Abstract
Mycobacterial UvrD2 is a DNA-dependent ATPase with 3′ to 5′ helicase activity. UvrD2 is an atypical helicase, insofar as its N-terminal ATPase domain resembles the superfamily I helicases UvrD/PcrA, yet it has a C-terminal HRDC domain, which is a feature of RecQ-type superfamily II helicases. The ATPase and HRDC domains are connected by a CxxC-(14)-CxxC tetracysteine module that defines a new clade of UvrD2-like bacterial helicases found only in Actinomycetales. By characterizing truncated versions of Mycobacterium smegmatis UvrD2, we show that whereas the HRDC domain is not required for ATPase or helicase activities in vitro, deletion of the tetracysteine module abolishes duplex unwinding while preserving ATP hydrolysis. Replacing each of the CxxC motifs with a double-alanine variant AxxA had no effect on duplex unwinding, signifying that the domain module, not the cysteines, is crucial for function. The helicase activity of a truncated UvrD2 lacking the tetracysteine and HRDC domains was restored by the DNA-binding protein Ku, a component of the mycobacterial NHEJ system and a cofactor for DNA unwinding by the paralogous mycobacterial helicase UvrD1. Our findings indicate that coupling of ATP hydrolysis to duplex unwinding can be achieved by protein domains acting in cis or trans. Attempts to disrupt the M. smegmatis uvrD2 gene were unsuccessful unless a second copy of uvrD2 was present elsewhere in the chromosome, indicating that UvrD2 is essential for growth of M. smegmatis.
Bacterial DNA helicases are nucleic acid-dependent NTPases that play essential roles in DNA replication, recombination, and repair. Escherichia coli UvrD and Bacillus PcrA exemplify a distinctive clade of NTPase/helicases, within helicase superfamily I, defined by the following properties: (i) specificity for hydrolysis of ATP or dATP; (ii) 3′ to 5′ directionality of DNA translocation and unwinding; (iii) a conserved ATPase1 motor domain composed of four structural subdomains (I, Ia, II, and IIa); and (iv) participation in classical bacterial mismatch repair, nucleotide excision repair, and homologous recombination pathways in vivo (1-4). Recent studies have revealed that mycobacteria and other bacterial genera have an additional mechanism to repair double-strand breaks via nonhomologous end joining (NHEJ) (5-12). Mycobacterial NHEJ in vivo depends on the DNA end-binding protein Ku and dedicated ATP-dependent DNA ligases.
A connection between bacterial NHEJ and DNA helicases was suggested by the recent identification of mycobacterial UvrD1 as a binding partner for Mycobacterium tuberculosis Ku in a two-hybrid screen (13). UvrD1 is structurally homologous to PcrA and UvrD. Yet, despite having a vigorous DNA-dependent ATPase activity, UvrD1 is a feeble helicase (13, 14). The distinctive property of UvrD1 is that it requires Ku in order to catalyze efficient ATP-dependent unwinding of 3′-tailed duplex DNAs (13). UvrD1, Ku, and DNA form a stable ternary complex, suggesting that Ku might serve as a processivity factor for unwinding by UvrD1. Ablation of UvrD1 sensitizes Mycobacterium smegmatis to killing by ultraviolet and ionizing radiation, thereby attesting to its function in DNA repair (13). Mounting evidence implicates DNA repair pathways in mycobacterial pathogenesis and the accrual of mutations that confer antibiotic resistance (15-17). Indeed, Curti et al. (14) cite unpublished findings that deletion of the M. tuberculosis uvrD1 gene reduces bacterial persistence in a murine model of tuberculosis infection.
Previously, we identified a second mycobacterial UvrD/PcrA paralogue, UvrD2, in the proteomes of M. tuberculosis, M. smegmatis, Mycobacterium avium, and Mycobacterium leprae (13). The conservation of primary structure between the M. tuberculosis UvrD1 and UvrD2 paralogues embraces the N-terminal 698 aa of UvrD1 and the N-terminal 599 aa of UvrD2, wherein there are 240 positions of side chain identity. However, the C-terminal segments of UvrD1 and UvrD2 have no apparent structural similarity. Thus, UvrD1 and UvrD2 differ principally within the C-terminal domain that, in the case of UvrD1, interacts with Ku (13). Purification and characterization of M. smegmatis UvrD2 (MsmUvrD2) revealed its similarity to UvrD1 with respect to DNA-dependent ATPase activity and a monomeric quaternary structure. The distinctive feature of UvrD2 versus UvrD1 is that UvrD2 is an efficient 3′ to 5′ helicase per se and does not rely on Ku for duplex unwinding (13).
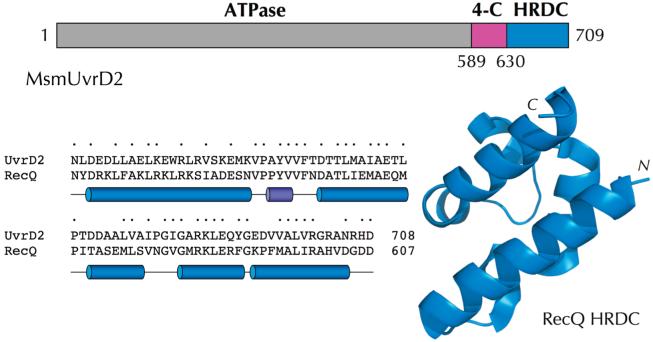
Here we focus on the unique C-terminal domain architecture of mycobacterial UvrD2 and its role in facilitating the autonomous helicase activity of UvrD2. The N-terminal motor domain of MsmUvrD2 is a typical UvrD/PcrA-type ATPase (depicted in gray in Figure 1). Fused to the distal end of the ATPase domain are two distinctive modules not usually present in UvrD-like helicases: (i) a tetracysteine domain (aa 589-630; a putative Zn finger) that is a signature feature of a UvrD2 subfamily and (ii) an HRDC domain (helicase and RNase D C-terminal) that is characteristic of many members of the RecQ helicase clade (18-23), which is a branch of helicase superfamily II (Figure 1). The crystal structure of the E. coli RecQ HRDC domain (18) reveals it to be composed of five α-helices and a short 310 helix located between α1 and α2 (Figure 1). The corresponding HRDC domain of MsmUvrD2 displays extensive primary structure similarity to the RecQ HRDC domain (Figure 1), and we presume it adopts a similar tertiary structure. It is remarkable that UvrD2, the motor domain of which belongs to superfamily I, is fused to an HRDC module derived from a superfamily II enzyme.
Figure 1.
Domain organization of M. smegmatis UvrD2. The 709-aa UvrD2 polypeptide is depicted as a horizontal bar composed of a large N-terminal ATPase domain (gray) flanked by downstream tetracysteine (magenta) and HRDC (blue) domains. The amino acid sequence of the UvrD2 HRDC domain is aligned to that of the E. coli RecQ HRDC domain; positions of side chain identity are indicated by •. The tertiary structure of the RecQ HRDC domain is shown at the right, and the secondary structure elements are depicted below the amino acid sequence at the left.
By studying deleted and mutated versions of MsmUvrD2, we show that the HRDC domain is dispensable for ATPase and helicase activities in vitro. However, deletion of the tetracysteine domain abolishes duplex unwinding without impacting the ATPase. The instructive result is that the helicase activity of the isolated motor domain of UvrD2 that lacks the tetracysteine and HRDC modules can be restored by mycobacterial Ku. These results signify that coupling of NTP hydrolysis to DNA unwinding can be performed by protein domains acting in cis or trans. We also provide genetic evidence that UvrD2 is essential for growth of M. smegmatis.
EXPERIMENTAL PROCEDURES
UvrD2 Proteins
pET28-His10Smt3-UvrD2 encodes the full-length MsmUvrD2 polypeptide fused to an N-terminal His10Smt3 tag (13). C-Terminal deletions of the HRDC domain or the tetracysteine plus HRDC domain and amino acid substitution mutations of full-length MsmUvrD2 were introduced by PCR amplification with mutagenic primers. The inserts of the MsmUvrD2 plasmids were sequenced to exclude the acquisition of unwanted coding changes during amplification or cloning. The expression plasmids were transformed into E. coli BL21(DE3). The wild-type and mutant His10Smt3-MsmUvrD2 proteins were produced by IPTG induction at 17 °C and purified from soluble lysates by Ni-agarose and DEAE-Sephacel chromatography as described previously (13). The His10Smt3 tag was then removed by digestion of the preparation with the Smt3-specific protease Ulp1 for 3 h at 4 °C (at a UvrD2:Ulp1 ratio of 1000:1). The tag-free MsmUvrD2 proteins were separated from His10-Smt3 by passage of the digest over a Ni-agarose column. MsmUvrD2 was recovered in the flow-through.
UvrD2 C-Terminal Domains
pET28-His10Smt3-UvrD2-(603-709) and pET28-His10Smt3-UvrD2-(589-709) encode His10Smt3-tagged versions of the isolated C-terminal HRDC domain and dual tetracysteine-HRDC domain, respectively. The recombinant proteins were produced in E. coli and isolated from soluble extracts by Ni-agarose chromatography as described above. The His10Smt3 tags were removed by treatment with Ulp1, and the tag-free domains were recovered in the flow-through fractions during a second Ni-agarose chromatography step. The HRDC and tetracysteine-HRDC domains were purified further by Sephacryl-100 gel filtration chromatography. The single discrete peaks of protein from the included volumes of the columns were concentrated by centrifugal ultrafiltration to final concentrations of 14-17 mg/mL in 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 5 mM DTT.
Nucleoside Triphosphatase Assay
Reaction mixtures containing (per 10 μL) 20 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 1 mM [γ-32P]ATP (Perkin-Elmer Life Sciences), 50 ng of salmon sperm DNA, and UvrD2 as specified were incubated for 5 min at 37 °C. An aliquot (2 μL) of the mixture was applied to a polyethylenimine-cellulose TLC plate, which was developed with 0.45 M ammonium sulfate. The radiolabeled material was visualized by autoradiography, and 32Pi formation was quantified by scanning the TLC plate with a Fujix BAS2500 imager.
Helicase Assay
The 5′-32P-labeled strand was prepared by reaction of a synthetic oligodeoxynucleotide with T4 polynucleotide kinase and [γ-32P]ATP. The labeled DNA was purified by electrophoresis through a native 18% polyacrylamide gel. The labeled strand was annealed to 2-fold excess of cDNA strands to form the tailed duplex helicase substrates shown in the figures. Helicase reaction mixtures (10 μL) containing 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 0.5 pmol (50 nM) of radiolabeled DNA, and proteins as specified were preincubated for 5 min on ice. The reaction was initiated by adding 1 mM ATP and 5 pmol of an unlabeled oligonucleotide corresponding to the labeled strand of the helicase substrate. Addition of excess of unlabeled strand was necessary to prevent the spontaneous reannealing of the unwound 32P-labeled DNA strand. The reaction mixtures were incubated for 5 min at 37 °C and then quenched by adding 2 μL of a solution containing 2% SDS, 200 mM EDTA, 40% glycerol, and 0.3% bromophenol blue. A control reaction mixture containing no protein was heated for 5 min at 95 °C. The reaction products were analyzed by electrophoresis through a 15 cm 15% polyacrylamide gel in 89 mM Tris-borate and 2.5 mM EDTA. The products were visualized by autoradiography and quantified by scanning the gel with a Fujix BAS-2500 imaging apparatus.
Deletion of uvrD2 from the M. smegmatis Chromosome
A targeting construct for M. smegmatis uvrD2 (MSmeg_1952) was constructed by amplifying the 5′ and 3′ flanking genomic regions by PCR. The 5′ flanking DNA included 618 nucleotides upstream and 6 nucleotides downstream of the start codon. The 3′ flanking DNA included 54 nucleotides upstream and 564 nucleotides downstream of the stop codon. Fusion of these two fragments via an overlapping SpeI site produced an in-frame deleted uvrD2 locus encoding a polypeptide of only 23 amino acids. This strategy avoided potential polar effects on flanking genes. The ΔuvrD2 cassette was cloned into suicide plasmid pmsg350, which contains a hygromycin-resistance marker and a sacB gene. The resulting plasmid, p1952-KO, was used in efforts to delete uvrD2 from the chromosome by two-step allelic exchange (24). Because we were unable to recover secondary recombinants of wild-type M. smegmatis that contained only the ΔuvrD2 locus, we constructed a strain in which a second copy of uvrD2 was integrated at the chromosomal attB locus. The attB-targeting plasmid was a derivative of pMV306kan into which was inserted a wild-type uvrD2 cassette comprising 119 nucleotides upstream of the start codon plus the entire open reading frame.
RESULTS
Benign Effects of Deleting the HRDC Domain of UvrD2
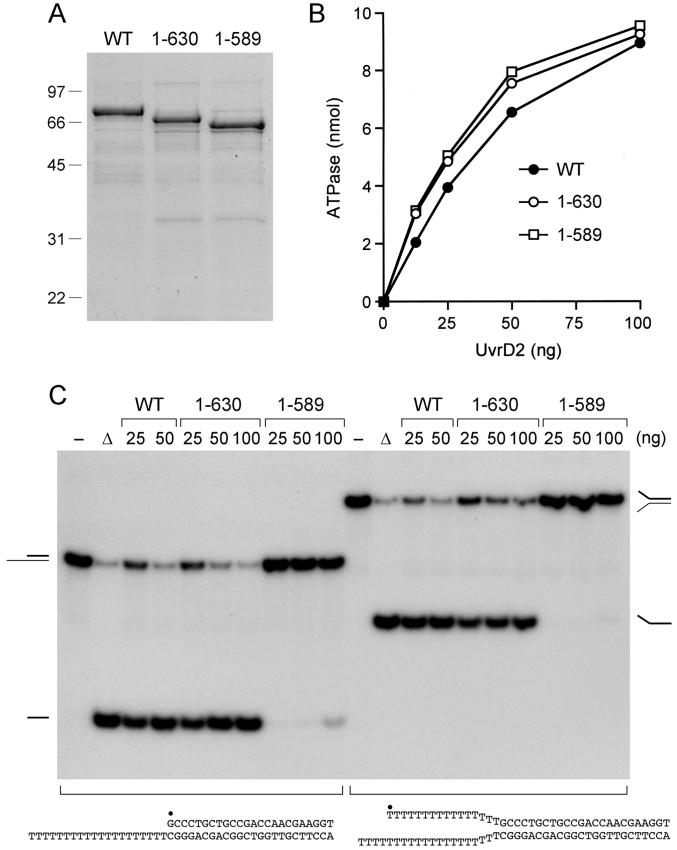
Purified recombinant wild-type UvrD2 and a truncated version, UvrD2-(1-630), that lacks the C-terminal HRDC domain displayed the expected size difference when analyzed by SDS-PAGE (Figure 2A). The two proteins displayed virtually identical specific activity for ATP hydrolysis in the presence of salmon sperm DNA (Figure 2B), and they were equally adept at unwinding two types of helicase substrates: (i) a 3′-tailed DNA composed of a 24-bp duplex segment and a 3′-T20 tail and (ii) a forked DNA composed of the 24-bp duplex with 3′-T20 and 5′-T15 tails on one end (Figure 2C). Thus, the HRDC domain is dispensable for the UvrD2 ATPase and helicase activities in vitro.
Figure 2.
Effects of deleting the HRDC and tetracysteine domains. (A) Aliquots (5 μg) of recombinant wild-type (WT) UvrD2 and truncation mutants UvrD2-(1-630) and UvrD2-(1-589) were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the left. (B) ATPase. Reaction mixtures (10 μL) containing 20 mM Tris-HCl, pH 8.0, 1 mM [γ-32P]ATP, 2 mM MgCl2, 50 ng of salmon sperm DNA, and wild-type or mutant UvrD2 as specified were incubated for 5 min at 37 °C. (C) Helicase. Reaction mixtures (10 μL) containing 5 mM MgCl2, 50 nM 3′-tailed or forked duplex DNA substrates (depicted at bottom with the 5′-32P label denoted by •), 1 mM ATP, and 25, 50, or 100 ng of wild-type or mutant UvrD2 as specified were incubated for 5 min at 37 °C. The products were analyzed by native PAGE and visualized by autoradiography. A control reaction without added protein is included in lane -; a control reaction lacking enzyme that was heat denatured prior to PAGE is shown in lane Δ.
Deletion of the Tetracysteine Domain Abolishes Helicase Activity
The UvrD2-(1-589) protein lacks the tetracysteine domain and the C-terminal HRDC domain (Figure 2A). The specific activity of UvrD2-(1-589) as a DNA-dependent ATPase was undiminished compared to that of wild-type UvrD2 and the ΔHRDC mutant (Figure 2B), yet the helicase activity of UvrD2-(1-589) was effectively ablated (Figure 2C). Thus, deletion of the tetracysteine domain uncouples ATP hydrolysis from duplex unwinding.
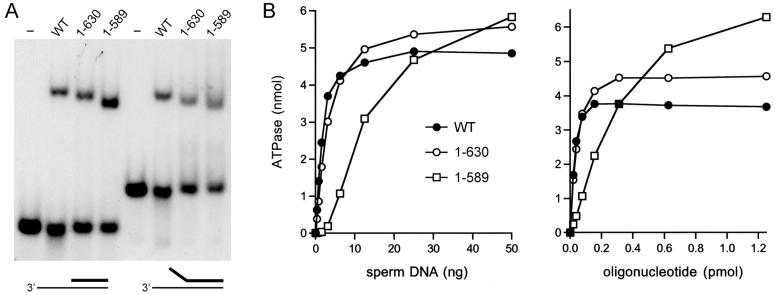
The ability of wild-type UvrD2 and the ΔHRDC and UvrD2-(1-589) mutants to bind to the helicase substrates was gauged by native gel electrophoresis (Figure 3A). ATP was omitted from the binding mixtures in order to preclude unwinding. All three of the UvrD2 proteins formed a discrete protein-DNA complex of retarded electrophoretic mobility, and each incremental deletion from the C-terminus of UvrD2 elicited an incremental increase in the mobility of the UvrD2-DNA complex (Figure 3A). Thus, we surmise that (i) neither the HRDC domain nor the tetracysteine domain is necessary for initial binding to the helicase substrate and (ii) the DNA-bound UvrD2-(1-589) protein is unable to use the energy of ATP hydrolysis to plow through the duplex segment.
Figure 3.
DNA binding and DNA dependence of ATP hydrolysis. (A) DNA binding. Reaction mixtures (10 μL) containing 20 mM Tris-HCl, pH 8.0, 2 mM MgCl2, 5% glycerol, 0.5 pmol of 32P-labeled 3′-tailed or forked duplex DNAs as specified, and 100 ng of wild-type or mutant UvrD2 were incubated at 37 °C for 10 min. Control reactions lacking UvrD2 are shown in lanes -. The mixtures were adjusted to 10% glycerol and then analyzed by electrophoresis through a 4% native polyacrylamide gel containing 45 mM Tris-borate and 1.2 mM EDTA. The gels were run at 100-200 V in the cold room. The gel was transferred to DEAE paper and dried under vacuum. The free DNAs and slower migrating protein-DNA complexes were visualized by autoradiography. (B) DNA titrations. ATPase reaction mixtures (10 μL) containing 20 mM Tris-HCl, pH 8.0, 1 mM [γ-32P]ATP, 2 mM MgCl2, 25 ng of wild-type or mutant UvrD2, and increasing amounts of salmon sperm DNA (or a 44-mer DNA oligonucleotide) were incubated for 5 min at 37 °C. The extents of ATP hydrolysis are plotted as a function of input salmon sperm DNA (left panel) or 44-mer oligonucleotide (right panel).
To obtain a more nuanced picture of the possible effects of C-terminal domain deletions on the DNA interactions of UvrD2, we measured ATP hydrolysis as a function of the concentration of the DNA cofactor (Figure 3B). Wild-type UvrD2 and UvrD2-(1-630) displayed a typical hyperbolic dependence of ATP hydrolysis on input DNA, whether it be salmon sperm DNA or a single-stranded 42-mer DNA oligonucleotide. The notable finding was that deletion of the tetracysteine module elicited a shift to the right in the DNA titration curve, which became sigmoidal in shape in the case of salmon sperm DNA, yet saturated at the same or slightly higher level of ATP hydrolysis than that seen with the two longer versions of UvrD2 (Figure 3B). Note that the ATPase assays that revealed no significant difference in specific activity of the wild-type and deletion mutants (Figure 2B) were performed at saturating levels of salmon sperm DNA. From the titration curve of the 42-mer oligonucleotide in Figure 3B, we estimate that deletion of the tetracysteine domain elicited about an 8-fold decrement in the functional affinity of single-stranded DNA for the “activation site” of the UvrD2 ATPase.
The Tetracysteine Domain Is a Signature of the Actinomycetales UvrD2 Clade
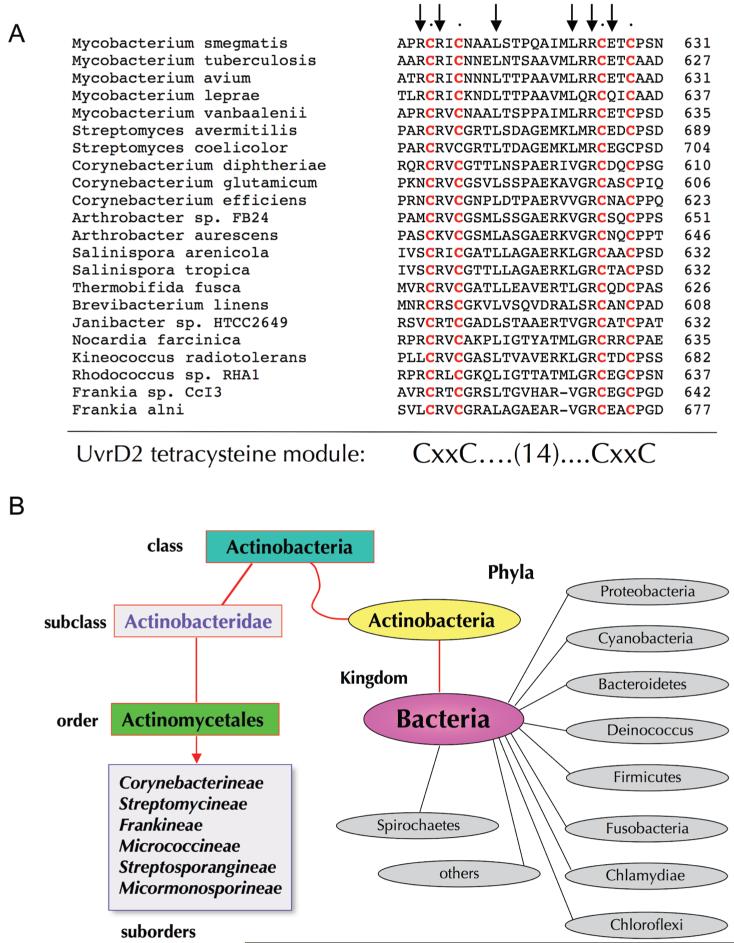
The amino acid sequence of the MsmUvrD2 tetracysteine module is shown in Figure 4A. A blast search of the NCBI database revealed that the close homologues of this module were found exclusively among a novel clade of bacterial helicase-like proteins. In every instance the tetracysteine module was fused to the distal end of the ATPase domain. An alignment of the bacterial UvrD2-like tetracysteine modules highlights a consensus CxxC-(14)-CxxC primary structure in 20 of the bacterial helicases shown, the only deviation being a 13-aa spacer between the CxxC motifs in the homologues from two Frankia species (Figure 4A). The alignment highlights several conserved non-cysteine residues of the domain, particularly two invariant arginines located within or preceding the putative Zn-binding CRxC and RCxxC motifs (Figure 4A).
Figure 4.
The UvrD2 tetracysteine module. (A) The amino acid sequence of the tetracysteine domain of M. smegmatis UvrD2 is aligned to the sequence of homologous helicase-like polypeptides from the indicated bacterial species. Side chains conserved in all of the aligned proteins are indicated by •. The putative Zn-binding cysteines are highlighted in red. (B) The UvrD2 tetracysteine module is the signature of an Actinomycetales helicase clade. The taxonomy of the suborders of bacteria that have a UvrD2-type tetracysteine domain helicase is shown. The major phyla of the bacterial kingdom that do not have homologues of the UvrD2-type helicase are shown in gray.
The striking feature of the bacteria that have the UvrD2-type tetracysteine helicases is that they belong to the same taxon (Figure 4B). Specifically, they are members of one of the indicated suborders of the order Actinomycetales of the Actinobacteridae subclass of the Actinobacteria class of the Actinobacteria phylum (Figure 4B). Mycobacteria belong to the Corynebacterineae suborder. It is worth stressing that the tetracysteine module of the UvrD2 clade is completely different from the RecQ Zn-binding module, which has the primary structure C-(16aa)-CxxCxxC and a distinctive tertiary structure in which the first and second cysteines are separated by an α-helix and loop (19).
Helicase Activity of UvrD2-(1-589) Is Restored by Ku
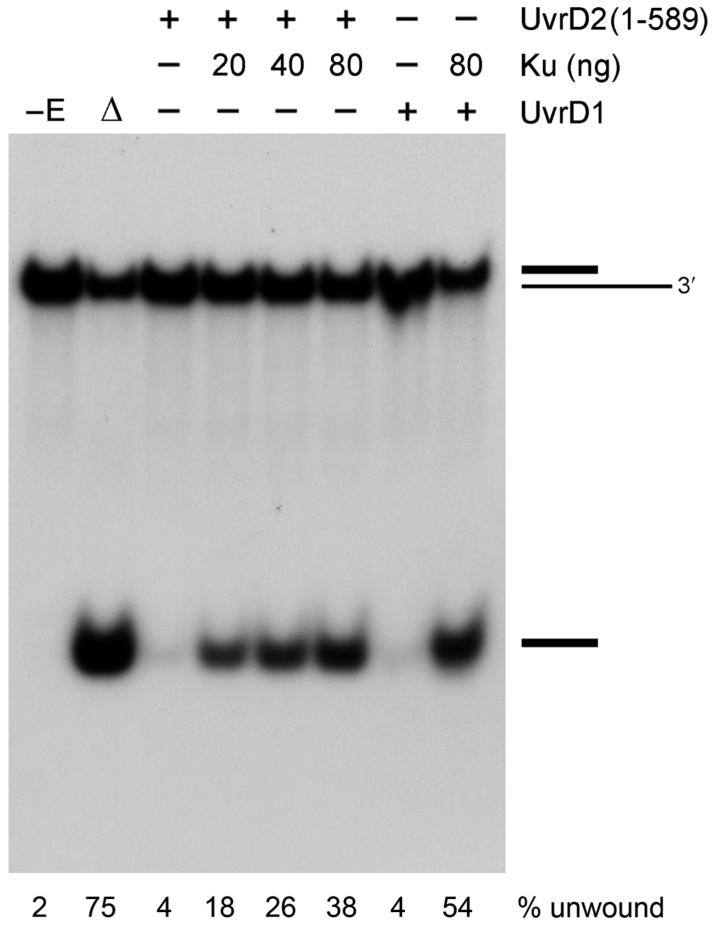
The attenuation of UvrD2 helicase function caused by deleting the tetracysteine module mimicked the natural state of its UvrD1 paralogue. This analogy was underscored by the finding that Ku, a stimulant of unwinding by UvrD1 (13), could compensate for the deficit of the UvrD2-(1-589) protein and revive its DNA unwinding activity (Figure 5). The extent of unwinding of 50 nM 3′-tailed DNA by 100 nM UvrD2-(1-589) increased with the amount of added Ku in the range of 20-100 nM Ku homodimer. We speculate that Ku enables UvrD2-(1-589) to translocate through the duplex segment of the 3′-tailed DNA and displace the radiolabeled strand.
Figure 5.
Ku restores the helicase activity of UvrD2-(1-589). Helicase reaction mixtures (10 μL) containing 5 mM MgCl2, 50 nM 3′-tailed duplex DNA substrate, 1 mM ATP, either 100 ng of UvrD2-(1-589) or 100 ng of UvrD1 (where indicated by +), and Ku as specified were incubated for 5 min at 37 °C. The products were analyzed by native PAGE and visualized by autoradiography. A control reaction without added protein is included in lane -E; a control reaction lacking enzyme that was heat denatured prior to PAGE is shown in lane Δ. The fraction of the input radiolabeled DNA corresponding to the unwound single strand was quantified by scanning the gel; the values are shown below each lane.
The Tetracysteine Domain, but Not the Four Cysteines, Is Essential for Helicase Activity
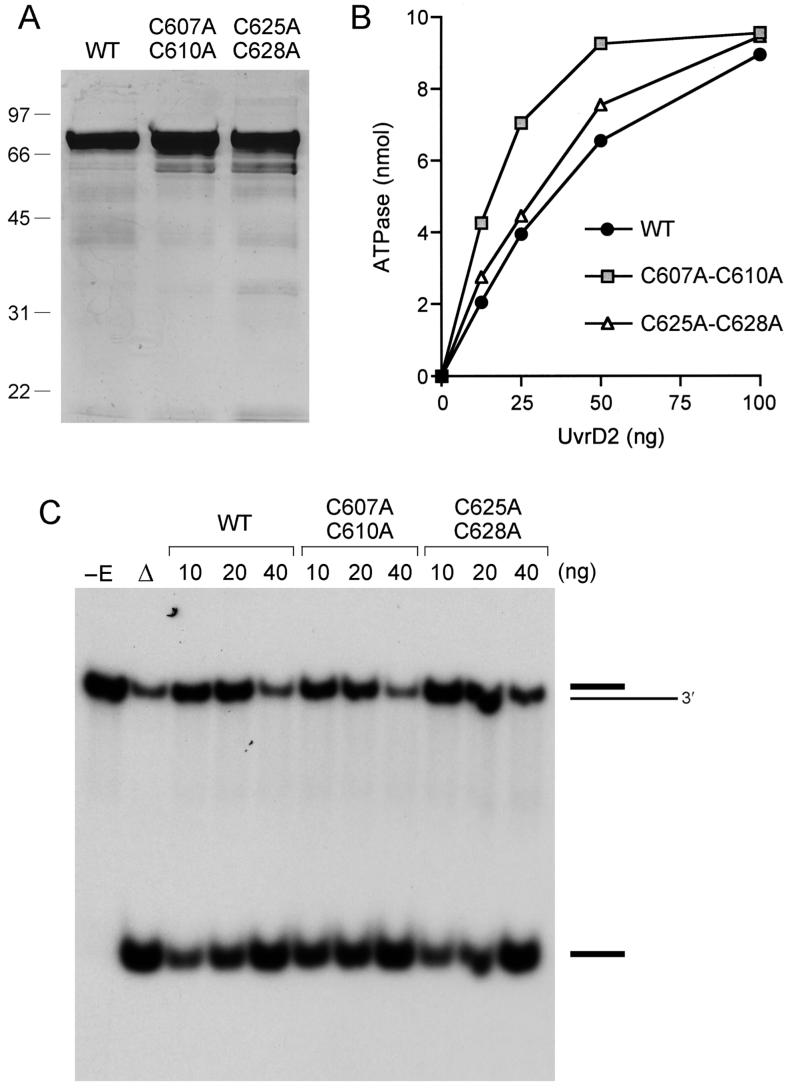
The potential role of Zn binding in the DNA unwinding reaction of UvrD2 was probed by replacing each of the CxxC motifs with a double-alanine variant AxxA, with the expectation that simultaneous loss of two of the four zinc ligands would preclude formation of a tetrahedral zinc complex by UvrD2. The full-length mutant proteins UvrD2-(C607A-C610A) and UvrD2-(C625A-C628A) were purified in parallel with wild-type UvrD2 (Figure 6A). The paired cysteine mutations had no deleterious effect on DNA-dependent ATPase specific activity (Figure 6B); this result was expected, given that a complete deletion of the tetracysteine module also had no impact on ATPase specific activity (Figure 2B). The C607A-C610A and C625A-C628A mutants displayed a hyperbolic dependence of ATP hydrolysis on the concentration of input salmon sperm DNA typical of wild-type UvrD2 (with half-maximal ATP hydrolysis attained at DNA concentrations similar to wild-type UvrD2); the cysteine mutants did not phenocopy the shift-to-the-right and sigmoidal character of the DNA titration curve seen for the truncated UvrD2-(1-589) mutant that lacked the tetracysteine domain (data not shown). The salient finding was that the two double-cysteine mutants retained helicase activity (Figure 6C). We surmise that the tetracysteine domain contributes non-cysteine functional groups important for coupling ATP hydrolysis to DNA unwinding. It is unlikely that zinc binding per se is critical for helicase activity.
Figure 6.
The CxxC motifs are not required for helicase activity. (A) Aliquots (5 μg) of recombinant wild-type (WT) UvrD2 and mutants C607A-C610A and C625A-C628A were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the left. (B) ATPase. Reaction mixtures (10 μL) containing 20 mM Tris-HCl, pH 8.0, 1 mM [γ-32P]ATP, 2 mM MgCl2, 50 ng of salmon sperm DNA, and wild-type or mutant UvrD2 as specified were incubated for 5 min at 37 °C. (C) Helicase. Reaction mixtures (10 μL) containing 5 mM MgCl2, 50 nM 3′-tailed duplex DNA substrate, 1 mM ATP, and 10, 20, or 40 ng of wild-type or mutant UvrD2 as specified were incubated for 5 min at 37 °C. The products were analyzed by native PAGE and visualized by autoradiography. A control reaction without added protein is included in lane -E; a control reaction lacking enzyme that was heat denatured prior to PAGE is shown in lane Δ.
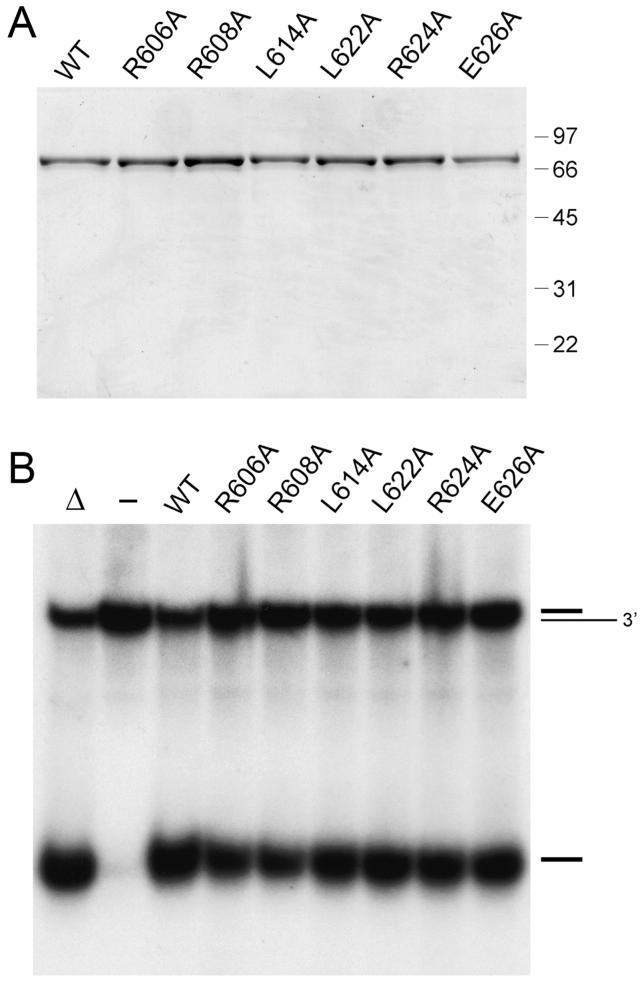
To probe whether individual non-cysteine side chains of the tetracysteine domain might be essential for DNA unwinding, we introduced alanine singly in lieu of Arg606, Arg608, Leu614, Leu622, Arg624, and Glu626 (residues denoted by arrows in Figure 4A). The UvrD2-Ala mutants were produced in E. coli and purified in parallel with wild-type UvrD2 (Figure 7A). The alanine mutants retained DNA-dependent ATPase activity (not shown) and helicase activity (Figure 7B). Conspicuous among the residues defined by the alanine scan as nonessential for UvrD2 function in vitro are Arg608 and Arg624, which are conserved as basic side chains in each of the Actinomycetales UvrD2 homologues shown in Figure 4A, and Leu614 and Leu622, which are conserved as bulky aliphatic side chains.
Figure 7.
Alanine scanning of the tetracysteine module. (A) Aliquots (2.5 μg) of recombinant wild-type (WT) UvrD2 and the indicated Ala mutants were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the right. (B) Helicase reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 50 nM 3′-tailed duplex DNA substrate, 1 mM ATP, and 50 ng of wild-type or mutant UvrD2 as specified were incubated for 5 min at 37 °C. The products were analyzed by native PAGE and visualized by autoradiography. A control reaction without added protein is included in lane -; a control reaction lacking enzyme that was heat denatured prior to PAGE is shown in lane Δ.
Isolated HRDC and Tetracysteine-HRDC Domains
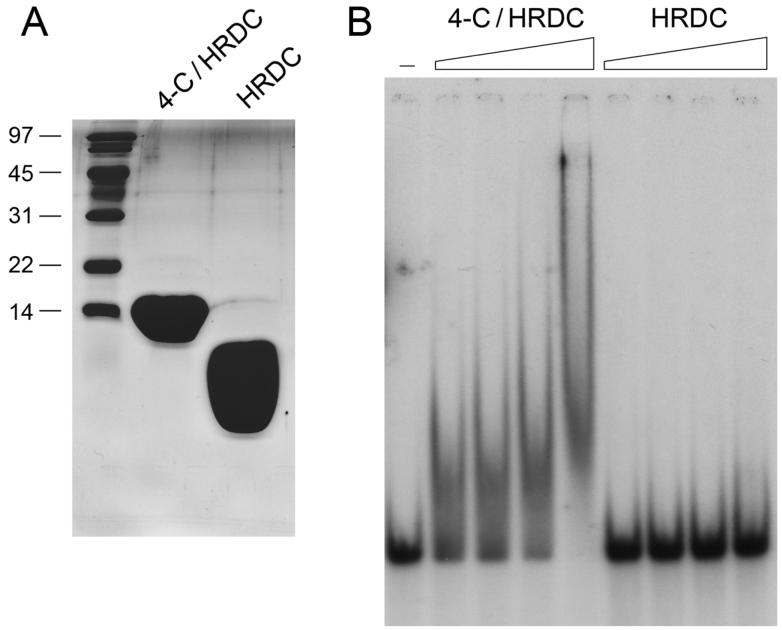
We produced the UvrD2 HRDC domain (aa 630-709) and the dual tetracysteine-HRDC domain (aa 589-709) in E. coli as His10Smt3 fusions, isolated the fusion proteins from soluble extracts by Ni-affinity chromatography, removed the tags by digestion with Ulp1, and then purified the tag-free domains by sequential Ni-agarose and gel-filtration chromatography steps (Figure 8A). Neither the HRDC nor tetracysteine-HRDC domain could restore helicase activity to the truncated UvrD2-(1-589) protein when added to standard helicase reaction mixtures at concentration ranges from 7- to 700-fold molar excess (for tetracysteine-HRDC) or 11- to 1100-fold excess (for HRDC) over the motor domain UvrD2-(1-589) (not shown). Electrophoretic mobility shift assays revealed no stable binding of the HRDC domain to the 3′-tailed helicase substrate at HRDC concentrations up to 2100-fold excess over the input DNA (Figure 8B). The dual tetracysteine-HRDC domain elicited a diffuse mobility shift of the helicase substrate when the domain was present at ≥25-fold molar excess over the DNA ligand (Figure 8B). No DNA mobility shift by the tetracysteine-HRDC domain was observed at the lower DNA and protein concentrations at which full-length UvrD2 or the N-terminal motor domain formed discrete protein-DNA complexes (i.e., the conditions used in Figure 3A; data not shown).
Figure 8.
C-Terminal domains of UvrD2. (A) Aliquots (24 μg) of recombinant tag-free HRDC and tetracysteine-HRDC domains of MsUvrD2 were analyzed by SDS-PAGE (17.5% acrylamide). The Coomassie blue-stained gel is shown. The sizes (kDa) and positions of marker proteins are indicated on the left. (B) DNA-binding reaction mixtures (10 μL) containing 20 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 5% glycerol, 170 nM 3′-tailed duplex DNA, and increasing concentrations of tetracysteine-HRDC domain (4.4, 8.8, 17.6, and 220 μM, proceeding from left to right) or HRDC domain (7.3, 14.6, 29.2, and 366 μM, proceeding from left to right) were incubated for 10 min at 37 °C. Protein was omitted from the control reaction in lane -. The mixtures were analyzed by electrophoresis through a 6% native polyacrylamide gel as described in the legend to Figure 3A. An autoradiogram of the gel is shown.
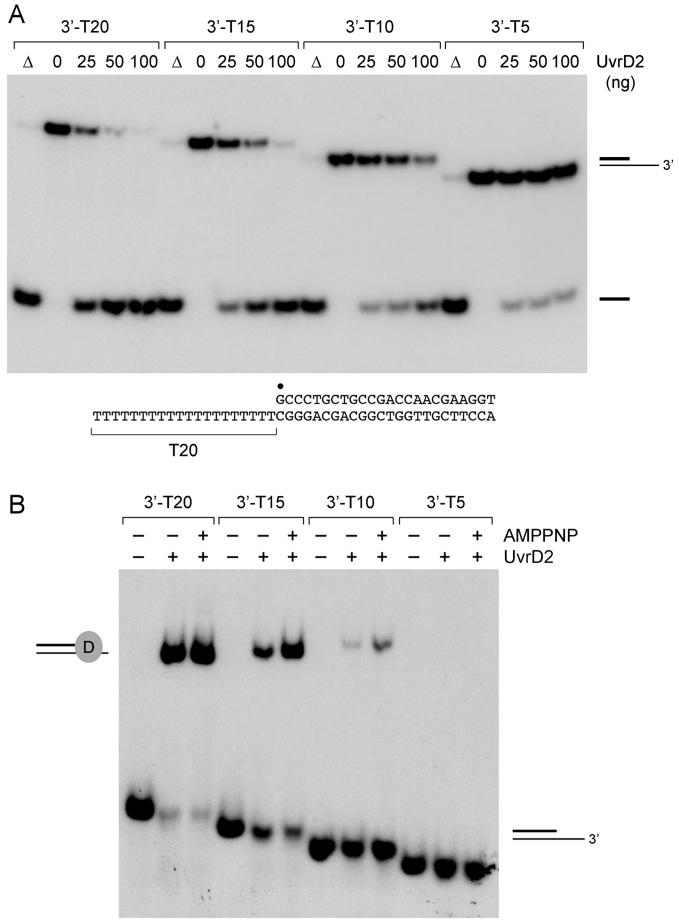
Influence of 3′-Tail Length on DNA Unwinding by UvrD2
Prevailing models of DNA unwinding by a UvrD/PcrA-type helicase posit that the enzyme loads onto the 3′ single-stranded tail and migrates to the ss-duplex junction, where it contacts a five-nucleotide segment of the 3′ single-stranded tail (which tracks across subdomains 1A and 2A) and a 14-16-bp duplex DNA segment (which interacts with subdomains 1B and 2B) (2, 3). To probe the 3′-tail requirement for duplex unwinding by UvrD2, we tested a series of helicase substrates with 3′-oligo(dT) tails of varying length (20, 15, 10, or 5 nucleotides) attached to an identical 24-bp duplex segment. UvrD2-mediated displacement of the radiolabeled 24-mer strand was optimal when the 3′-tail length was 20 nucleotides (Figure 9A). Helicase specific activity decreased by factors of 2 and 4 when the 3′ tail was retracted to 15 and 10 nucleotides, respectively (Figure 9A). Unwinding was inhibited severely when the tail was shortened to only 5 nucleotides. Analysis of UvrD2 binding to these helicase substrates by native gel electrophoresis revealed a sharp decrement in the yield of a stable UvrD2-DNA binary complex as the 3′ tail was shortened from 15 to 10 nucleotides (Figure 9B). Inclusion of 1 mM AMPPNP in the binding reaction mixtures enhanced the stability of the UvrD2-DNA complex and increased the yield of complex formation on the helicase substrates with T15 and T10 3′ tails (Figure 9B) However, AMPPNP addition did not result in detectable complex formation on the T5-tailed DNA (Figure 9B). Thus, the dependence of UvrD2 helicase function on 3′-tail length correlates with a length requirement for stable loading of UvrD on the substrate.
Figure 9.
Effect of 3′-tail length on helicase activity and DNA binding. (A) Helicase activity. Reaction mixtures (10 μL) containing 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 50 nM duplex DNA substrate with 3′-T20, 3′-T15, 3′-T10, or 3′-T5 tails, 1 mM ATP, and 0, 25, 50, or 100 ng of UvrD2 were incubated for 5 min at 37 °C. The products were analyzed by native PAGE and visualized by autoradiography. A control reaction lacking enzyme that was heat denatured prior to PAGE is shown in lane Δ. The 3′-T20 substrate is shown at the bottom with the 5′-32P label denoted by β. (B) DNA binding. Reaction mixtures (10 μL) containing 20 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 5% glycerol, 0.5 pmol of 32P-labeled 3′-tailed DNAs as specified, and UvrD2 (200 ng) or 1 mM AMPPNP (where indicated by +) were incubated at 37 °C for 10 min. The mixtures were adjusted to 10% glycerol and then analyzed by native gel electrophoresis. The free DNAs and UvrD2-DNA complexes were visualized by autoradiography.
Evidence That UvrD2 Is Essential for Growth of M. smegmatis
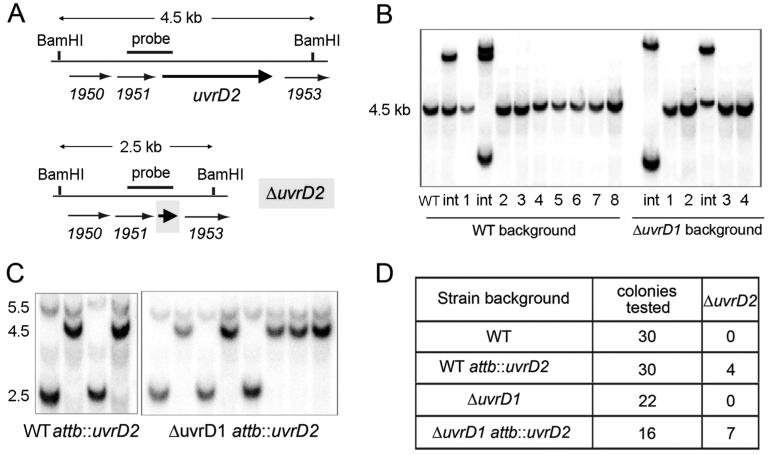
To gauge the role of UvrD2 in mycobacterial physiology, an attempt was made to delete the uvrD2 gene of M. smegmatis by removing the bulk of the open reading frame and rejoining the 5′ and 3′ termini with maintenance of the translation frame. The ΔuvrD2 cassette was cloned into a mycobacterial suicide plasmid containing hygR and sacB markers, which was then used to transform M. smegmatis mc2155 (wild-type) and M. smegmatis ΔuvrD1 (13) to hygromycin resistance. The hygR integrants were tested for sensitivity to sucrose to verify the presence of the sacB marker. The hygR sucS ΔuvrD2 uvrD2+ merodiploid strains were genotyped by Southern blotting to verify targeted integration at uvrD2 (Figure 10B, lanes int). The merodiploids were then grown in sucrose to select for recombination between the tandem uvrD2 and ΔuvrD2 genes and loss of the intervening sacB marker. Individual sucR isolates were screened for sensitivity to hygromycin. All of the resulting sucR hygS segregants that were genotyped by Southern blotting (n = 30 for wild type and n = 22 for ΔuvrD1 strain) retained the wild-type uvrD2 locus and lost the ΔuvrD2 knockout cassette (Figure 10B). However, the chromosomal uvrD2 locus could be disrupted in a strain of M. smegmatis in which a second allele of uvrD2 was preintegrated at the attB locus (Figure 10C). These results are consistent with uvrD2 being essential for viability of M. smegmatis.
Figure 10.
Evidence that UvrD2 is essential in M. smegmatis. (A) Schematic maps of the wild-type M. smegmatis genomic uvrD2 locus (MSmeg_1952), indicated by the thick arrow, with flanking ORFs depicted as thin arrows. BamHI restriction sites and the genomic segment used as a radiolabeled probe for genotyping are indicated. A map of the ΔuvrD2 locus generated by targeted insertion of an in-frame deletion of the uvrD2 ORF is shown below the wild-type locus map. (B) BamHI digests of genomic DNA from wild-type M. smegmatis and the ΔuvrD1 mutant, hygR sucS merodiploids generated by integration of the ΔuvrD2 cassette at the uvrD2 locus (int), and individual hygS sucR segregants derived from the merodiploids were resolved by agarose gel electrophoresis, transferred to a membrane, and hybridized to a 32P-labeled DNA probe generated by amplification of the segment depicted in panel A. All of the segregants retained the 4.5-bp BamHI fragment corresponding to the wild-type uvrD2 locus. (C) BamHI digests of genomic DNA from individual hygS sucR segregants derived from the merodiploids of attB::uvrD2 and ΔuvrD1 attB::uvrD2 strains of M. smegmatis containing the ΔuvrD2 cassette integrated at the uvrD2 locus were genotyped by Southern blotting as described in panel B. Segregants containing only the 2.5 kb ΔuvrD2 locus were recovered when a second uvrD2 allele was preintegrated at the genomic attB site. (D) Summary of the outcomes of attempts to delete uvrD2 in M. smegmatis strains that do or do not have an extra copy of uvrD2 integrated into the chromosomal attB locus.
DISCUSSION
UvrD2 is one of two UvrD-like paralogues in mycobacteria. The present study highlights features of UvrD2 that not only distinguish it from UvrD1 but underscore the uniqueness of UvrD2 among bacterial helicases. UvrD2 has a “hybrid” structure composed of domains characteristic of helicases in both superfamily I (the N-terminal ATPase domain) and superfamily II (the C-terminal HRDC domain). The ATPase and HRDC domains are linked by a tetracysteine module found only in mycobacterial UvrD2 and its homologues from other Actinomycetales species.
The N-terminal motor domains of UvrD1 and UvrD2 are conserved, and their ATPase activities are similar. The disparity that UvrD1 is a feeble helicase while UvrD2 is adept at duplex unwinding (13) can now be attributed to the distinct C-terminal structure of UvrD2. Deletion of the tetracysteine module cripples UvrD2 as an autonomous helicase, without grossly affecting ATP hydrolysis or initial binding to the 3′-tailed helicase substrate. The remarkable finding is that the latent motor activity of the N-terminal ATPase domain can be revived in the presence of Ku. Thus, UvrD2 deprived of its tetracysteine domain becomes UvrD1-like in its reliance on Ku for duplex unwinding.
Deleting the HRDC domain of UvrD2 had no apparent effect on any of the biochemical activities tested. Our results are in accord with previous reports that deleting the HRDC domains of E. coli RecQ had no impact on ATP hydrolysis, RecQ-catalyzed unwinding of a 3′-tailed DNA duplex, or the dependence of ATP hydrolysis on the concentration of the DNA cofactor (22). The observation that removal of the RecQ HRDC domain suppressed the ability of RecQ to bind to the 3′-tailed helicase substrate in a gel-shift assay was interpreted as indicating a role for HRDC in “stable DNA binding” (18). By contrast, loss of the UvrD2 HRDC domain did not preclude stable UvrD2 binding to a 3′-tailed DNA; moreover, the isolated UvrD2 HRDC domain by itself did not bind stably to the 3′-tailed DNA. Note that the RecQ HRDC domain by itself can bind single-stranded DNA; on this basis it was proposed that HRDC domains might target RecQ-family proteins to specific DNA structures (18). Consistent with this idea, the ability of the BLM helicase (a eukaryal RecQ homologue) to dissolve double Holliday junctions depends on its HRDC domain, although unwinding of a 3′-tailed duplex by the BLM helicase is unaffected by an HRDC deletion (21). Studies of a Deinococcus radiodurans RecQ helicase suggest that HRDC domains can have either stimulatory or suppressive effects on helicase functions (23), signifying that inferences about the possible regulatory role of an HRDC domain may not be portable from one helicase to another.
The tetracysteine domain of mycobacterial UvrD2 emerges from this study as a key factor in coupling ATP hydrolysis to DNA unwinding. As discussed above, the CxxC-(14aa)-CxxC motif is a signature of UvrD2 and is clearly different from the C-(16aa)-CxxCxxC Zn-finger module found in RecQ family helicases. A double mutation of two cysteines in the RecQ-like BLM helicase that abolished Zn2+ binding resulted in global defects in ATP hydrolysis, DNA binding, and helicase activity of the mutant protein (25). Because removal of Zn2+ from preformed wild-type BLM protein by chelation had no deleterious effect on any of these activities, but increased sensitivity of BLM to thermal inactivation and limited proteolysis, it was inferred that Zn2+ is not essential for catalysis by BLM but plays a critical role in attaining and stabilizing an active protein fold (25). In the case of UvrD2, the tetracysteine domain is clearly not essential to generate a vigorous DNA-dependent ATPase. Deletion of the tetracysteine module did cause a shift in the DNA concentration dependence of ATP hydrolysis, consistent with reduced affinity for the ATPase-activating DNA ligand, but the ATPase activity of UvrD2-(1-589) at saturating DNA cofactor concentration was equivalent to full-length UvrD2. It is conceivable that the modest effects of tetracysteine domain deletion on “functional” DNA affinity in an ATPase assay are amplified during the DNA unwinding reaction when, according to the inchworm model (2), the weakening of one of the two transient DNA interfaces could lead to premature dissociation of the helicase from the DNA substrate prior to its transit through the duplex segment. In other words, the processivity of UvrD2 could be diminished by loss of the tetracysteine domain. Consistent with this idea, the isolated tetracysteine-HRDC domain was able to bind to the helicase substrate, a property not observed for the HRDC domain alone.
The UvrD2 tetracysteine domain is a plausible candidate to be a Zn finger. Therefore, we tested the potential role of Zn2+ binding by pairwise mutations of the CxxC motifs. The notable finding was that elimination of two cysteines at a time had no impact on UvrD2 ATPase or helicase activity. Thus, the domain, rather than the four cysteines, is essential for DNA unwinding by UvrD2. Although we have not assayed whether the recombinant wild-type UvrD2 protein contains a single Zn2+ atom, we regard it as unlikely that the C607A-C610A and C625A-C628A mutants could still bind Zn2+ after losing two of the four metal ligands. We do not rule out positive contributions of the cysteines to UvrD2 stability or to other biochemical attributes. For example, the cysteines might form disulfide bonds that could allow for redox regulation of UvrD2 function. Nonetheless, our inference from the mutational data is that Zn2+ binding is not required for the ATPase and helicase activities of UvrD2. Alanine scanning mutagenesis of six of the non-cysteine side chains of the tetracysteine module (including four that are most highly conserved in the UvrD2 clade) revealed no impairment of ATPase or helicase activities of UvrD2. Such findings suggest that this module plays a structural role, rather than contributing individual side chains that are essential for DNA unwinding.
The loss of processive unwinding upon deletion of the tetracysteine domain is rectified in the presence of mycobacterial Ku, a homologue of the eukaryal DNA end-binding protein that forms an O-shaped topological clamp around duplex DNA (26). Whereas Ku is required for DNA unwinding by UvrD1, wild-type UvrD2 is not stimulated by Ku, because it is inherently adept at unwinding the substrates tested (13). The avidity of UvrD1 for Ku is mediated by its C-terminal segment, which was identified as a Ku-binding partner in a two-hybrid screen (13). UvrD2 lacks a counterpart of the Ku-binding segment of UvrD1. Although the helicase assays hint that a latent Ku-binding function of UvrD2 might be unmasked when the tetracysteine and HRDC domains are deleted, we were unable to detect a stable UvrD2-(1-589)-DNA-Ku ternary complex when the three components were mixed in the absence of ATP and then analyzed by native gel electrophoresis (not shown). Nonetheless, the functional data imply that Ku and the tetracysteine domain offer alternative pathways for coupling ATPase and helicase activity. In the case of Ku, unwinding is enabled by protein interactions in trans, whereas the tetracysteine module functions in cis. A simple unifying model would be that the UvrD2 tetracysteine domain aids in forming a C-shaped clamp around the helicase substrate, analogous to the clamp-forming role of the tetracysteine domain of E. coli DNA ligase (27).
Finally, UvrD1 and UvrD2 appear to play distinct roles in mycobacterial physiology. UvrD1 is not essential for viability of M. smegmatis, although deletion of uvrD1 sensitizes cells to killing by DNA-damaging agents (13). Our attempts here to delete the chromosomal uvrD2 locus were unsuccessful. The fact that the uvrD2 locus could be deleted if a second copy of the uvrD2 gene had been integrated elsewhere on the chromosome provides genetic evidence that UvrD2 is essential in M. smegmatis. Mycobacterial UvrD1 is reminiscent of E. coli UvrD, a nonessential DNA repair enzyme, while UvrD2 appears comparable to PcrA, which is essential for viability of Bacillus subtilis and Staphylococcus aureus (28, 29). The observations that E. coli UvrD is essential for growth in the absence of the paralogous Rep helicase and that PcrA can restore viability to an E. coli rep- uvrD- double mutant (29, 30) suggest that there is functional overlap between bacterial UvrD/PcrA-like helicases, with PcrA being uniquely able to supply all essential activities. Thus, we speculate that UvrD1 and UvrD2 might have overlapping functions, with UvrD2 being pluripotent with respect to essential tasks. Our findings recommend UvrD2, which is conserved in pathogenic bacteria that cause human tuberculosis, leprosy, diphtheria, and nocardiosis, as a potential target for anti-infective drug discovery. Determining which DNA transactions are affected in mycobacteria when UvrD2 is inactivated, either singly or in a ΔuvrD1 back-ground, will require more sophisticated genetic manipulations, including the isolation of temperature-sensitive uvrD2 strains and/or achievement of tight regulation of uvrD2 transcription sufficient to rapidly elicit a conditional null phenotype.
Footnotes
This research was supported by Grant AI64693 from the National Institutes of Health.
- EDTA
- ethylenediaminetetraacetic acid
- DTT
- dithiothreitol
- TLC
- thin-layer chromatography
- ATPase
- adenosinetriphosphatase
- PCR
- polymerase chain reaction
- HRDC
- helicase and RNase D C-terminal
- AMPPNP
- adenylylβ,γ-imidotriphosphate
REFERENCES
- 1.Matson SW, Robertson AB. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res. 2006;34:4089–4097. doi: 10.1093/nar/gkl450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structure of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;87:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- 3.Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomko EJ, Fischer CJ, Niedziela-Majka A, Lohman TM. A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell. 2007;26:335–347. doi: 10.1016/j.molcel.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 6.Gong C, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS. Mechanism of nonhomologous end-joining in mycobacteria: a low fidelity repair system driven by Ku, ligase D and ligase C. Nat. Struct. Mol. Biol. 2005;12:304–312. doi: 10.1038/nsmb915. [DOI] [PubMed] [Google Scholar]
- 7.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, di Fagagna F, Devine KM, Bowater RP, Jeggo PA, Jackson SP, Doherty AJ. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 2002;297:1686–1689. doi: 10.1126/science.1074584. [DOI] [PubMed] [Google Scholar]
- 8.Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 9.Moeller R, Stackebrandt E, Reitz G, Berger T, Rettberg P, Doherty AJ, Horneck G, Nicholson WL. Role of DNA repair by nonhomologous end joining in Bacillus subtilis spore resistance to extreme dryness, mono- and polychromatic UV, and ionizing radiation. J. Bacteriol. 2007;189:3306–3311. doi: 10.1128/JB.00018-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephanou NC, Gao F, Bongiorno P, Ehrt S, Schnappinger D, Shuman S, Glickman MS. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J. Bacteriol. 2007;189:5237–5246. doi: 10.1128/JB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitcher RS, Green AJ, Brzostek A, Korycka-Machala M, Dziadek J, Doherty AJ. NHEJ protects mycobacteria in stationary phase against the harmful effects of dessication. DNA Repair. 2007;6:1271–1276. doi: 10.1016/j.dnarep.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinha KM, Stephanou NC, Gao F, Glickman MS, Shuman S. Mycobacterial UvrD1 is a Ku-dependent DNA helicase that plays a role in multiple DNA repair events, including double-strand break repair. J. Biol. Chem. 2007;282:15114–15125. doi: 10.1074/jbc.M701167200. [DOI] [PubMed] [Google Scholar]
- 14.Curti E, Smerdon SJ, Davis EO. Characterization of the helicase activity and substrate specificity of Mycobacterium tuberculosis UvrD. J. Bacteriol. 2007;189:1542–1555. doi: 10.1128/JB.01421-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darwin KH, Nathan CF. Role for nucleotide excision repair in virulence of Mycobacterium tuberculosis. Infect. Immun. 2005;73:4581–4587. doi: 10.1128/IAI.73.8.4581-4587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boschoff HI, Reed MB, Barry CE, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 17.Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE. Inhibition of mutation and combating the evolution of antibiotic resistance. PLOS Biol. 2005;3:1024–1033. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein DA, Keck JL. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13:1173–1182. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein DA, Zittel MC, Keck JL. High-resolution structure of the E. coli RecQ helicase core. EMBO J. 2003;22:4910–4921. doi: 10.1093/emboj/cdg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitano K, Yoshihara N, Hakoshima T. Crystal structure of the HRDC domain of human Werner syndrome protein WRN. J. Biol. Chem. 2007;282:2717–2728. doi: 10.1074/jbc.M610142200. [DOI] [PubMed] [Google Scholar]
- 21.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. EMBO J. 2005;24:2679–2687. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein DA, Keck JL. Domain mapping of Escherichia coli RecQ defines the roles of conserved N- and C-terminal regions in the RecQ family. Nucleic Acids Res. 2003;31:2778–2785. doi: 10.1093/nar/gkg376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killoran MP, Keck JL. Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J. Biol. Chem. 2006;281:12849–12857. doi: 10.1074/jbc.M600097200. [DOI] [PubMed] [Google Scholar]
- 24.Braunstein M, Brown AM, Kurtz S, Jacobs WR. Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 2001;183:6979–6990. doi: 10.1128/JB.183.24.6979-6990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo R, Rigolet R, Zargarian L, Fermandjian, Xi XG. Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom’s syndrome helicase. Nucleic Acids Res. 2005;33:3109–3124. doi: 10.1093/nar/gki619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 27.Nandakumar J, Nair PA, Shuman S. Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate. Mol. Cell. 2007;26:257–271. doi: 10.1016/j.molcel.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Iordanescu I. Characterization of the Staphylococcus aureus chromosomal gene pcrA, identified by mutations affecting plasmid pT181 replication. Mol. Gen. Genet. 1993;241:185–192. doi: 10.1007/BF00280216. [DOI] [PubMed] [Google Scholar]
- 29.Petit MA, Dervyn E, Rose M, Entian KD, McGovern S, Ehrlich SD, Braund C. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling circle replication. Mol. Microbiol. 1998;29:261–283. doi: 10.1046/j.1365-2958.1998.00927.x. [DOI] [PubMed] [Google Scholar]
- 30.Petit MA, Ehrlich D. Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J. 2002;21:3137–3147. doi: 10.1093/emboj/cdf317. BI800725Q. [DOI] [PMC free article] [PubMed] [Google Scholar]