NILM indicates no intraepithelial lesion or malignancy; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion.
FIGURE 1.
Distribution of gene methylation in combined negative for intraepithelial lesion or malignancy (NILM)/low-grade squamous intraepithelial lesions (LSIL) and high-grade SIL (HSIL) Pap test samples. The relative level of methylation (percentage of methylation, %M) for each of the 4 genes (DAPK1, IGSF4, SPARC, and TFPI2) is shown for combined NILM/LSIL (N/L; n = 60) and HSIL (H; n = 39) samples. The black bar indicates the mean %M.
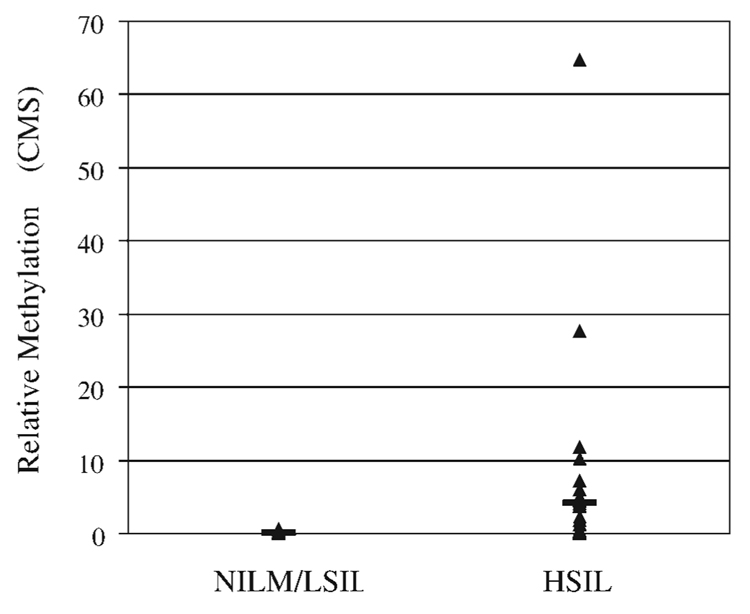
To determine the total methylation in each Pap test sample, we calculated a cumulative methylation score, which was defined as the sum of %M for each of the 4 individual genes tested.22 The range of cumulative methylation scores were 0 to 64.74 (mean, 4.210) in HSIL and 0 to 0.66 (mean, 0.059) in combined NILM/LSIL samples (P = .0256; Fig. 2). The difference between the median cumulative methylation scores for HSIL compared with NILM/LSIL samples was highly significant (P <.0001, Mann-Whitney test).
FIGURE 2.
Cumulative methylation of DAPK1, IGSF4, SPARC, and TFPI2 in combined negative for intraepithelial lesion or malignancy (NILM)/low-grade squamous intraepithelial lesions (LSIL) and high-grade SIL (HSIL) Pap test samples. The cumulative methylation score (CMS), defined as the sum of percentage methylation for each of 4 genes tested, is shown for combined NILM/LSIL (n = 60) and HSIL (n = 39) samples. The black bar indicates the mean CMS.
Using the relative level of methylation (%M) for each individual gene and cumulative methylation scores for the 4 genes in combination, we performed ROC analysis and determined the AUC as a measure of test performance to determine if each gene, and the genes in combination, could distinguish HSIL from combined NILM/LSIL Pap test samples. As shown in Table 2, the AUC for each individual gene was significantly greater than 0.5, demonstrating that each gene has the ability to distinguish HSIL from combined NILM/LSIL samples. The AUC for the 4 genes in combination was significantly greater than 0.5 and greater than each of the individual genes, supporting the concept that methylation profiles of genes in combination can provide a better test.
TABLE 2.
Gene Methylation Distinguishes HSIL From NILM/LSIL Pap Tests
| Gene | Area under the ROC curve |
P* | 95% CI of area |
|---|---|---|---|
| DAPK1 | 0.61 | .0012 | 0.54 to 0.69 |
| IGSF4 | 0.61 | .0023 | 0.53 to 0.69 |
| SPARC | 0.67 | .0002 | 0.58 to 0.77 |
| TFPI2 | 0.60 | .0028 | 0.53 to 0.67 |
| All 4 genes | 0.76 | <.0001 | 0.67 to 0.85 |
NILM indicates no intraepithelial lesion or malignancy; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; ROC, receiver operating characteristic; CI,confidence interval.
P values represent statistically significant differences from .5.
To determine if the level of methylation was associated with histologic outcome, the total methylation in Pap test samples was compared with histologic outcome. There were significant differences in median cumulative methylation scores for Pap test samples with histologic follow-up of CIN1 versus CIN2 (P =.0002) and CIN 1 versus CIN3 (P <.0001; Table 3). However, the difference in median cumulative methylation scores between CIN2 and CIN3 was not significant (P =.6969; Table 3)
TABLE 3.
Cumulative Methylation in Pap Tests Stratified by Histologic Outcome
| Cumulative methylation score* | ||||
|---|---|---|---|---|
| Histologic outcome | Mean | 95% CI of mean | Median† | 95% CI of median |
| CIN1, n=30 | 0.051 | −0.008 to 0.111 | 0.000 | 0.0 to 0.0 |
| CIN2, n=12 | 2.576 | 0.355 to 4.797 | 0.475 | 0.0 to 5.970 |
| CIN3, n=27 | 4.936 | −0.294 to 10.167 | 0.170 | 0.0 to 3.730 |
CIN1 indicates cervical intraepithelial neoplasia 1; CIN2, cervical intraepithelial neoplasia 2; CIN3,cervical intraepithelial neoplasia 3; CI, confidence interval.
Cumulative methylation score is the sum of percentage methylation for each of 4 genes tested.
CIN1 vs CIN2, P =.0002; CIN1 vs CIN3, P <.0001; CIN2 vs CIN3, P =.6969 (Mann-Whitney test)
To determine if there was an association between gene methylation and HPV, detection and genotyping of HPV in Pap test samples was carried out using a PCR-based method (Table 4).19 Overall, HPV was detected in 10 (33%) of 30 NILM Pap tests compared with 29 (97%) of 30 Pap tests with LSIL (P < .0001) and 38 (97%) of 39 Pap tests with HSIL (P < .0001). Oncogenic HPV types were identified in 4 (13%) of 30 NILM Pap tests, 27 (90%) of 30 Pap tests with LSIL, and 37 (95%) of 39 Pap tests with HSIL. Given that HPV-16 is the most common oncogenic type and is present in approximately 50% of cervical cancers, we further stratified Pap test samples into those that were positive for HPV-16. In this cohort of samples, HPV-16 was detected in 3 (10%) of 30 Pap tests with LSIL and 23 (59%) of 39 Pap tests with HSIL (P < .0001). The frequency of detection of multiple HPV types was similar in Pap tests with LSIL and HSIL. There were no significant associations between the presence or types of HPV and methylation status of any gene.
TABLE 4.
Pap Test Samples Stratified by HPV Test Results
| No. of samples (%) | |||
|---|---|---|---|
| HPV Category | NILM, n=30 | LSIL, n=30 | HSIL, n=39 |
| Negative* | 20 (67) | 1 (3) | 1 (3) |
| Nononcogenic | 6 (20) | 2 (7) | 1 (3) |
| Oncogenic | 4 (13) | 27 (90) | 37 (95) |
| HPV-16† | 0 (0) | 3 (10) | 23 (59) |
| Multiple types | 2 (7) | 25 (83) | 28 (72) |
NILM indicates no intraepithelial lesion or malignancy; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; HPV, human papillomavirus.
NILM vs LSIL and NILM vs HSIL, P <.0001 (Fisher exact test).
LSIL vs HSIL, P <.0001 (Fisher exact test).
DISCUSSION
Detection and treatment of HSIL (CIN2 and particularly precancerous CIN3) or more severe lesions is the main emphasis of cervical cancer screening programs. Despite the success of the Pap test as a screening test for cervical cancer, limitations in its reproducibility and its sensitivity for the detection of HSIL have led to the search for clinically useful ancillary tests to aid in management of women with cervical cytologic abnormalities. Aberrant DNA methylation of tumor suppressor genes is a molecular test that could potentially serve as a useful biomarker for early detection and/or risk of progression to cervical cancer.23,24 In the present study we examined the methylation status of 4 genes, individually and in combination, in a cohort of biopsy-confirmed HSIL and LSIL, and cytologically negative residual liquid-based Pap tests using real-time QMSP. Our results demonstrate that methylation of each of the 4 genes (DAPK1, IGSF4, SPARC, and TFPI2) occurs more frequently and at a higher relative level in HSIL compared with NILM and LSIL Pap tests. Using quantitative methylation data and ROC analysis, we found that each gene and the 4 genes in combination could distinguish HSIL from combined NILM/LSIL Pap tests. The total methylation was significantly greater in samples with histologic follow-up of CIN2 or CIN3 compared with CIN1; however, there was no significant difference between samples with follow-up of CIN2 versus CIN3. Detection of aberrant DNA methylation was not associated with the presence or type of HPV
In this study, DAPK1 methylation was detected in 10 (25.6%) of 39 HSIL Pap tests compared with 2 (3.3%) of 60 combined NILM/LSIL Pap tests. Methylation of IGSF4 was also detected in 10 (25.6%) of 39 HSIL Pap tests compared with 3 (5%) of 60 combined NILM/LSIL Pap tests. Previous studies that have examined the methylation status of DAPK1 and IGSF4 in cervical tissues or Pap tests with intraepithelial lesions have shown similar findings of more frequent methylation of both tumor suppressor genes in HSIL (CIN2/3) compared with LSIL (CIN1) or negative samples. In the most comprehensive study to date, DAPK1 was found to be methylated in 12 (52.2%) of 23 exfoliated samples with histologic outcomes of CIN3, 4 (17.4%) of 23 with CIN2, 3 (7.7%) of 39 with CIN1, and 3 (2.1%) of 140 with negative histologic outcomes.14 A study by Steenbergen et al.12 showed that IGSF4 was methylated in 7 (35%) of CIN2/3 but absent in 12 CIN1 lesions and 9 negative tissue samples. We previously examined DAPK1 and IGSF4 methylation in a feasibility study using residual liquid-based Pap tests and a multiplex, nested methylation-specific PCR approach and found that DAPK1 and IGSF4 were methylated in approximately 64% of HSIL Pap tests but absent in LSIL and NILM Pap tests.17 The increased frequency of methylation detected in HSIL in our previous study compared with the current findings may, in part, be due to differences in detection methods (multiplex, nested MSP vs real-time QMSP) and/or the location of the primers within the promoters of these 2 genes.
In the present study, SPARC was methylated in 19 (48.7%) of 39 Pap tests with HSIL compared with 10 (16.7%) of 60 Pap tests with NILM/LSIL. TFPI2 was methylated in 9 (23.1%) of 39 Pap tests with HSIL compared with 2 (3.3%) of 60 Pap tests with NILM/LSIL. SPARC and TFPI2 were recently identified as novel genes frequently methylated in cervical carcinoma through the use of global demethylation and microarray expression analysis in cervical cancer cell lines.15 Using the quantitative MethylLight assay for detection of methylation,25 those authors found that SPARC was methylated in 20 (91%) of 22 exfoliated cervical samples with invasive cervical carcinoma compared with 1 (5%) of 21 normal control samples; TFPI2 was methylated in 18 (82%) of 22 invasive cervical carcinoma samples compared with 8 (38%) of 21 normal control samples. The differences in the frequency of SPARC and TFPI2 methylation detected in normal/negative samples in our study may be due, in part, to differences in primer and/or probe sequences. To our knowledge, the methylation status of SPARC and TFPI2 have not been previously examined in precursor squamous intraepithelial lesions. Our findings, which show more frequent methylation of SPARC and TFPI2 in HSIL compared with NILM/LSIL samples, suggest that these genes may play a role in cervical carcinogenesis.
In addition to qualitative assessments for the presence or absence of DNA methylation, real-time QMSP provides a quantitative assessment of the relative level of methylation in the sample compared with a reference gene, such as ACTB. Using quantitative data and area under the ROC curve as a measure of test performance, we determined that each individual gene could distinguish HSIL from combined NILM/LSIL Pap test samples, and that the combination of 4 genes improved test performance. In addition, quantitative analysis of relative levels of methylation in samples allows for the establishment of cutoff values for a positive test. However, selection of the appropriate cutoff value for a positive test should be based on cost-benefit analysis and cutoffs may differ depending on the population tested and prevalence of the disease (ie, its use as a screening test vs as an adjunct to Pap tests with abnormal findings). Based on a cutoff of >0% total methylation in the sample (ie, methylation detected for any of the 4 genes), the assay has a clinical sensitivity and specificity of 64.1% and 80%, respectively, for the detection of HSIL. Increasing the cutoff for a positive test to >0.66% methylation would increase the specificity of the assay for the detection of HSIL to 100%; however, the sensitivity would be reduced to 43.6%. Because Pap test samples usually consist of heterogeneous mixtures of abnormal and normal squamous cells, the danger in setting a cutoff value that results in 100% specificity is that a low level of detectable methylation may be a significant finding if it represents limited sampling of a high-grade lesion. Although our results demonstrate that aberrant DNA methylation of DAPK1, IGSF4, SPARC, and TFPI2 can distinguish between HSIL and NILM/LSIL samples and that analysis of the genes in combination improves test performance, analysis of these genes by QMSP would not likely provide the sensitivity and specificity necessary for a clinically useful test. Thus, the identification of additional novel genes or combination of genes that are methylated in HSIL but not in NILM/LSIL samples is necessary to improve clinical performance.
Comparison of total methylation in Pap test samples with histologic outcome revealed that the median cumulative methylation score was significantly greater in both CIN2 and CIN3 compared with CIN1. These findings provide additional evidence to support the 2-tiered Bethesda System for cytological reporting and the biological differences between LSIL (CIN1) and HSIL (CIN2/3).1 Although total methylation was greater in HSIL Pap tests, our results showed that it was not significantly different between Pap tests with histologic outcomes of CIN2 versus CIN3, which may, in part, be due to the relatively small sample size. The increased risk of progression of CIN2/3 lesions to invasive cervical cancer is associated with persistent HR-HPV infection and the accumulation of genetic and epigenetic alterations.26 Given this, our findings raise the possibility that HSIL samples with aberrant DNA methylation and histologic outcomes of CIN2 or CIN3 may represent a subset of lesions that are at increased risk of progression. Further studies that address aberrant DNA methylation as a molecular biomarker of increased risk of progression to cervical cancer would be of interest. However, given the ethical considerations, an animal model would be necessary for such studies.
In contrast to the differences in the frequency of aberrant DNA methylation in HSIL and LSIL Pap tests, detection and typing of HPV by a PCR-based method showed that the prevalence of oncogenic HPV types was similar in HSIL (95%) and LSIL (90%) Pap test samples. Further stratification of samples based on the identification of the most common oncogenic type, HPV-16, revealed that the prevalence of HPV-16 was significantly greater in HSIL (59%) compared with LSIL (10%; P < .0001). Despite these differences, we found no significant association between HPV prevalence or type and aberrant DNA methylation. Our results for HPV-16 typing are similar to data from the ALTS that showed that HPV-16 was present in 49% of women with HSIL and 21.1% of women with LSIL.2,27 Importantly, findings from the ALTS, as well as a large study performed in a screening population, showed that the risk of progression to CIN3 or cancer is significantly greater among women with HPV-16, and possibly HPV-18, compared with other oncogenic types.27,28 Thus, HPV typing may prove to be clinically useful to further stratify women at risk for underlying high-grade cervical lesions.
In summary, detection of DNA methylation in liquid-based Pap tests by QMSP allows for quantitation of relative levels of methylation in samples and the establishment of an appropriate cutoff value for a positive test. Aberrant DNA methylation of DAPK1, IGSF4, SPARC, and TFPI2 can each distinguish HSIL from NILM/LSIL Pap tests. The combination of these 4 genes improves test performance; however, the overall sensitivity is relatively low. Although testing for DNA methylation holds promise as an adjunct to the Pap test, further studies are needed to identify additional genes that are selectively methylated in HSIL and to determine the optimal combination of genes that will provide increased clinical sensitivity while maintaining high specificity.
Acknowledgments
Supported by NIH/NCI CA98252 (Career Development Award to K.S.G.) and CA123612 (to K.S.G.).
This work was presented in part at the 96th Annual Scientific United States and Canadian Academy of Pathology Meeting, March 24–30, 2007, San Diego, California.
The authors thank Frances H. Burroughs for technical assistance.
Footnotes
The authors have no conflicts of interest.
REFERENCES
- 1.Solomon D, Davey D, Kurman R, et al. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA. 2002;287:2114–2119. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 2.Sherman ME, Castle PE, Solomon D. Cervical cytology of atypical squamous cells-cannot exclude high-grade squamous intraepithelial lesion (ASC-H): characteristics and histologic outcomes. Cancer. 2006;108:298–305. doi: 10.1002/cncr.21844. [DOI] [PubMed] [Google Scholar]
- 3.Wright TC, Jr, Massad LS, Dunton CJ, Spitzer M, Wilkinson EJ, Solomon D. 2006 consensus guidelines for the management of women with abnormal cervical cancer screening tests. Am J Obstet Gynecol. 2007;197:346–355. doi: 10.1016/j.ajog.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 4.ASCUS-LSIL Triage Studt (ALTS) Group. Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 2003;188:1383–1392. doi: 10.1067/mob.2003.457. [DOI] [PubMed] [Google Scholar]
- 5.The Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J Natl Cancer Inst. 2000;92:397–402. doi: 10.1093/jnci/92.5.397. [DOI] [PubMed] [Google Scholar]
- 6.Dunne EF, Unger ER, Sternberg M, et al. Prevalence of HPV infection among females in the United States. JAMA. 2007;297:813–819. doi: 10.1001/jama.297.8.813. [DOI] [PubMed] [Google Scholar]
- 7.Insinga RP, Glass AG, Rush BB. Diagnoses and outcomes in cervical cancer screening: a population-based study. Am J Obstet Gynecol. 2004;191:105–113. doi: 10.1016/j.ajog.2004.01.043. [DOI] [PubMed] [Google Scholar]
- 8.Syrjanen KJ. Spontaneous evolution of intraepithelial lesions according to the grade and type of the implicated human papillomavirus (HPV) Eur J Obstet Gynecol Reprod Biol. 1996;65:45–53. doi: 10.1016/0028-2243(95)02303-a. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hyper-methylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 10.Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7:1982–1986. [PubMed] [Google Scholar]
- 11.Narayan G, Arias-Pulido H, Koul S, et al. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Mol Cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steenbergen RD, Kramer D, Braakhuis BJ, et al. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- 13.Reesink-Peters N, Wisman GB, Jeronimo C, et al. Detecting cervical cancer by quantitative promoter hypermethylation assay on cervical scrapings: a feasibility study. Mol Cancer Res. 2004;2:289–295. [PubMed] [Google Scholar]
- 14.Feng Q, Balasubramanian A, Hawes SE, et al. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97:273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- 15.Sova P, Feng Q, Geiss G, et al. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:114–123. doi: 10.1158/1055-9965.EPI-05-0323. [DOI] [PubMed] [Google Scholar]
- 16.Virmani AK, Muller C, Rathi A, Zoechbauer-Mueller S, Mathis M, Gazdar AF. Aberrant methylation during cervical carcinogenesis. Clin Cancer Res. 2001;7:584–589. [PubMed] [Google Scholar]
- 17.Gustafson KS, Furth EE, Heitjan DF, Fansler ZB, Clark DP. DNA methylation profiling of cervical squamous intraepithelial lesions using liquid-based cytology specimens: an approach that utilizes receiver-operating characteristic analysis. Cancer. 2004;102:259–268. doi: 10.1002/cncr.20425. [DOI] [PubMed] [Google Scholar]
- 18.Gustafson KS. Locked nucleic acids can enhance the analytical performance of quantitative methylation-specific polymerase chain reaction. J Mol Diagn. 2008;10:33–42. doi: 10.2353/jmoldx.2008.070076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gravitt PE, Peyton CL, Apple RJ, Wheeler CM. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36:3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz N, Bosch FX, de Sanjose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 21.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 22.Fackler MJ, McVeigh M, Mehrotra J, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64:4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 23.Duenas-Gonzalez A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer. 2005;4:38. doi: 10.1186/1476-4598-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7:11–22. doi: 10.1038/nrc2050. [DOI] [PubMed] [Google Scholar]
- 25.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208:152–164. doi: 10.1002/path.1866. [DOI] [PubMed] [Google Scholar]
- 27.Castle PE, Solomon D, Schiffman M, Wheeler CM. Human papillomavirus type 16 infections and 2-year absolute risk of cervical precancer in women with equivocal or mild cytologic abnormalities. J Natl Cancer Inst. 2005;97:1066–1071. doi: 10.1093/jnci/dji186. [DOI] [PubMed] [Google Scholar]
- 28.Khan MJ, Castle PE, Lorincz AT, et al. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J Natl Cancer Inst. 2005;97:1072–1079. doi: 10.1093/jnci/dji187. [DOI] [PubMed] [Google Scholar]