Summary
In this study, the function of two established Fe-S cluster biogenesis pathways, Isc (Iron sulfur cluster) and Suf (Sulfur mobilization), was compared under aerobic and anaerobic growth conditions by measuring the activity of the Escherichia coli global anaerobic regulator FNR. A [4Fe-4S] cluster is required for activity of FNR under anaerobic conditions. Assaying expression of FNR-dependent promoters in strains containing various deletions of the iscSUAhscBAfdx operon, revealed that under anaerobic conditions FNR activity was reduced by 60% in the absence of the Isc pathway. In contrast, a mutant lacking the entire Suf pathway had normal FNR activity, although overexpression of the suf operon fully rescued the anaerobic defect in FNR activity in strains lacking the Isc pathway. Expression of the sufA promoter and levels of SufD protein were upregulated 2–3 fold in Isc− strains under anaerobic conditions, suggesting that increased expression of the Suf pathway may be partially responsible for the FNR activity remaining in strains lacking the Isc pathway. In contrast, use of the O2-stable [4Fe-4S] cluster FNR variant, FNR-L28H, showed that overexpression of the suf operon did not restore FNR activity to strains lacking the Isc pathway under aerobic conditions. In addition, activity of FNR-L28H was more impaired under aerobic conditions compared to anaerobic conditions. The greater requirement for the Isc pathway under aerobic conditions was not due to a change in the rate of Fe-S cluster acquisition by FNR-L28H between aerobic and anaerobic conditions as shown by 55Fe labelling experiments. Using 35S-methionine pulse-chase assays, we observed that the Isc pathway, but not the Suf pathway, is the major pathway required for conversion of O2-inactivated apo-FNR to [4Fe-4S]-FNR upon the onset of anaerobic growth conditions. Taken together, these findings indicate a major role for the Isc pathway in FNR Fe-S cluster biogenesis under both aerobic and anaerobic conditions.
Keywords: FNR, Fe-S cluster biogenesis, Isc, Suf, oxygen
Introduction
Fe-S clusters are cofactors of proteins that function in diverse and vital roles in multiple cellular processes such as respiration, photosynthesis, gene regulation and nitrogen fixation.1 How these Fe-S cluster cofactors are synthesized and inserted into the relevant target proteins is not well understood. In Escherichia coli, the two gene clusters that are involved in Fe-S biogenesis are iscSUAhscBAfdx, encoding the proteins of the Isc pathway, and sufABCDSE, encoding the proteins of the Suf pathway. While characterization of strains lacking one or the other pathway has led to the conclusion that the Isc proteins function as the major housekeeping Fe-S cluster biogenesis system of E. coli, strains lacking both pathways are inviable2; 3; 4, raising the question of the interplay between these two pathways. In addition, even though some Fe-S clusters are sensitive to O2 or reactive oxygen species, a systematic comparison of the requirements of Fe-S biogenesis under aerobic and anaerobic growth conditions has not been investigated. In this study, we examine the contributions of the Isc and Suf pathways to Fe-S cluster biogenesis under aerobic and anaerobic growth conditions using the transcription factor FNR as a model system.
In the housekeeping Isc system, IscS is a cysteine desulfurase5; 6; 7 that provides the sulfur for Fe-S cluster biogenesis. IscU serves as the scaffold upon which Fe-S clusters are assembled.8; 9 HscB and HscA are homologs of the molecular chaperones DnaJ and DnaK, respectively. However, unlike DnaJ and DnaK, HscA and HscB are “specialized chaperones” that specifically interact with IscU.10; 11 The roles of IscA and Fdx in Fe-S cluster assembly are less clear; however, it has been proposed that E. coli IscA functions as an alternate Fe-S scaffolding protein12 or as an Fe donor for the assembly process.13 The E. coli Suf Fe-S cluster biogenesis pathway includes proteins that show functional similarity to those of the Isc pathway. For example, SufS is also a cysteine desulfurase14 and SufA exhibits sequence similarity to IscA and can function as an Fe-S cluster scaffolding protein.15; 16 SufC has ATPase activity17; 18 as found with HscA of the Isc system.11 SufC also forms a complex with SufB and SufD, which further increases the cysteine desulfurase activity of SufS in the presence of SufE in vitro.19 However, components of the Isc and Suf systems do not appear to be interchangeable in E. coli since deletions of any of the suf genes, with the exception of sufA, were shown to be synthetically lethal with the iscS deletion.4; 20 Apparently, the cysteine desulfurase, SufS, cannot directly substitute for IscS and most likely requires the other Suf accessory proteins. In addition, persulfide bound IscS acts as a sulfur donor in other sulfur transferase reactions, a function apparently not present in SufS.21 Nevertheless, the reduced activities of the Fe-S proteins glutamate synthase and succinate dehydrogenase observed in a strain lacking the Isc system could be restored when expression of the suf operon was elevated, suggesting that the two systems have redundant functions in overall Fe-S biogenesis.3
The roles of the Isc and Suf pathways have largely been examined under aerobic and oxidative stress conditions because of the extreme sensitivity of some Fe-S clusters to oxidation by oxygen, hydrogen peroxide, and superoxide.22; 23 The involvement of the Isc proteins in Fe-S cluster biogenesis under aerobic growth conditions has been established for several Fe-S proteins.24; 25; 26 In addition, expression of both the Isc and Suf pathways are upregulated upon exposure to hydrogen peroxide, suggesting that both pathways may contribute to Fe-S cluster assembly under oxidative stress conditions.27 However, a comparison to anaerobic growth conditions, where clusters are expected to be more stable, has not been systematically investigated, although expression of both the isc and suf operons is known to be lower under these conditions.28
To address whether there are functional differences in the Isc or Suf pathways between aerobic and anaerobic conditions, we examined changes in activity of the anaerobic regulator FNR and a constitutively active mutant derivative, FNR-L28H. Under anaerobic conditions, FNR regulates the expression of genes involved in anaerobic metabolism and its activity is dependent on the presence of a [4Fe-4S] cluster, which promotes dimerization and subsequent DNA binding.29 Under aerobic conditions, the [4Fe-4S] cluster is destroyed by O2, leading to the formation of inactive, apo-FNR.30; 31; 32; 33; 34 Previous studies demonstrated that IscS is involved in Fe-S cluster assembly for FNR since FNR activity was impaired in strains lacking IscS grown under anaerobic conditions.24 We report here the role of the other Isc proteins and the Suf system in providing Fe-S clusters for FNR under anaerobic growth conditions. To directly compare Fe-S cluster biogenesis under anaerobic and aerobic conditions, we monitored the activity of the mutant FNR-L28H, which contains an O2-resistant [4Fe-4S] cluster35, in strains lacking Isc or Suf. In addition, we directly compared the rate of Fe-S cluster acquisition under aerobic and anaerobic conditions by measuring the steady-state rate of 55Fe incorporation into FNR-L28H. Finally, we examined the involvement of the Isc and Suf pathways in the reactivation of apo-FNR to [4Fe-4S]-FNR upon O2-deprivation.
Results
FNR activity is only partially decreased in anaerobic cells lacking the Isc pathway
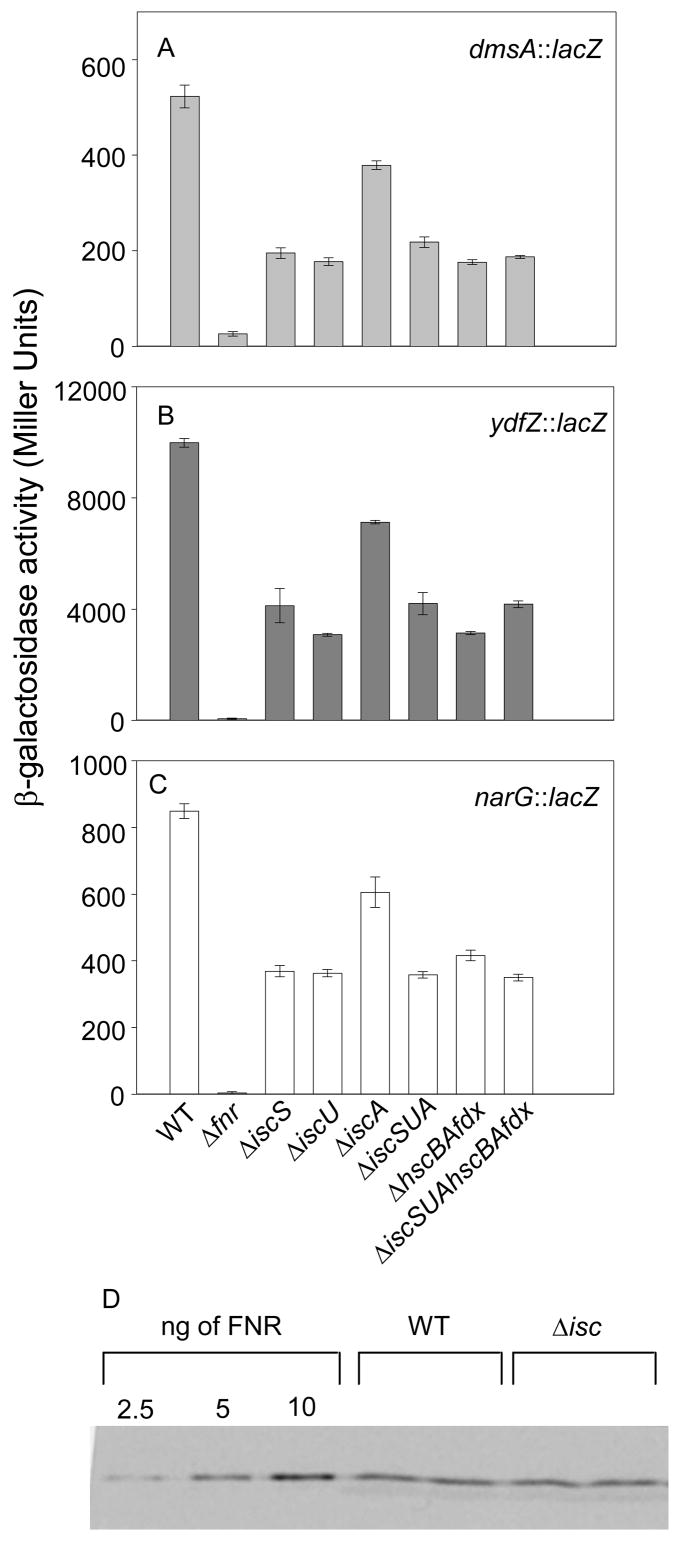
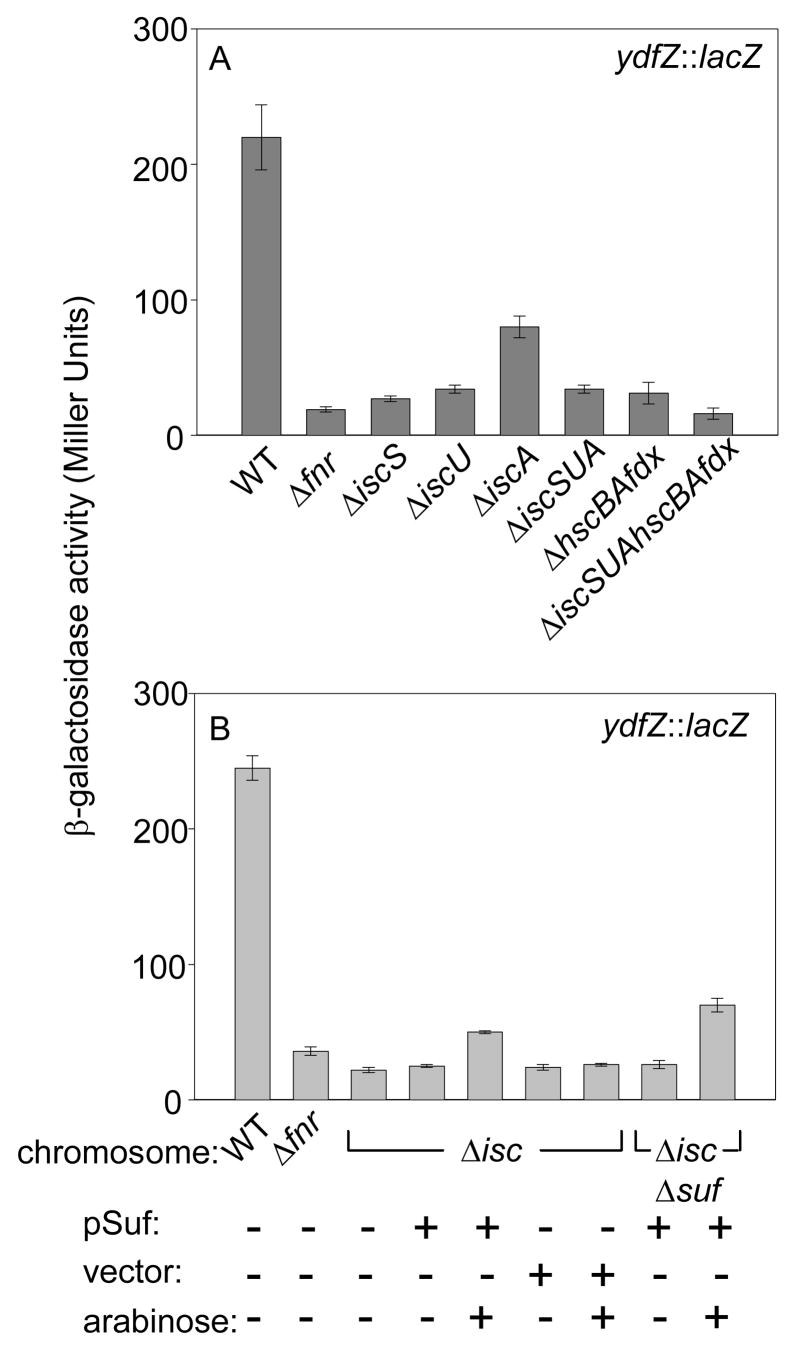
Deletion of the genes encoding the entire Isc pathway, ΔiscSUAhscBAfdx, reduced FNR activity by ~60% (Figure 1). The magnitude of the decrease in FNR activity was similar for several FNR-activated promoters analyzed (dmsA, ydfZ, and narG) (Figure 1) and was not due to changes in FNR protein levels in wild-type and ΔiscSUAhscBAfdx strains as shown by Western blot analysis (Figure 1d). These results confirm that the Isc pathway is the major route for FNR Fe-S biogenesis under anaerobic conditions and show that for the Isc directed portion of FNR Fe-S biogenesis, no other cysteine desulfurase substitutes for IscS in agreement with previous studies.4; 20 Furthermore, with the exception of IscA, we found that strains lacking individual components such as IscS or IscU yielded a similar phenotype as strains lacking IscS, IscU and IscA (ΔiscSUA) or HscA, HscB and Fdx (ΔhscBAfdx), suggesting that all of these proteins contribute a necessary function to FNR Fe-S biogenesis.
Figure 1.
FNR-dependent activation of the A) dmsA, B) ydfZ, and C) narG promoters in anaerobically grown strains containing various deletions of the iscSUAhscBAfdx operon measured by β-galactosidase assays. Strains were grown anaerobically in M9 minimal glucose medium containing nicotinic acid (12.5 μg ml−1). Strains containing the PnarG::lacZ fusion (C) were also grown in the presence of 20 mM KNO3. The error bars represent the standard errors of triplicate samples. D) Western blot analysis of FNR protein assayed from anaerobically grown wild-type and ΔiscSUAhscBAfdx (Δisc) strains. The lanes represent the signals obtained from duplicate samples of 250 μl of cells (WT or Δisc) grown to an OD600 of 0.4 and purified FNR protein (2.5 ng, 5 ng and 10 ng). Quantification of the FNR protein bands from three independently grown samples (data not shown) showed that wild-type and ΔiscSUAhscBAfdx strains contained 34 ± 2 and 35 ± 2 pg of FNR protein / μl of cells at OD600 of 0.4, respectively.
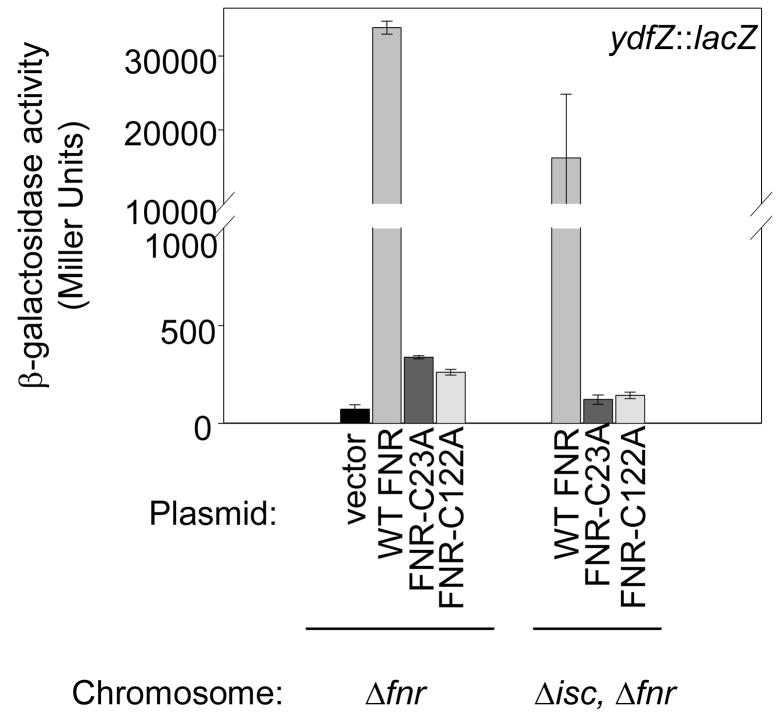
The FNR activity (~40%) remaining in the ΔiscSUAhscBAfdx strain was shown to be dependent on the [4Fe-4S] cluster by assaying FNR variants previously shown to be specifically defective in ligating the Fe-S cluster of FNR (FNR-C23A, FNR-C122A).29; 36; 37; 38; 39 Substitution of either FNR Fe-S cluster ligand, C23 or C122, eliminated expression of PydfZ in the strain lacking the Isc pathway (Figure 2), indicating that the FNR activity present in the ΔiscSUAhscBAfdx strain still depends on the [4Fe-4S] cluster of FNR. Thus, these data show that another Fe-S cluster biogenesis pathway must be functional under anaerobic conditions.
Figure 2.
Anaerobic expression of the ydfZ promoter in Δfnr and Δfnr, ΔiscSUAhscBAfdx strains expressing either wild-type FNR, FNR-C23A or FNR-C122A from the vector pACYC184. Strains were grown under anaerobic conditions in M9 minimal glucose medium containing chloramphenicol (20 μg ml−1) and nicotinic acid (12.5 μg ml−1).
Other known Fe-S biogenesis proteins are not necessary for FNR activity under anaerobic conditions when the Isc pathway is functional
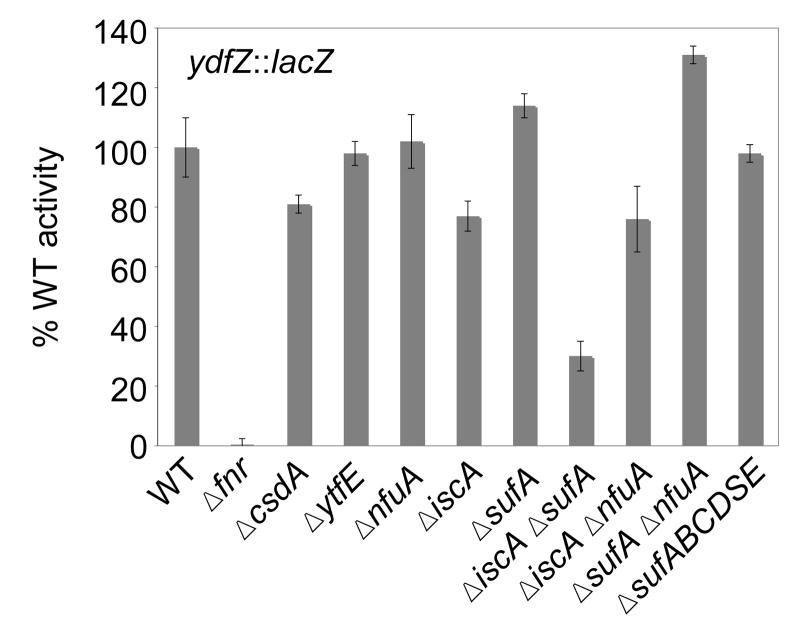
To test whether the Suf pathway provides [4Fe-4S] clusters for FNR under anaerobic conditions, FNR activity was measured in a strain deleted for the sufABCDSE operon. FNR-dependent activation of the ydfZ (Figure 3), dmsA, and narG promoters (data not shown) in the ΔsufABCDSE strain was similar to that of the parent suf+ strain, indicating that the Suf pathway is not necessary for FNR Fe-S cluster assembly under anaerobic growth conditions when the Isc pathway is present. However, since previous studies have shown that SufA can functionally replace IscA3; 4; 20, we investigated whether the function of SufA explained the minor (less than 2 fold) effect of ΔiscA strains on FNR activity. By comparing ydfZ expression in ΔiscA ΔsufA or ΔsufA strains, we found that only strains that lacked both IscA and SufA reduced ydfZ expression to levels observed with the deletion of the entire Isc pathway, indicating that SufA and IscA function interchangeably under anaerobic growth conditions (Figure 3). Furthermore, FNR activity was not decreased in strains lacking either CsdA, a third cysteine desulfurase40, or NfuA, a protein containing an N-terminal region similar to IscA and a C-terminal region similar to that of NifU41; 42 (Figure 3). Strains lacking IscA and NfuA or SufA and NfuA also showed no additional decrease in ydfZ expression compared to any of the single mutants, indicating the absence of functional redundancy that was observed between SufA and IscA (Figure 3). Finally, deletion of ytfE, whose gene product has recently been implicated in Fe-S cluster assembly and/or repair43, also did not affect FNR activity (Figure 3). Thus, in the presence of the Isc pathway, other known Fe-S assembly proteins are not necessary for FNR Fe-S cluster biogenesis under anaerobic conditions.
Figure 3.
β-galactosidase activity from the FNR-activated ydfZ promoter in anaerobically grown strains containing single or double gene deletions of fnr, csdA, ytfE, nfuA, iscA, and sufA or deletion of the entire the sufABCDSE operon. Strains were grown in M9 minimal glucose medium containing nicotinic acid (12.5 μg ml−1). β-galactosidase activities are normalized to FNR activity in the WT strain (100%).
In the absence of the Isc pathway, higher levels of the Suf Fe-S cluster biogenesis proteins can restore FNR activity
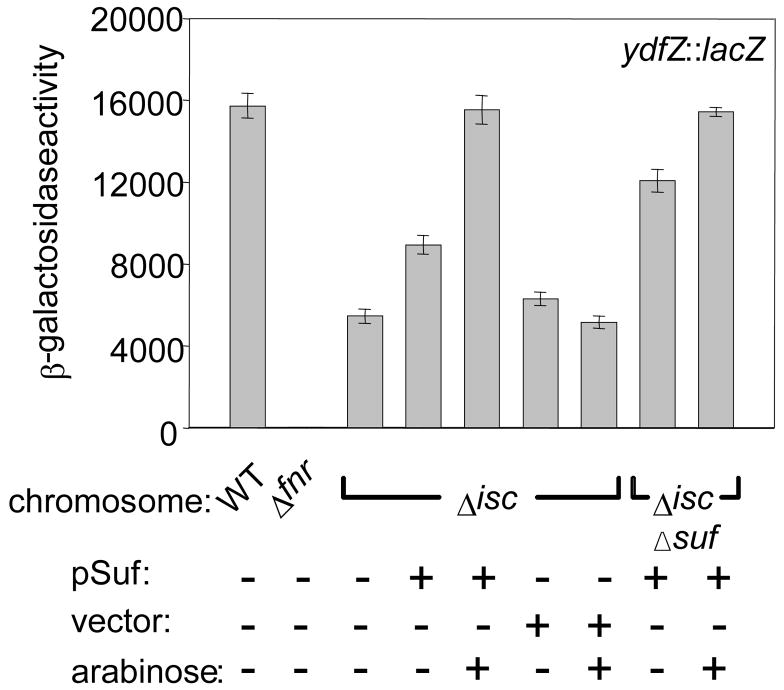
Overexpression of the sufABCDSE operon (suf) has previously been shown to restore the defects in activities of other Fe-S proteins, e.g. glutamate synthase and succinate dehydrogenase, in strains lacking the Isc pathway.3 These results raised the question of whether the defect in FNR activity observed in Isc− strains under anaerobic conditions could also be rescued by overexpression of suf. To answer this question, FNR activity was measured in strains that contained a plasmid with the sufABCDSE operon under the control of the arabinose-inducible PBAD promoter (Figure 4) and contained ΔiscSUAhscBAfdx or ΔiscSUAhscBAfdx, ΔsufABCDSE. As expected, the strain lacking both the Isc and Suf pathways (ΔiscSUAhscBAfdx, ΔsufABCDSE) was viable only when the suf plasmid, not the vector, was present.2; 3; 4 Even when no arabinose was present in the growth medium, strains containing the suf plasmid had a 9-fold increase in SufD protein levels (measured by Western blots using antibodies to SufD; data not shown) and a ~2-fold increase in FNR activity relative to the parent suf+ strain. When 0.2% arabinose was present in the growth medium, the defect in FNR activity observed in the Isc− strain was completely rescued (Figure 4) by the ~ 30-fold overexpression (data not shown) of the suf operon; a similar level of ydfZ expression was observed in the Isc−Suf− strain upon arabinose induction of the suf plasmid. This restoration of FNR activity can be attributed to the increased levels of the Suf proteins since FNR activity was not increased in the ΔiscSUAhscBAfdx strain transformed with the vector alone. Taken together, these data suggest that the Suf pathway is biochemically sufficient to replace the Isc pathway in FNR Fe-S cluster assembly when expressed at elevated levels under anaerobic conditions.
Figure 4.
β-galactosidase activity from the ydfZ promoter in anaerobically grown wild-type, Δfnr, and ΔiscSUAhscBAfdx (Δisc), and ΔiscSUAhscBAfdx, ΔsufABCDSE (Δisc, Δsuf) strains. The ΔiscSUAhscBAfdx strain derivatives contained either the plasmid expressing the sufABCDSE operon from the arabinose-inducible PBAD promoter (pSuf), the vector alone, or no vector. All cultures were grown anaerobically in M9 minimal glucose medium containing nicotinic acid (12.5 μg ml−1). The plasmid-containing strains were also grown in the presence of ampicillin (final concentration of 50 μg ml−1) and, where indicated, 0.2% arabinose.
Expression of the suf operon is upregulated in strains lacking the Isc pathway
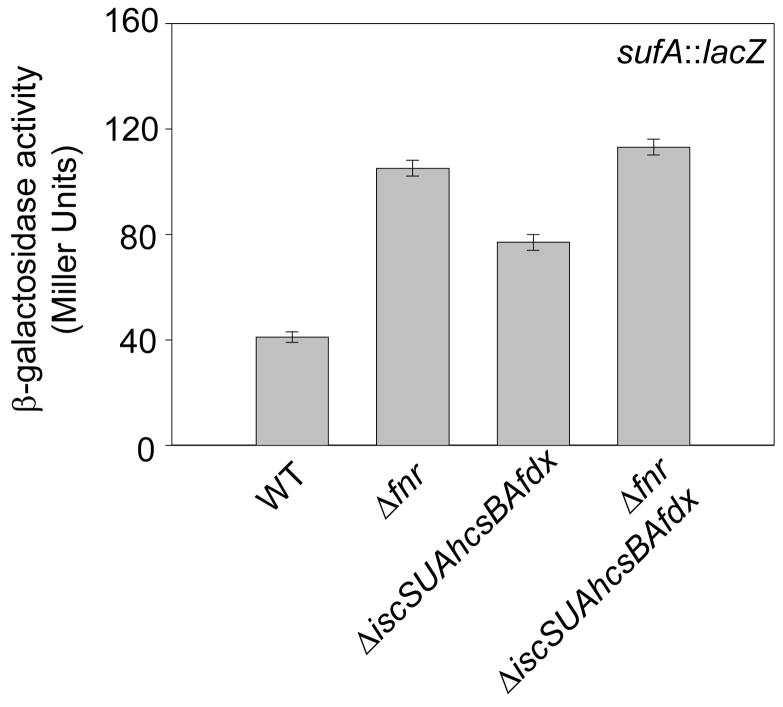
Since overexpression of the Suf pathway can restore FNR activity to wild-type levels in strains completely lacking the Isc pathway, we considered the possibility that the FNR activity (40%) remaining in strains lacking the Isc pathway might result from Suf-directed Fe-S biogenesis if Suf protein levels are upregulated. Expression of the suf operon is known to be repressed by FNR under anaerobic conditions44 and is activated by IscR lacking an Fe-S cluster28; thus a change in activity of either regulator, resulting from the absence of the Isc proteins, could increase sufA promoter expression. Indeed, anaerobic expression of PsufA::lacZ was ~2 fold higher in the ΔiscSUAhscBAfdx strain compared to that of wild-type (Figure 5). In addition, Western blot analysis using α-SufD antibodies revealed that Isc− strains contained ~2–3-fold more SufD protein than Isc+ strains (data not shown), consistent with the observed increase in PsufA expression. Thus, elevated levels of the Suf proteins may explain some or all of the remaining FNR activity under anaerobic conditions in cells lacking the Isc pathway.
Figure 5.
Anaerobic β-galactosidase activity from the sufA promoter in strains lacking fnr, the iscSUAhscBAfdx operon (Δisc), or both. Cultures were grown anaerobically in M9 minimal glucose medium containing a final concentration of nicotinic acid (12.5 μg ml−1).
The requirement for the Isc pathway in FNR Fe-S cluster assembly is greater under aerobic growth conditions
A key unanswered question is whether there are differences in the requirements for the Isc and Suf pathways in FNR Fe-S cluster assembly between aerobic and anaerobic growth conditions. This question is particularly relevant since both pathways are upregulated under aerobic conditions, when some Fe-S clusters are known to be less stable. However, it is difficult to measure wild-type FNR Fe-S cluster formation under aerobic conditions because the [4Fe-4S] cluster in FNR is unstable to O2, and the protein is predominately in the apo-form, resulting in undetectable regulation of most FNR-dependent promoters. To circumvent this problem, two approaches were used. First, we examined the aerobic activation of an unusually strong FNR regulated promoter, that of ydfZ, which still has measurable, FNR-dependent activity under aerobic conditions, albeit at levels ~60-fold less than in anaerobic grown cultures. Second, we utilized an FNR mutant (FNR-L28H) in which the Fe-S cluster is resistant to disruption by O2.35 Therefore, [4Fe-4S]-FNR-L28H accumulates under aerobic conditions, in contrast to wild-type FNR, which would be largely in the apo-form as a result of cluster turnover.
The first approach takes advantage of the observation that aerobic activation of PydfZ is still dependent on the [4Fe-4S] form of FNR (data not shown), indicating that some low level of holo-FNR is present under aerobic conditions. While aerobic ydfZ promoter activity was not affected in ΔsufABCDSE, ΔcsdA, ΔytfE or ΔnfuA strains (data not shown), we found that the isc genes were absolutely required (Figure 6a). Activity of the ΔiscSUAhscBAfdx strain was decreased 10-fold, the same magnitude as the fnr deletion strain (Figure 6a), in remarkable contrast to the just 60% reduction in ydfZ expression observed under anaerobic conditions (Figure 1b). Importantly, overexpression of the Suf pathway was not able to rescue the defects in FNR activity observed in the ΔiscSUAhscBAfdx strain (Figure 6b), in stark contrast to what was observed under anaerobic conditions (Figure 4). When expression of the suf operon, under the control of PBAD, was induced with arabinose under aerobic conditions to levels similar to those under anaerobic conditions (~30 fold; data not shown), only a small increase in wild-type FNR activation of ydfZ was observed in either the ΔiscSUAhscBAfdx or ΔiscSUAhscBAfdx, ΔsufABCDSE strains (Figure 6b).
Figure 6.
Activation of the ydfZ promoter by wild-type FNR in aerobically grown cultures as measured by β-galactosidase assays. A) Aerobic FNR activity in strains containing various deletions of the ΔiscSUAhscBAfdx operon. B) Aerobic FNR activity in wild-type, Δfnr, and ΔiscSUAhscBAfdx (Δisc), and ΔiscSUAhscBAfdx, ΔsufABCDSE (Δisc, Δsuf) strains. The ΔiscSUAhscBAfdx strain derivatives contained either the plasmid expressing the sufABCDSE operon from the arabinose-inducible PBAD promoter (pSuf), the vector alone, or no vector. All cultures were grown aerobically in M9 minimal glucose medium supplemented with nicotinic acid (12.5 μg ml−1). The plasmid-containing strains were also grown in the presence of ampicillin (final concentration of 50 μg ml−1) and, where indicated, 0.2% arabinose.
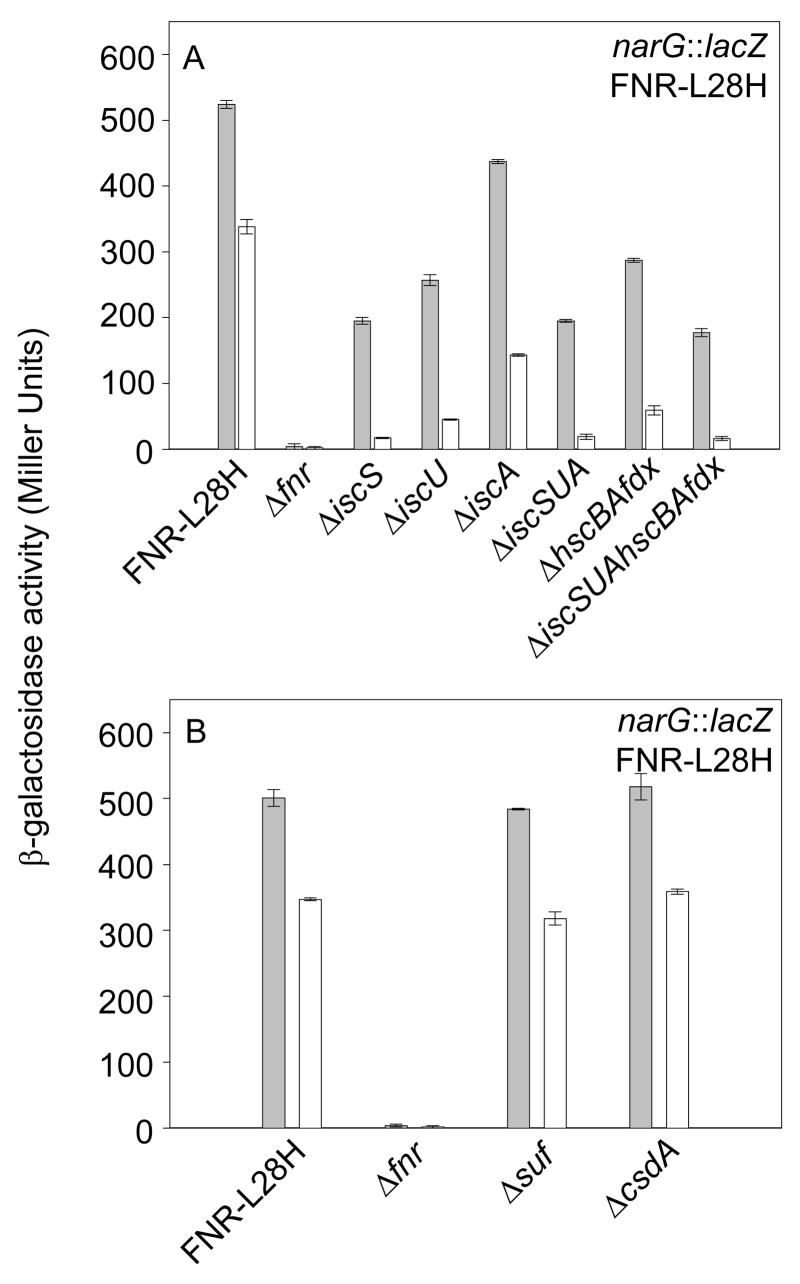
The second approach utilized a mutant of FNR containing a Leu to His substitution at position 28 (FNR-L28H) that renders the [4Fe-4S] cluster relatively resistant to destruction by O2.35 Furthermore, similar levels of [4Fe-4S]-FNR-L28H accumulate in aerobically and anaerobically grown cells35; 45, thus providing a unique system to compare Fe-S biogenesis requirements under aerobic and anaerobic conditions. Similar to that observed for narG expression by wild-type FNR (Figure 1), FNR-L28H function was reduced ~2–3 fold under anaerobic growth conditions in the various isc deletion strains except the ΔiscA strain (Figure 7a). However, under aerobic growth conditions, FNR-L28H activity was decreased more than 20-fold in strains lacking the entire Isc pathway or only IscS, 10-fold for the strains lacking IscU or HscB/HscA/Fdx and ~3-fold in the ΔiscA strain. Similar to that observed for wild-type FNR, neither the Suf pathway nor the CsdA protein were required for Fe-S cluster assembly of FNR-L28H under either anaerobic or aerobic growth conditions (Figure 7b).
Figure 7.
Activity of the mutant FNR-L28H, which contains an O2-stable [4Fe-4S] cluster, in anaerobic (gray bars) and aerobic (white bars) cultures grown in M9 minimal glucose medium containing a final concentration of 20 mM KNO3. All strains lacked the wild-type chromosomal copy of fnr but expressed FNR-L28H in single copy from the chromosome except Δfnr. A) Activation of PnarG by FNR-L28H in strains containing various deletions of the iscSUAhscBAfdx operon. Cultures also contained nicotinic acid (12.5 μg ml−1) in the medium. B) Activation of PnarG by FNR-L28H in strains lacking the sufABCDSE operon (Δsuf) or csdA.
Taken together, these results indicate that only the Isc pathway functions in FNR Fe-S cluster synthesis under aerobic growth conditions. Furthermore, even when the Suf machinery was overexpressed under aerobic conditions to the same levels as under anaerobic conditions, it could not functionally substitute for the Isc machinery suggesting that the Suf machinery shows a difference in substrate selectivity or activity under aerobic conditions.
The rate of Fe-S cluster acquisition by FNR-L28H is similar between aerobic and anaerobic conditions
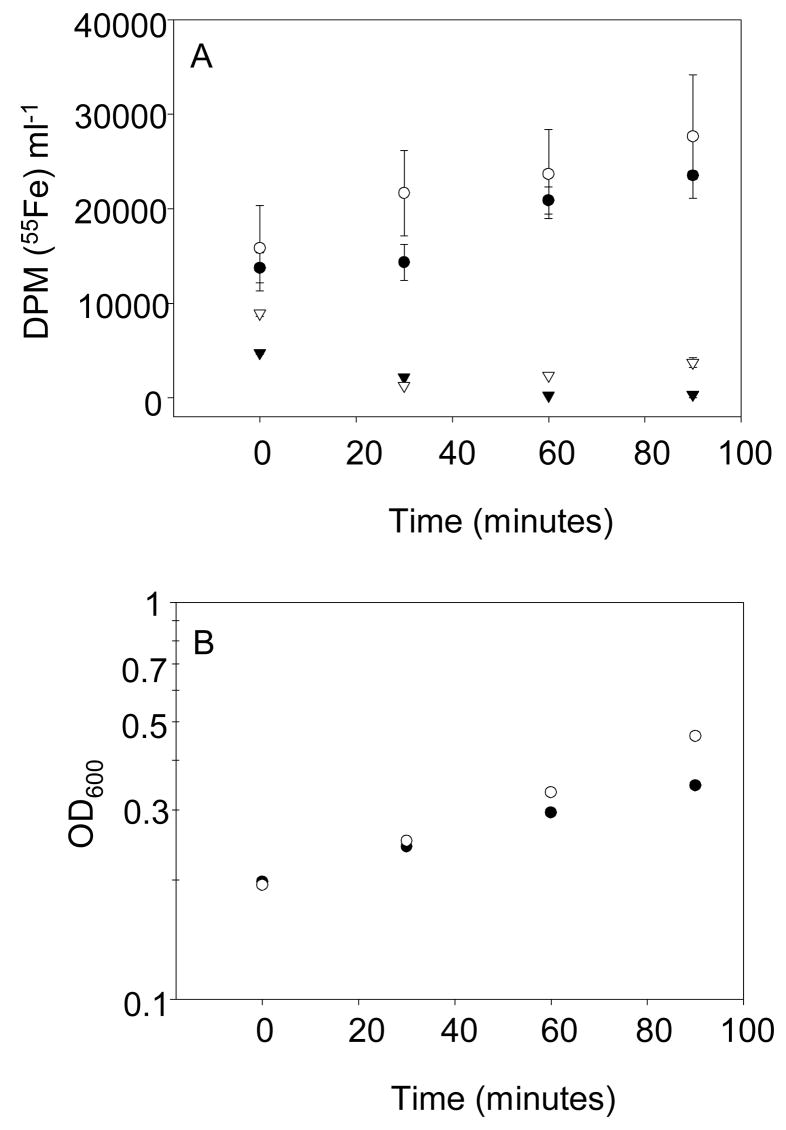
Previous studies indicated that expression of both the Isc and Suf pathways is decreased under anaerobic growth conditions28, thus raising the question as to whether a reduction in the levels of Fe-S cluster biogenesis proteins under anaerobic conditions is rate-limiting for FNR Fe-S cluster synthesis in vivo. To address this issue, we developed an in vivo assay to directly measure the rate of 55Fe incorporation into plasmid-derived FNR-L28H. FNR-L28H is not targeted for protein degradation46 and so the rate of 55Fe incorporation should reflect the rate of [4Fe-4S] acquisition by newly synthesized apo-FNR-L28H. We found that 55Fe was incorporated into immunoprecipitated FNR-L28H in a linear fashion from steady state labelled cultures grown under either aerobic or anaerobic growth conditions (Figure 8). Furthermore, the rate of 55Fe incorporation into FNR-L28H from equal volumes of cell culture was similar for aerobic and anaerobic conditions (125 ± 17 and 120 ± 26 DPM 55Fe ml−1 min−1 or 8.2 ± 1.1 × 109 and 7.9 ± 1.1 × 109 [4Fe-4S] clusters ml−1 min−1, respectively). Since Western blot analysis revealed that there is less than a 2-fold difference in total FNR-L28H protein levels in aerobically and anaerobically grown cells (data not shown), these data suggest that the rate of Fe-S cluster acquisition by FNR-L28H was similar under both conditions. The incorporation of 55Fe was specific to the cluster-containing form of FNR, since little 55Fe incorporation was detected when the experiment was performed with a strain expressing FNR-L28H-C122S (Figure 8), a mutant that is unable to ligate a Fe-S cluster.47
Figure 8.
A) In vivo incorporation of 55Fe into [4Fe-4S] cluster-containing FNR-L28H (circles) and clusterless FNR-L28H-C122S (triangles). pACYC184-derived FNR-L28H and FNR-L28H-C122S were immunoprecipitated from 1 ml aliquots of aerobically (open symbols) or anaerobically (closed symbols) grown cultures. The total DPM per ml for 55Fe was calculated as described in the Methods. The error bars represent the standard errors of triplicate samples. B) Growth measured by the optical density at 600 nm, of the aerobically (open circles) or anaerobically (closed circles) 55Fe-labelled cultures expressing FNR-L28H.
The plasmid-derived levels of FNR-L28H, which were ~6–7-fold higher than chromosomally produced wild-type FNR levels31, did not perturb normal Fe-S metabolism, as measured by changes in IscS protein levels, expressed from the isc operon. Western blot analysis showed no difference in IscS protein levels between the parent and plasmid containing strain and IscS levels were still oxygen regulated (data not shown), consistent with the reported ~ 7-fold increase in expression of the isc operon under aerobic growth conditions.28 Taken together, these data indicate that the differences in the levels of the Isc machinery under aerobic and anaerobic conditions do not affect the rate of Fe-S cluster acquisition by FNR-L28H and supports the notion that the greater role of the Isc pathway for FNR Fe-S biogenesis under aerobic conditions likely reflects changes in the function of the Suf pathway.
Reactivation of apo-FNR to [4Fe-4S]-FNR requires the Isc pathway
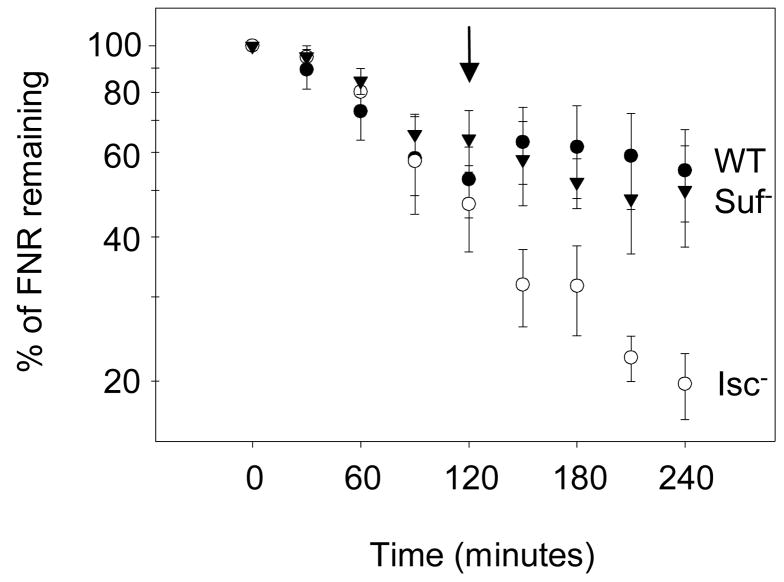
A recent study demonstrated that the population of apo-FNR present in aerobic cells is reactivated to [4Fe-4S]-FNR upon a shift to anaerobic growth conditions.46 To determine what Fe-S biogenesis machinery is necessary to mediate this process, we measured apo-FNR conversion to [4Fe-4S]-FNR in cells lacking either the Isc or Suf pathway. Reactivation of apo-FNR was determined by monitoring the FNR degradation rate via 35S-methionine pulse-chase assays since apo-FNR is targeted for degradation, whereas [4Fe-4S] cluster-containing FNR is resistant to proteolysis. In this assay, an aerobically grown culture is pulse-labelled with 35S-methionine for one minute, excess unlabelled methionine is added, and the amount of immunoprecipitated 35S-met-FNR is quantified over time. As observed previously for the wild-type strain46, FNR was degraded under aerobic growth conditions, but upon shifting the culture to anaerobic growth conditions at 120 minutes following addition of the methionine chase, degradation of FNR was impaired (Figure 9). As suggested previously46, these data indicate that the existing apo-FNR present in aerobic cells acquires a [4Fe-4S] cluster upon the switch to anaerobic growth conditions and thus escapes protein degradation. Similar results were observed in the Δsuf strain, suggesting that reactivation of apo-FNR to [4Fe-4S]-FNR does not require the Suf pathway. In contrast, the labelled population of apo-FNR in cells lacking the Isc proteins continued to decrease after switching the culture to anaerobic growth conditions. These data indicate that the Isc machinery is necessary for the reactivation of apo-FNR that previously lost its cluster. Thus the Isc pathway functions in de novo Fe-S biogenesis of FNR and in rebuilding Fe-S clusters in O2-inactivated FNR protein.
Figure 9.
Reactivation of apo-FNR to [4Fe-4S]-FNR upon switching aerobic cultures to anaerobic growth conditions. Methionine was added after 1 min of labelling of aerobically grown cells with 35S-methionine, followed by immunoprecipitation of FNR. At 120 minutes after adding the unlabeled methinonine, wild-type (filled circles), Isc− (open circles) or Suf−(filled triangles) cultures were switched to anaerobic growth conditions (indicated by the arrow). The amount of 35S-met-FNR remaining over time was calculated by normalizing to the amount of 35S-met-FNR present upon addition of the unlabeled methionine at time zero.
Discussion
This study examined the roles of the Isc and Suf pathways in FNR Fe-S cluster biogenesis. Our findings demonstrate that the Isc pathway is the major pathway for FNR Fe-S biogenesis under both aerobic and anaerobic conditions. However, the ability of the alternative Fe-S biogenesis pathway, Suf, to support FNR Fe-S biogenesis was inhibited by aerobic growth conditions even though Suf functions in the biogenesis of other Fe-S proteins under aerobic conditions.4; 48 These results open new areas of inquiry regarding how Fe-S proteins are targeted to either the Isc or Suf pathways during acquisition of Fe-S clusters and the role of O2 in regulating usage of these pathways.
Aerobic growth conditions increase the demand for Fe-S biogenesis
The demand for overall Fe-S biogenesis is expected to be greater under aerobic growth conditions because the general rate of Fe-S cluster turnover is increased under these conditions. For example, it has been reported that the rate constants with which Fe-S clusters of dehydratases react with superoxide and hydrogen peroxide are extremely high under aerobic conditions: 3 × 106 M−1 s−1 and 4 × 103 M−1 s−1, respectively.22; 23 Although E. coli synthesizes enzymes such as superoxide dismutase, catalase, and peroxidase to combat these oxidants, it is predicted that even micromolar concentrations of superoxide and hydrogen peroxide are sufficient to destroy the FeS clusters of some classes of Fe-S proteins.22; 23 Thus, under aerobic growth conditions, Fe-S clusters are most likely continually being destroyed by these oxidants, which would increase the demand for Fe-S biogenesis or repair. In this case, the Isc machinery and/or the Suf pathway would be expected to provide Fe-S clusters to a greater population of substrate proteins, providing a rationale for the increased expression of both the suf and isc operons under aerobic conditions as compared to anaerobic conditions.28
The major role of the Isc pathway for FNR Fe-S biogenesis under both aerobic and anaerobic conditions further supports the housekeeping function described for the Isc pathway
Previous studies have shown that the Isc pathway is required for the activity of many Fe-S proteins although a systematic investigation of all the pathway components has not been undertaken.24; 25; 26 Our studies show that all of the Isc components, with the exception of IscA, are necessary for FNR Fe-S cluster assembly under both aerobic and anaerobic conditions. In particular, we found that HscB and HscA are as important for Fe-S cluster assembly under anaerobic conditions as IscS and IscU, despite the fact that many obligate anaerobic microbes lack hscB- and hscA-like genes.3
In addition, we found that the rate of FNR Fe-S cluster biosynthesis is the same under both aerobic and anaerobic conditions suggesting that the Isc pathway is not limiting for FNR Fe-S cluster synthesis, which supports its role as the housekeeping Fe-S biogenesis pathway. Consistent with this notion, there was no further increase in FNR-L28H activity when the levels of the Isc proteins were increased (data not shown). Thus, our results show that, even for an abundant protein like FNR, the Isc machinery is present at sufficient levels to accommodate Fe-S biogenesis. The Isc machinery was also found to promote synthesis of Fe-S clusters in O2-inactivated FNR, suggesting that the determinants used for targeting FNR to the machinery are similar for both the apo-form of FNR generated from cluster loss and newly synthesized FNR.
Under anaerobic conditions, usage of Fe-S biogenesis pathways may be more promiscuous
While the Isc pathway is the major pathway for FNR Fe-S cluster synthesis under anaerobic conditions, analysis of strains lacking the Isc pathway under anaerobic conditions revealed an additional Fe-S cluster insertion activity(s). Our finding that the Suf pathway, when overexpressed, could functionally replace the Isc pathway under anaerobic conditions suggests that the Suf pathway may be responsible for FNR Fe-S cluster assembly in the absence of the Isc pathway. However, since strains lacking both pathways are not viable2; 3; 4, it is experimentally difficult to resolve this question. For similar technical reasons, we were unable to determine whether these minor Fe-S insertion activities are always active in anaerobic cells or are implemented only in the absence of the Isc pathway. Whatever the explanation, these minor activities are not sufficient under aerobic conditions since we found that only the Isc pathway was utilized for FNR Fe-S cluster assembly in the presence of O2. Given the large number of anaerobic respiratory enzymes that contain Fe-S clusters, it will be important to determine whether the utilization of the Fe-S biogenesis pathways is similar to FNR. As far as we are aware, a systematic study has not been carried out to investigate the magnitude of the defects in activity of other anaerobic Fe-S enzymes in strains lacking the Isc pathway.
Why does the Suf pathway not function in FNR Fe-S cluster synthesis under aerobic conditions?
We were surprised to find that under aerobic growth conditions, overexpression of the Suf pathway to the same level as under anaerobic conditions does not restore FNR Fe-S biogenesis. It seems less likely that the inability of the Suf pathway to restore Fe-S cluster synthesis is due to a decrease in the specific activity of the Suf pathway under aerobic conditions since this pathway has been shown previously to be active under these growth conditions as well as under conditions of oxidative stress.4; 48 Rather, it seems more likely that, under aerobic conditions, FNR no longer competes efficiently with other substrates to engage the Suf machinery (Figure 10). Aerobic and oxidative stress conditions are known to damage Fe-S clusters, thus increasing the demand for Fe-S cluster synthesis, and accordingly, more specific substrates to compete with FNR. Since FNR is an abundant protein at several thousand molecules per cell, our result suggests that other Fe-S protein substrates must interact with the Suf pathway with greater affinity than FNR, thereby limiting FNR cluster acquisition. Regardless of the specific mechanism, these results raise the question of how apoproteins are targeted to the Isc pathway versus the Suf pathway. Further experiments will be necessary to determine if substrate proteins contain specific targeting sequences that direct them to particular Fe-S biogenesis pathways and/or if there are other intrinsic differences in the activities of the Suf and Isc pathways. In addition, this study shows that growth under aerobic conditions is a sufficient perturbant to study differences in Fe-S biogenesis pathway function.
Figure 10.
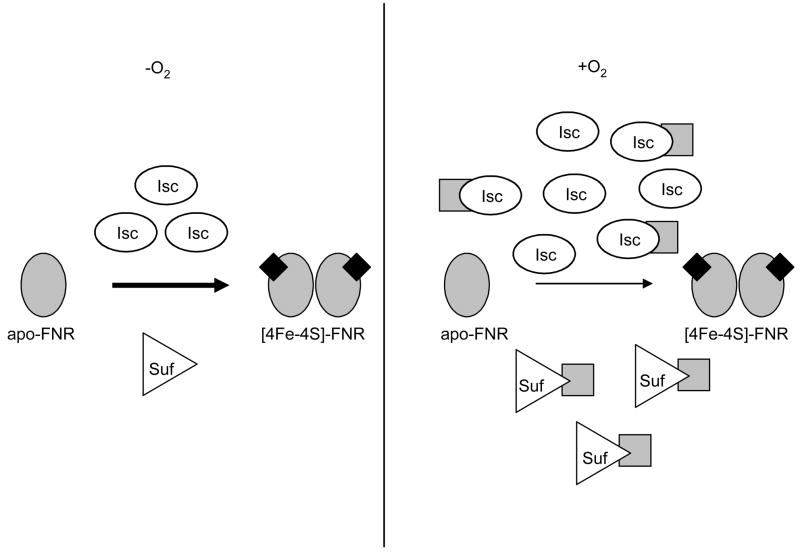
Model for FNR Fe-S cluster biogenesis requirements under aerobic and anaerobic conditions. Under both conditions, monomeric, apo-FNR acquires Fe-S clusters (represented as black diamonds) primarily from the Isc pathway. Under anaerobic conditions, Suf can provide up to 40% of Fe-S clusters for FNR when the Isc pathway is absent. Although expression of both the Isc pathway and the Suf pathway is increased under aerobic conditions compared to anaerobic conditions, Suf does not appear to provide Fe-S clusters to FNR in the absence of Isc. Furthermore, the demand for FNR Fe-S cluster biogenesis via the Isc pathway is greater under aerobic conditions compared to anaerobic conditions. This increased demand for Fe-S cluster biogenesis may be due to the higher rate Fe-S cluster turnover under aerobic conditions, and thus increased levels of apo-protein substrates (gray squares).
Differences in the roles of the IscA homologs between aerobic and anaerobic conditions
Although previous studies have demonstrated that IscA can serve as an Fe-S cluster scaffolding protein in vitro12, we found that IscA was not necessary for FNR Fe-S cluster assembly under either aerobic or anaerobic growth conditions. Rather, we found that SufA can functionally replace IscA under anaerobic growth conditions for FNR Fe-S cluster assembly. We assume that this is also true under aerobic conditions, but we could not test this genetically because the plating efficiency of a ΔiscA ΔsufA strain was reduced 100-fold under aerobic conditions and we were unable to obtain linear growth in liquid cultures to assay FNR activity. We were also surprised that strains lacking both SufA and IscA are viable under anaerobic conditions (albeit with a 2-fold slower growth rate on glucose minimal media; data not shown), since deletion of both the entire isc and suf operons is lethal under anaerobic conditions (data not shown). This suggests that either the function of IscA and SufA is not absolutely required for Fe-S cluster biogenesis under anaerobic conditions or that the third IscA homolog, ErpA, functions under these conditions.49 ErpA is essential for growth by either aerobic or anaerobic respiration but not fermentation, because it has a specialized role in maturation of Fe-S proteins required for synthesis of precursors of quinones, required for respiration. Since ErpA is not required for growth under the anaerobic fermentative conditions used in this study, and erpA is expressed under these conditions49, it is possible that ErpA could support both growth and partial activity of FNR in the strain lacking IscA and SufA. The more dramatic phenotype of strains lacking IscA and SufA under aerobic conditions is consistent with a recent report by Lu et al20 and underscores a critical role of all IscA homologs under aerobic growth conditions.
In summary, we propose that the specificity and/or activity of the Suf and Isc Fe-S biogenesis pathways may be optimized to respond to both the particular environmental conditions that can damage Fe-S clusters and to changes in the levels of specific substrates. Overall, such a strategy would allow cells to maintain the function of Fe-S proteins even when cells find themselves in a hostile environment. Future work will address whether the properties of the Isc and Suf pathways acquired by studying FNR as a model, extend to other Fe-S proteins.
Materials and Methods
Construction of strains
The strains and plasmids used in this study are listed in Table 1. An in frame ΔiscS deletion strain was constructed by replacing the coding region of iscS (codons 2-404) with a CmR resistance cassette flanked by FRT (FLP recognition target) sites from plasmid pKD32 in strain BW25993 / pKD46 as described.50 In frame ΔiscU (codons 2-128), ΔiscA (codons 2-107), ΔiscSUA (codon 2 of iscS through codon 107 of iscA), ΔhscBAfdx (codon 2 of hscB through codon 111 of fdx), ΔiscSUAhscBAfdx (codon 2 of iscS through codon 111 of fdx), ΔsufA (codons 2-122), ΔsufABCDSE (codon 2 of sufA through codon 138 of sufE), ΔnfuA (codons 2-191), ΔompT (codons 2-317), Δ lacY (codons 2-417), ΔiscR (codons 2-120)51 and ΔcsdA (codons 2-401) deletion strains were constructed in a similar manner; in some cases the coding regions were replaced with the KnR cassette (instead of CmR) flanked by FRT sites from plasmid pKD13.50 Transduction with P1 vir was used to move the CmR or KnR alleles to strains containing the dmsA (PK3292), ydfZ (PK8202), sufA (PK6879, PK6898), or narG (RZ7350, PK910) promoter-lacZ fusions to produce the corresponding strain derivatives listed in Table 1. In strains containing one or more isc gene deletions, it was confirmed by Western blot analysis that the CmR or KnR alleles did not decrease expression of downstream genes within the isc operon. In some cases, the CmR cassette was deleted by transforming strains with pCP20 encoding FLP recombinase50 and screening for Cm sensitivity. All gene deletions were confirmed by colony PCR.
Table 1.
E. coli strains and plasmids used in this work.
| Construct | Relevant genotype | Reference / Source |
|---|---|---|
| Strains
| ||
| MG1655 | F−, λ−, rph-1 | This laboratory |
| PK4811 | MG1655 but ΔfnrΩSpr / Smr | This laboratory |
| PK8431 | PK4811 but ΔompT::kan | This study |
| PK8436 | Same as PK8431 but with | This study |
| pPK434 | ||
| PK8499 | Same as PK8431 but with pPK437 | This study |
| WO19 | ΔsufABCDSE19::kan | Storz/Imlay laboratories |
| WO231 | MG1655 ΔsufD | F.W. Outten |
| JOEY18 | MG1655 ΔytfE::kan | S. Spiro |
| RZ4500 | MG1655 but lacZΔ145 | 58 |
| PK3292 | RZ4500 but λPC25 | 24 |
| PK3293 | PK3292 but ΔfnrΩSpr / Smr | 59 |
| PK7692 | PK3292 but ΔiscS::cat | This study |
| PK7694 | PK3292 but ΔiscU::kan | This study |
| PK7696 | PK3292 but ΔiscA:: kan | This study |
| PK7680 | PK3292 but ΔiscSUA::cat | This study |
| PK7690 | PK3292 but ΔhscBAfdx::cat | This study |
| PK7682 | PK3292 but Δ iscSUAhscBAfdx::cat | This study |
| PK6463 | PK3292 but ΔcsdA::cat | This laboratory |
| PK6465 | PK3292 but ΔsufABCDSE19:: kan | This study |
| RZ7350 | RZ4500 but narG234::MudI1734 | 60 |
| PK8245 | RZ7350 but ΔiscS::cat | This study |
| PK8246 | RZ7350 but ΔiscU::cat | This study |
| PK8247 | RZ7350 but ΔiscA::cat | This study |
| PK8248 | RZ7350 but ΔiscSUA::cat | This study |
| PK8249 | RZ7350 but ΔhscBAfdx::cat | This study |
| PK8250 | RZ7350 but ΔiscSUAhscBAfdx::cat | This study |
| PK8474 | RZ7350 but ΔsufABCDSE::cat | This study |
| PK8479 | RZ7350 but ΔcsdA::cat | This study |
| RZ8480 | RZ7350 but ΔfnrΩSpr / Smr | 39 |
| PK910 | RZ8480 but λfnr-L28H | 47 |
| PK8228 | PK910 but ΔiscS::cat | This study |
| PK8229 | PK910 but ΔiscU::cat | This study |
| PK8230 | PK910 but ΔiscA::cat | This study |
| PK8231 | PK910 but ΔiscSUA::cat | This study |
| PK8232 | PK910 but ΔhscBAfdx::cat | This study |
| PK8233 | PK910 but ΔiscSUAhscBAfdx::cat | This study |
| PK8478 | PK910 but ΔsufABCDSE::cat | This study |
| PK8482 | PK910 but ΔcsdA::cat | This study |
| PK8202 | MG1655 (−233 to +13 relative to translational start site) PydfZ::lacZ | This study |
| PK8222 | PK8202 but ΔiscS::cat | This study |
| PK8223 | PK8202 but ΔiscU::cat | This study |
| PK8224 | PK8202 but ΔiscA::cat | This study |
| PK8225 | PK8202 but ΔiscSUA::cat | This study |
| PK8226 | PK8202 but ΔhscBAfdx::cat | This study |
| PK8227 | PK8202 but iscSUAhscBAfdx::cat | This study |
| PK8428 | Same as PK8227 but with pGS0164 | This study |
| PK8445 | Same as PK8227 but with pBADmychisC | This study |
| PK8405 | PK8202 but ΔnfuA | This study |
| PK8407 | PK8202 but ΔytfE | This study |
| PK8476 | PK8202 but ΔsufABCDSE::cat | This study |
| PK8605 | PK8476 but CmS | This study |
| PK8607 | PK8605 but with pGS0164 | This study |
| PK8611 | PK8607 but Δ iscSUAhscBAfdx::cat | This study |
| PK8481 | PK8202 but ΔcsdA::cat | This study |
| PK8835 | PK8202 but ΔsufA | This laboratory |
| PK8836 | PK8202 but ΔsufA ΔnfuA | This laboratory |
| PK8865 | PK8202 but ΔsufA ΔiscA | This laboratory |
| PK8544 | PK8202 but ΔiscA ΔnfuA::cat | This laboratory |
| PK8203 | PK8202 but ΔfnrΩSpr / Smr | This study |
| PK8457 | PK8203 but ΔompT Δlacy | This study |
| PK8601 | PK8457 but with pPK434 | This study |
| PK8234 | Same as PK8203 but with pRZ7411 | This study |
| PK8235 | Same as PK8203 but with pACYC184 | This study |
| PK8240 | Same as PK8203 but with pPK6928 | This study |
| PK8241 | Same as PK8203 but with pPK6929 | This study |
| PK8259 | PK8203 but ΔiscSUAhscBAfdx | This study |
| PK8270 | Same as PK8259 but with pRZ7411 | This study |
| PK8271 | Same as PK8259 but with pACYC184 | This study |
| PK8272 | Same as PK8259 but with pPK6928 | This study |
| PK8273 | Same as PK8259 but with pPK6929 | This study |
| PK6879 | MG1655 (−200 to +40 relative to transcriptional start site) PsufA::lacZ | 28 |
| PK8452 | PK6879 but ΔiscSUAhscBAfdx::cat | This study |
| PK6898 | PK6879 but ΔfnrΩSpr / Smr | This laboratory |
| PK8453 | PK6898 but ΔiscSUAhscBAfdx::cat | This study |
| BW25993 | lacIq Δ lacZWJ16 hsdR514ΔaraBADAH33Δ rhaBADLD78 | 50 |
| Plasmids | ||
| pBAD/Myc-hisC | Apr | Invitrogen |
| pGS0164 | pBAD/Myc-hisC but with sufABCDSE in NcoI and EcoRI sites | 19 |
| pRZ7411 | Cmr, HindIII-BamHI of fnr, -521 to +1115 of fnr in pACYC184 | 39 |
| pACYC184 | Cmr | 61 |
| pPK6928 | Same as pRZ7411 but fnr-C23A | 46 |
| pPK6929 | Same as pRZ7411 but fnr-C122A | 46 |
| pCP20 | Apr | 50 |
| pKD46 | Phage λ gam-bet-exo genes under ParaB control | B. L. Wanner |
| pKD32 | FRT-cat-FRT | B. L. Wanner |
| pKD13 | FRT-kan-FRT | 50 |
| pPK434 | HindIII-BamHI of fnr-L28H (−521 to +1115 of fnr-LH28) in pACYC184 | This laboratory |
| pPK437 | Same as pPK434 but with fnr-L28H-C122S | This laboratory |
β-galactosidase assays
Levels of β-galactosidase were measured in strains containing dmsA, ydfZ, narG, and sufA promoter-lacZ fusions as described.52; dmsA and narG encode for a subunit of dimethyl sulfoxide reductase and nitrate reductase, respectively53; 54; ydfZ encodes for a putative selenium-binding protein.55 Cultures were grown aerobically or anaerobically to an OD600 of ~0.2 in M9 minimal medium with 0.2% (w/v) glucose, 10 μM ferric ammonium citrate, 2 μg ml−1 thiamine, and 0.2 μM ammonium molybdate as previously described.56 Final concentrations of nicotinic acid (12.5 μg ml−1), KNO3 (20 mM), chloramphenicol (20 μg ml−1), or ampicllin (50 μg ml−1) were added to the medium where indicated. To terminate cell growth and any further protein synthesis, either chloramphenicol (final concentration 20 μg ml−1) or tetracycline (final concentration 10 μg ml−1) was added, and cells were placed on ice until assayed for β-galactosidase activity.52 β-galactosidase assays were repeated at least three times and error bars represent the standard errors of triplicate samples. The β-galactosidase activity was normalized to account for the difference in cell number per ml of culture as determined via viable plating assays.31 At an OD600 of 0.4, aerobic and anaerobic cultures contained (2.6 ± 0.2) × 108 and (4 ± 0.3) × 108 cells ml−1, respectively. Therefore, β-galactosidase activity was normalized by multiplying the aerobic values by a factor of 1.5.
Western blots
Western blots using α-FNR antibodies or α-IscS antibodies were performed as previously described.31 Western blots using α-SufD antibodies were carried out in a similar manner except that the α-SufD antibodies were precleared using a SufD− cell lysate57 and HRP-conjugated goat anti-rabbit secondary antibodies (Santa Cruz Biotechnology) were used. After treating the blots with Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer), chemiluminescence was quantified using a UVP Laboratory Products Epi Chem II Darkroom and Molecular Dynamics Image Quant software.
35S-methionine pulse-chase assays followed by immunoprecipitation of FNR
35S-methinonine pulse-chase assays followed by immunoprecipitation of FNR were carried out as previously described.46
55Fe labelling followed by immunoprecipitation of FNR-L28H under non-denaturing conditions
An overnight culture of a strain expressing FNR-L28H or FNR-L28H-C122S from a plasmid (PK8436 or PK8499, respectively) was grown aerobically in M9 minimal medium containing final concentrations of 0.2% (w/v) glucose, 2 μM ferric ammonium citrate, 2 μg ml−1 thiamine, 0.2 μM ammonium molybdate, and 20 μg ml−1 chloramphenicol. The overnight culture was diluted 1:100 into fresh media, which in addition to containing 2 μM ferric ammonium citrate, also contained 2.6 μCi of [55Fe]ferric ammonium citrate (specific activity 93.76 mCi/mg) per milliliter of culture; the final Fe concentration was 2.5 μM. ([55Fe]ferric ammonium citrate was obtained by treating 5 mCi of [55Fe]ferric chloride in 0.5 M HCl (GE Healthcare) with 100 μl of 0.3 M potassium citrate and 20 μl of 14.5 M ammonium hydroxide.) The culture was sparged with either an aerobic or anaerobic gas mixture as previously described.56 At an OD600 of ~0.2, aliquots (1 ml) of the culture were collected at various times by centrifugation (2 min at 13,600 ×g) in Eppendorf microfuge tubes. The cell pellets were washed once in 1 ml of fresh media containing 2 μM of unlabelled ferric ammonium citrate, centrifuged (2 min at 13,600 ×g), and frozen on a dry ice/ethanol bath. For immunoprecipitation of 55Fe radiolabelled FNR-L28H (or FNR-L28H-C122S), cell pellets were thawed and lysed using the EasyLyse Bacterial Protein Extraction Solution (Epicentre) by resuspending each pellet in 200 μl of a mixture containing 5 mM Tris-HCl [pH 7.5], 0.5% Triton X-100, 2 mM MgCl2 plus 0.2 μl of Epicentre Enzyme mix. After incubating the samples at room temperature for 5 min, the samples were centrifuged (2 min at 13,600 × g) and the supernatant was transferred to 200 μl of a solution containing pre-cleared α-FNR antibodies57 in 50 mM Tris-HCl [pH 7.9], 10% glycerol, 500 mM NaCl, 1% Triton X-100 that were pretreated with 10 μl of UltraLink Protein A beads (Pierce). (The antibodies were pretreated with Protein A beads by incubating for one hour at room temperature with gentle rocking.) After gently rocking the samples at room temperature for one hour, the beads were pelleted, washed twice with 50 mM Tris-HCl [pH 7.9], 10% glycerol, 500 mM NaCl, 1% Triton X-100, and resuspended in 50 μl of double-distilled water. The samples were transferred as a slurry of beads to scintillation vials containing 3 mls of BioSafe II scintillation cocktail (Research Products International) and counted for 1 min on a Packard Tri-Carb 2100TR Liquid Scintillation Analyzer. Counting regions A and B were set at 0–5 keV and 5–180 keV, respectively. Using the previously determined counting efficiencies for 55Fe in the two counting regions (9.5% in region A, 1.2% in region B), the total DPM for 55Fe was determined. The number of [4Fe-4S] clusters was determined from the DPM by dividing by the specific activity of added 55Fe [1.905 × 10−8 dpm 55Fe molecule−1], multiplying by 5 for the dilution with unlabelled Fe and dividing by 4 for the number of Fe per cluster.
Acknowledgments
We would like to thank Dr. Stephen Spiro for providing E. coli strain JOEY18 and Dr. Roland Lill for advice on Fe55 labelling and immunoprecipitation. We also acknowledge Dr. Gary Roberts, Dr. Hwan Youn, and members of the Kiley laboratory for critical reading of this manuscript. This work was supported by NIH grant GM045844 (to P.J.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–81. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–3. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- 3.Tokumoto U, Kitamura S, Fukuyama K, Takahashi Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J Biochem (Tokyo) 2004;136:199–209. doi: 10.1093/jb/mvh104. [DOI] [PubMed] [Google Scholar]
- 4.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–72. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 5.Flint DH. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J Biol Chem. 1996;271:16068–74. [PubMed] [Google Scholar]
- 6.Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–4. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- 7.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. Transfer of sulfur from IscS to IscU during Fe/S Cluster Assembly. J Biol Chem. 2001;276:44521–44526. doi: 10.1074/jbc.M106907200. [DOI] [PubMed] [Google Scholar]
- 8.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:7790–5. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK. IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry. 2000;39:7856–62. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- 10.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J Biol Chem. 2002;277:27353–9. doi: 10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 11.Vickery LE, Silberg JJ, Ta DT. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Protein Sci. 1997;6:1047–56. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry. 2001;40:14069–80. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- 13.Ding H, Clark RJ, Ding B. IscA mediates iron delivery for assembly of iron-sulfur clusters in IscU under the limited accessible free iron conditions. J Biol Chem. 2004;279:37499–504. doi: 10.1074/jbc.M404533200. [DOI] [PubMed] [Google Scholar]
- 14.Mihara H, Maeda M, Fujii T, Kurihara T, Hata Y, Esaki N. A nifS-like gene, csdB, encodes an Escherichia coli counterpart of mammalian selenocysteine lyase. Gene cloning, purification, characterization and preliminary x-ray crystallographic studies. J Biol Chem. 1999;274:14768–72. doi: 10.1074/jbc.274.21.14768. [DOI] [PubMed] [Google Scholar]
- 15.Ollagnier-de Choudens S, Nachin L, Sanakis Y, Loiseau L, Barras F, Fontecave M. SufA from Erwinia chrysanthemi. Characterization of a scaffold protein required for iron-sulfur cluster assembly. J Biol Chem. 2003;278:17993–8001. doi: 10.1074/jbc.M300285200. [DOI] [PubMed] [Google Scholar]
- 16.Sendra M, Ollagnier-de Choudens S, Lascoux D, Sanakis Y, Fontecave M. The SUF iron-sulfur cluster biosynthetic machinery : Sulfur transfer from the SUFS-SUFE complex to SUFA. FEBS Lett. 2007;581:1362–1368. doi: 10.1016/j.febslet.2007.02.058. [DOI] [PubMed] [Google Scholar]
- 17.Rangachari K, Davis CT, Eccleston JF, Hirst EM, Saldanha JW, Strath M, Wilson RJ. SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett. 2002;514:225–8. doi: 10.1016/s0014-5793(02)02369-4. [DOI] [PubMed] [Google Scholar]
- 18.Nachin L, Loiseau L, Expert D, Barras F. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. Embo J. 2003;22:427–37. doi: 10.1093/emboj/cdg061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Outten FW, Wood MJ, Munoz FM, Storz G. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem. 2003;278:45713–9. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Yang J, Tan G, Ding H. Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem J. 2008;409:535–43. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- 21.Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60:12–23. doi: 10.1007/s00253-002-1107-4. [DOI] [PubMed] [Google Scholar]
- 22.Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2006;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–76. [PubMed] [Google Scholar]
- 24.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:9009–14. doi: 10.1073/pnas.160261497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tokumoto U, Takahashi Y. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J Biochem (Tokyo) 2001;130:63–71. doi: 10.1093/oxfordjournals.jbchem.a002963. [DOI] [PubMed] [Google Scholar]
- 26.Djaman O, Outten FW, Imlay JA. Repair of oxidized iron-sulfur clusters in Escherichia coli. J Biol Chem. 2004;279:44590–9. doi: 10.1074/jbc.M406487200. [DOI] [PubMed] [Google Scholar]
- 27.Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, Storz G. DNA Microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol. 2001;183:4562–4570. doi: 10.1128/JB.183.15.4562-4570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol Microbiol. 2006;60:1058–75. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 29.Kiley PJ, Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev. 1999;22:341–52. doi: 10.1111/j.1574-6976.1998.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 30.Jordan PA, Thompson AJ, Ralph ET, Guest JR, Green J. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett. 1997;416:349–352. doi: 10.1016/s0014-5793(97)01219-2. [DOI] [PubMed] [Google Scholar]
- 31.Sutton VR, Stubna A, Patschkowski T, Münck E, Beinert H, Kiley PJ. Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli. Biochemistry. 2004;43:791–8. doi: 10.1021/bi0357053. [DOI] [PubMed] [Google Scholar]
- 32.Lazazzera BA, Beinert H, Khoroshilova N, Kennedy MC, Kiley PJ. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem. 1996;271:2762–2768. doi: 10.1074/jbc.271.5.2762. [DOI] [PubMed] [Google Scholar]
- 33.Green J, Bennett B, Jordan P, Ralph ET, Thomson AJ, Guest JR. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem J. 1996;316:887–892. doi: 10.1042/bj3160887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoroshilova N, Beinert H, Kiley PJ. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc Natl Acad Sci USA. 1997;92:2499–2503. doi: 10.1073/pnas.92.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bates DM, Popescu CV, Khoroshilova N, Vogt K, Beinert H, Münck E, Kiley PJ. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J Biol Chem. 2000;275:6234–40. doi: 10.1074/jbc.275.9.6234. [DOI] [PubMed] [Google Scholar]
- 36.Spiro S, Guest JR. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol Microbiol. 1988;2:701–7. doi: 10.1111/j.1365-2958.1988.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 37.Melville SB, Gunsalus RP. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–6. [PubMed] [Google Scholar]
- 38.Sharrocks AD, Green J, Guest JR. In vivo and in vitro mutants of FNR the anaerobic transcriptional regulator of E. coli. FEBS Lett. 1990;270:119–22. doi: 10.1016/0014-5793(90)81248-m. [DOI] [PubMed] [Google Scholar]
- 39.Lazazzera BA, Bates DM, Kiley PJ. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 1993;7:1993–2005. doi: 10.1101/gad.7.10.1993. [DOI] [PubMed] [Google Scholar]
- 40.Mihara H, Kurihara T, Yoshimura T, Soda K, Esaki N. Cysteine sulfinate desulfinase, a NIFS-like protein of Escherichia coli with selenocysteine lyase and cysteine desulfurase activities. Gene cloning, purification, and characterization of a novel pyridoxal enzyme. J Biol Chem. 1997;272:22417–24. doi: 10.1074/jbc.272.36.22417. [DOI] [PubMed] [Google Scholar]
- 41.Angelini S, Gerez C, Ollagnier-de Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J Biol Chem. 2008;283:14084–91. doi: 10.1074/jbc.M709405200. [DOI] [PubMed] [Google Scholar]
- 42.Dombrecht B, Tesfay MZ, Verreth C, Heusdens C, Napoles MC, Vanderleyden J, Michiels J. The Rhizobium etli gene iscN is highly expressed in bacteroids and required for nitrogen fixation. Mol Genet Genomics. 2002;267:820–8. doi: 10.1007/s00438-002-0715-0. [DOI] [PubMed] [Google Scholar]
- 43.Justino MC, Almeida CC, Goncalves VL, Teixeira M, Saraiva LM. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol Lett. 2006;257:278–84. doi: 10.1111/j.1574-6968.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- 44.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol. 2005;187:1135–60. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popescu CV, Bates DM, Beinert H, Münck E, Kiley PJ. Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc Natl Acad Sci USA. 1998;95:13431–13435. doi: 10.1073/pnas.95.23.13431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mettert EL, Kiley PJ. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J Mol Biol. 2005;354:220–32. doi: 10.1016/j.jmb.2005.09.066. [DOI] [PubMed] [Google Scholar]
- 47.Bates DM, Lazazzera BA, Kiley PJ. Characterization of FNR* mutant proteins indicates two distinct mechanisms for altering oxygen regulation of the Escherichia coli transcription factor FNR. J Bacteriol. 1995;177:3972–8. doi: 10.1128/jb.177.14.3972-3978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patzer SI, Hantke K. SufS is a NifS-like protein, and SufD is necessary for stability of the [2Fe-2S] FhuF protein in Escherichia coli. J Bacteriol. 1999;181:307–9. doi: 10.1128/jb.181.10.3307-3309.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de Mattos J, Fontecave M, Barras F. ErpA, an iron sulfur (Fe S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:13626–31. doi: 10.1073/pnas.0705829104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz CJ, Giel JL, Patschkowski T, Luther C, Ruzicka FJ, Beinert H, Kiley PJ. IscR, an Fe-S cluster-containing transcription factor, represses expression of Escherichia coli genes encoding Fe-S cluster assembly proteins. Proc Natl Acad Sci USA. 2001;98:14895–900. doi: 10.1073/pnas.251550898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 53.Cotter PA, Gunsalus RP. Oxygen, nitrate, and molybdenum regulation of dmsABC gene expression in Escherichia coli. J Bacteriol. 1989;171:3817–3823. doi: 10.1128/jb.171.7.3817-3823.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart V, Rabin RS. Two-Component Signal Transduction. American Society for Microbiology; Washington, D. C.: 1995. [Google Scholar]
- 55.Lacourciere GM, Levine RL, Stadtman TC. Direct detection of potential selenium delivery proteins by using an Escherichia coli strain unable to incorporate selenium from selenite into proteins. Proc Natl Acad Sci USA. 2002;99:9150–9153. doi: 10.1073/pnas.142291199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sutton VR, Kiley PJ. Techniques for studying the oxygen-sensitive transcription factor FNR from Escherichia coli. Methods Enzymol. 2003;370:300–12. doi: 10.1016/S0076-6879(03)70027-5. [DOI] [PubMed] [Google Scholar]
- 57.Harlow E, Lane D. Antibodies, A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- 58.Choe M, Reznikoff WS. Anaerobically expressed Escherichia coli genes identified by operon fusion techniques. J Bacteriol. 1991;173:6139–46. doi: 10.1128/jb.173.19.6139-6146.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamberg KE, Kiley PJ. FNR-dependent activation of the class II dmsA and narG promoters of Escherichia coli requires FNR-activating regions 1 and 3. Mol Microbiol. 2000;38:817–27. doi: 10.1046/j.1365-2958.2000.02172.x. [DOI] [PubMed] [Google Scholar]
- 60.Kiley PJ, Reznikoff WS. Fnr mutants that activate gene expression in the presence of oxygen. J Bacteriol. 1991;173:16–22. doi: 10.1128/jb.173.1.16-22.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–56. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]