Abstract
We have previously shown that the death receptor ligand TRAIL induces an increase in intracellular C16-ceramide in sensitive SW480 but not in resistant SW620 cells. Resistance in SW620 cells was overcome by exogenous ceramide leading us to propose that defective ceramide signaling contributes to TRAIL resistance. In this study we found that the increase in C16-ceramide in SW480 cells was inhibited by fumonisin B1, an inhibitor of ceramide synthases (CerS). Protein analysis revealed that TRAIL resistant SW620 cells expressed lower levels of ceramide synthase 6 (CerS6, also known as longevity assurance homologue 6), which prompted us to investigate the effect of CerS6 modulation on TRAIL phenotype. RNAi against CerS6 resulted in a specific and significant decrease of the C16-ceramide species, which was sufficient to inhibit TRAIL-induced apoptosis. In cells with decreased levels of CerS6, caspase-3 was activated but failed to translocate into the nucleus. CerS6 localized primarily to the perinuclear region, suggesting this enzyme may play a role in regulation of nuclear permeability. Moderate elevation in CerS6 expression was sufficient to reverse TRAIL resistance in SW620 cells. These results suggest that modulation of CerS6 expression may constitute a new therapeutic strategy to alter apoptotic susceptibility.
Keywords: TRAIL, ceramide, apoptosis, ceramide, synthase, longevity assurance homologue (LASS)
INTRODUCTION
TRAIL (tumor necrosis factor related apoptosis inducing ligand) is a death receptor ligand of the TNF superfamily that can selectively kill cancer cells without toxicity towards normal cells (de Jong et al., 2001; Wiley et al., 1995). Systemic administration of recombinant TRAIL has recently been deemed safe in a Phase I clinical trial (Ashkenazi & Herbst, 2008). Unfortunately, not all malignant cells are susceptible to the apoptotic effects of TRAIL and new strategies to enhance TRAIL-mediated killing of tumor cells are an area of intense investigation. Identification of mechanisms that lead to resistance has a critical two-fold purpose: (1) to facilitate stratification of tumors likely to respond to TRAIL therapy and (2) to develop therapeutic strategies to overcome TRAIL resistance. TRAIL induces apoptosis by binding to agonistic TRAIL receptors (DR4/TRAIL-R1 and/or DR5/TRAIL-R2) followed by activation of initiator caspases and subsequent (mitochondria-dependent or –independent) activation of effector caspases-3 and −7 (Koschny et al., 2007; MacFarlane, 2003). During apoptosis, caspase-3 translocates to the nucleus to cleave targets such as PARP (Widlak & Garrard, 2005; Wilson, 1998). Numerous proteins such as cFLIP, anti-apoptotic members of the Bcl-2 family, and IAP’s can negatively regulate the apoptotic signal at or upstream of caspase-3 activation (Dohi et al., 2004; Peter, 2004; Sharpe et al., 2004; Wiley et al., 1995).
Sphingolipids have also been shown to influence apoptotic responses. The sphingolipid ceramide in particular has been associated with antiproliferative responses such as growth arrest, senescence, differentiation and apoptosis (Ogretmen & Hannun, 2004). Data obtained by liquid chromatography-mass spectroscopy (LC-MS), which allows study of specific ceramide species suggests that generation of C16-ceramide is specifically involved in apoptotic signaling (Kroesen et al., 2003; Thomas et al., 1999). We have recently shown that the isogenic colon cancer cell lines SW480 and SW620 are sensitive and resistant to TRAIL, respectively (Voelkel-Johnson et al., 2005). Our data indicated that differences in TRAIL sensitivity may be related to differences in sphingolipid metabolism. TRAIL sensitive SW480 cells had higher basal levels of C16-ceramide, which further increased in response to TRAIL. We also demonstrated that exogenous C6-ceramide, which is most likely metabolized to C16-ceramide (Ogretmen et al., 2002), sensitized resistant SW620 cells to TRAIL-induced apoptosis but failed to further enhance TRAIL sensitivity in SW480 cells, suggesting that in these cells sufficient endogenous ceramide was available to achieve a maximal apoptotic response. We hypothesized that exogenous ceramide corrects a defect in sphingolipid metabolism present only in resistant and not sensitive cells (Voelkel-Johnson et al., 2005). Here we extended our previous study and identified ceramide synthase 6 (CerS6 also known as longevity assurance homolog 6/LASS6), which preferentially generates C16-ceramide, as a novel protein that can influence TRAIL susceptibility. RNAi against CerS6 resulted in a specific decrease in intracellular C16-ceramide and protected SW480 cells against TRAIL-mediated apoptosis while increasing CerS6 expression sensitized SW620 cells to TRAIL. Downregulation of CerS6 did not interfere with caspase activation but appears to inhibit translocation of activated caspase-3 into the nucleus. Our data suggest that CerS6 may regulate events at the nuclear membrane and allow late stage apoptotic signaling, exemplified by activated caspase 3, to proceed into the nucleus. This finding posits that CerS6 holds a novel position in the apoptotic pathway.
MATERIALS AND METHODS
Cell Lines and Culture
The cell lines SW480 and SW620 were purchased from the American Type Culture Collection (Rockville, MD). SW480 cells were originally isolated from a primary colon carcinoma, while the SW620 line was established from a metastasis that arose in the same patient one year later. Cell lines were cultured in Primaria plasticware (Falcon, Bedford, MA) and maintained in RPMI 1640 medium (Gibco/Invitrogen, Carlsbad, CA) supplemented with heat-inactivated 10% fetal bovine serum (Hyclone, Logan, UT). Cell cultures were maintained at 37oC in a 5% CO2 atmosphere.
Real Time PCR
SW480 cells and SW620 cells were plated 5 X 105 cells/well in 6 wells plates and allowed to grow for two days. RNA was isolated using QiaShredder columns and the RNAeasy kit from Qiagen (Valencia, CA). RNA was reverse transcribed with the RNAScript using OligodT primers (Ambion, Austin, TX). Real time PCR was performed on a BioRad iCycler. Primer sets for CerS (LASS) genes and cyclophilin internal control were purchased from Applied Biosystems (Warrington, UK). Power SYBR Green PCR Master Mix was used according to instructions from Applied Biosystems. Expression levels of CerS5 and CerS6 were normalized to cyclophilin using the 2^(-delta delta) computation.
RNAi
The CerS6 siRNA sequence used was 5’ AAG GTC TTC ACT GCA ATT ACA (Custom siRNA from Qiagen, Valencia, CA). A scrambled control siRNA was also purchased from Qiagen. SW480 cells were plated 2 X 105 per well in a 12-well plate and transfected the next day with Oligofectamine (Invitrogen, Carlsbad, CA). Briefly, for each well 6ul siRNA (20mM stock) and 9ul Oligofectamine were allowed to incubate for 5 minutes in 195ul OptiMEM. Cells were washed with PBS and siRNA complexes added to wells. After 4 hours, 1 ml of growth media was added, bringing siRNA concentration to 97nM. At 48 hours after siRNA transfection, cells were treated with 100 ng/ml TRAIL for 16 hours, unless stated otherwise. Samples for mass spectrophotometric analysis were scaled up for a 100mm dish and harvested 72 hours after transfection.
Plasmids expressing CerS6 shRNA (target sequence 5’ GAA CTG CTT CTG GTC TTA CTT) and GFP shRNA (control) were purchased from OpenBiosystems. After transfection, clones were selected in 5μg/ml puromycin and maintained in 3μg/ml puromycin.
LC/MS analysis
Sphingolipid analysis was performed as described previously (Voelkel-Johnson et al., 2005). Samples were normalized to phosphate (Pi) or cell number as indicated.
Antibodies and Western Blot Analysis
Western blotting was performed as described previously (Voelkel-Johnson et al., 2005). Antibody sources were as follows: Mouse anti-Cleaved PARP and rabbit anti-Caspase 9 were obtained from Cell Signaling (Danvers, MA). Mouse anti-Caspases-3, -8, and rat anti-Caspase-2 were obtained from Alexis (SanDiego, CA). Rabbit anti-Actin was obtained from Sigma (St. Louis, MO). Rabbit anti-PARP was obtained from Santa Cruz (Santa Cruz, CA). Mouse anti-CerS6 (LASS6) was obtained from Abnova (Taipei City, Taiwan). All primary antibodies were used at 1:1000 dilutions. Mouse and rabbit secondary antibodies conjugated to HRP were obtained from Santa Cruz and used at 1:5000 and 3:50000, respectively.
Caspase activity assay
SW480 cells were plated for siRNA treatment as described and transfected with scrambled siRNA or siRNA against CerS6. After 48 hours, cells were left untreated or treated with 100 ng/ml TRAIL for 16 hours. The Apo-ONE homogeneous Caspase 3/7 assay from Promega (Madison, WI) was performed as described previously (Voelkel-Johnson et al., 2005).
FLICA and Hoechst staining
SW480 cells were plated and treated as described under “RNAi”. After a 15-hr treatment with 100 ng/ml TRAIL, cells were stained with FLICA reagent (Immunohistochemistry Technologies, LLC, Bloomington, MN), incubated for 45 minutes at 37°C. Hoechst dye (Acros Organics, Belgium) at a final concentration of 10 μg/ml was added for an additional 10 minutes. The cells were then washed and visualized at 200X using a Zeiss fluorescent microscope with UV or Rhodamine filters.
Nuclear isolation and flow cytometry
SW480 cells were plated and treated as described under “RNAi.” After TRAIL treatment, cells were scraped, washed with PBS, and pelleted. Cells were resuspended in 1 ml cold nuclei extraction buffer (320 mM sucrose, 5 mM MgCl2, 10 mM HEPES, 1% Triton X at pH 7.4), vortexed for 10 seconds and incubated on ice for 10 minutes. Samples were centrifuged for 5 minutes at 3500 RPM at 4oC to obtain a pellet of nuclei. Nuclei were washed with nuclei wash buffer (320 mM sucrose, 5 mM MgCl2, 10 mM HEPES at pH 7.4). Nuclei yield and integrity were confirmed for a small sample by microscopic examination with Hoechst dye staining (as described above). Nuclei were resuspended in 90 μl labeling buffer (wash buffer with 1% BSA and 0.1% sodium azide added) and 10 μl FITC-conjugated anti-active caspase-3 antibody (Cell Signaling, Danvers, MA). After a 1-hour incubation the cells were washed with labeling buffer and resuspended in 200 μl labeling buffer for analysis at the MUSC flow cytometry core facility using a FACSCalibur with CellQuest Pro software (BD Biosciences, Franklin Lakes, NJ).
Confocal microscopy
SW480 cells (2×105) were plated overnight on 35-mm uncoated glass-bottomed dishes (MatTek, Ashland, MA) in RPMI supplemented with 10% FBS and then transfected with pFLAG-LASS6 using Lipofectamine (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations. pFLAG-LASS6 contains the CerS6/LASS6 sequences with an N-terminal FLAG-tag under control of the CMV promoter. This plasmid was provided by Dr. Besim Ogretmen, MUSC. After 36 hours cells were fixed, permeabilized with 0.1% Triton-X100 (Sigma), washed, blocked in 2% human serum for one hour, incubated with primary antibody diluted in 2% human serum for two hours at room temperature, washed, and incubated with secondary antibody in 2% serum for one hour at room temperature. Cells were stained with DRAQ5 nuclear stain (Axxora, San Diego, CA) diluted 1:1000 in PBS for 20 minutes, washed, and visualized by laser-scanning confocal microscopy. Antibody sources and dilutions were as follows: Mouse anti-flag (1:50 from Sigma), Goat anti-Lamin B (1:100 from Santa Cruz), rabbit anti-calreticulin (1:100, Sigma), Alexa-555-conjugated donkey anti-mouse and Alexa-488 conjugated donkey anti-goat (both 1:100 from Invitrogen).
Plasmids and transfection of SW620 cells
The FLAG-tagged LASS6/CerS6 sequence from pFLAG-LASS6 was subcloned into the XhoI and EcoRI sites of pIRES2-EGFP (Clontech, Mountain View, CA) yielding pCerS6-IRES2-EGFP. DNA was prepared using the Endo-free Midi kit (Qiagen). For transient transfections, cells were plated 1 X 106/well in a 6-well plate. The next day transfection with either pIRES2-EGFP from Clontech or pCerS6-IRES2-EGFP was performed with Lipofectamine (Invitrogen) according to manufacturer’s instructions. After 48 hours, cells were treated with 100 ng/ml TRAIL for 24 hours and GFP positive cells visualized at 200x magnification. For stable transfections, plasmids were diluted 1:50 in a recently developed polymer named EDGE3’3, a kind gift from Dr. Kaushal Rege (Arizona State University)(Barua et al., 2008), and the complexes applied to cells for 4 hours in OptiMem (Gibco, Carlsbad, CA). Media was then changed to normal growth media for two days before selection media containing 1 mg/ml G418 was added (Cellgro, Manassas, VA). After one week in selection media, cells were sorted to isolate the GFP positive population. Isolated cells were expanded for an additional two weeks in selection media before experiments were performed. For experiments, cells were plated at 2×106 cells/well in 6-well plates overnight and incubated in the absence or presence of 100 ng/ml TRAIL for 4 hours. Cell lysates were prepared for western blot analysis.
RESULTS
TRAIL-induced increases in C16-ceramide are accompanied by decreases in sphingosine and can be inhibited by fumonison B1
We have previously shown that clinically relevant concentrations of TRAIL selectively increase intracellular levels of C16-ceramide in sensitive SW480 but not resistant SW620 cells (Kelley et al., 2001; Voelkel-Johnson et al., 2005). In this study, we initially expanded the time course to determine whether changes in C16-ceramide can be detected at later time points in resistant cells. As shown in Figure 1A and Table 1, even 22 hours after TRAIL treatment, intracellular C16-ceramide levels remain unchanged in resistant SW620 cells. In contrast, C16-ceramide levels continue to increase in a time-dependent manner in TRAIL sensitive SW480 cells. In SW480 cells, the increase in C16-ceramide was accompanied by a decrease in intracellular sphingosine (Figure 1B, Table 1). These data indicated the possible involvement of ceramide synthases (CerS), which utilize sphinganine or sphingosine as substrates to generate dihydroceramide or ceramide in the de novo or salvage pathway of sphingolipid synthesis, respectively (Ogretmen & Hannun, 2004). To further investigate the possible role of CerS, we used the inhibitor fumonison B1 (FB1). As shown in Figure 1C, pretreatment with FB1 for 2 hours significantly inhibited the TRAIL-induced increase in C16-ceramide. We also observed accumulation of both sphinganine and sphingosine in SW480 cells treated with FB1 and TRAIL (data not shown). In summary, these data suggested that CerS are stimulated upon TRAIL treatment in SW480 cells.
Figure 1.
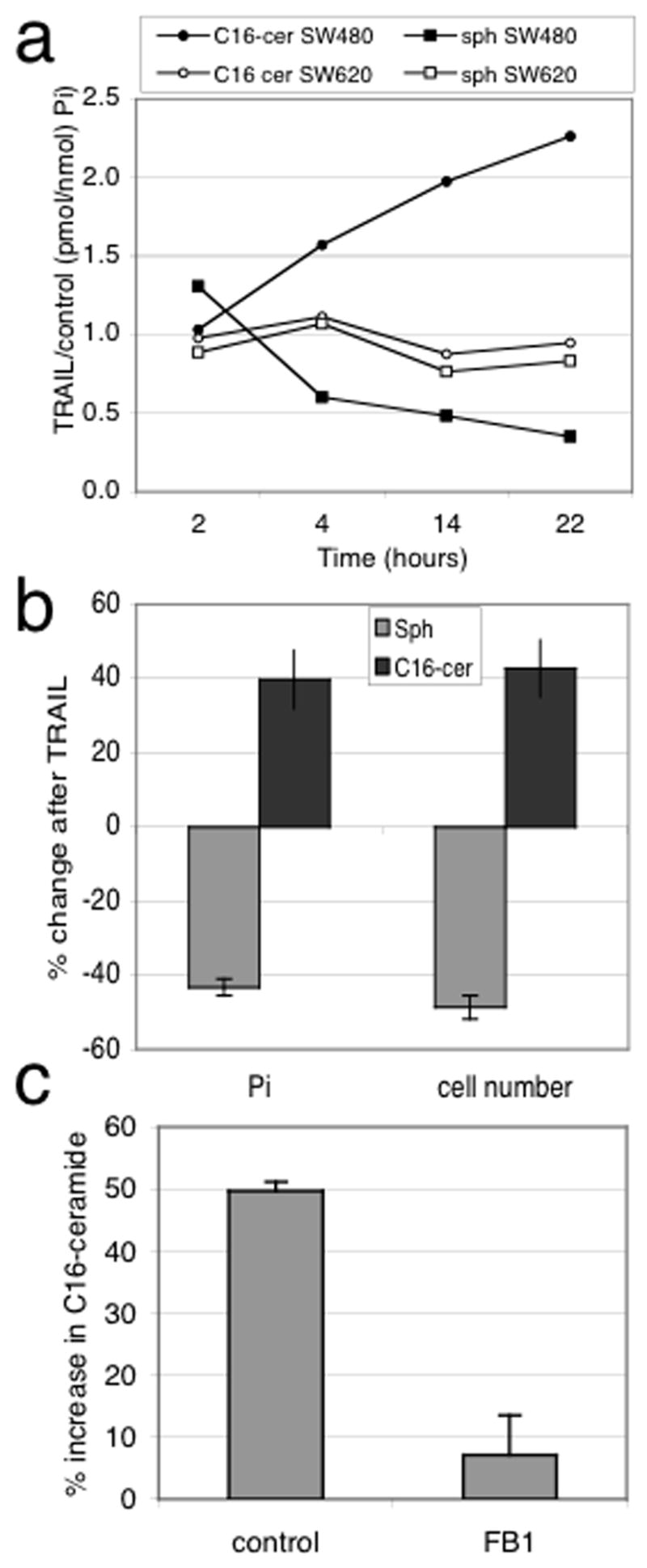
Sphingolipid responses to TRAIL. (A) Time course analysis of C16-ceramide and sphingosine in SW480 and SW620 cells. Cells were treated with 100 ng/ml TRAIL for the indicated times and duplicate samples analyzed by LC/MS. Data were normalized to Pi and TRAIL-induced changes expressed relative to untreated control cells. The mean ± sd for this experiment are shown in Table 1. (B) Changes in sphingosine levels in SW480 cells 4 hours after TRAIL treatment. Data was normalized to Pi and cell number and expressed as mean ± sd (n=3). C. SW480 cells were incubated in the absence or presence of 100 uM FB1 for 2 hours followed by exposure to 100 ng/ml TRAIL for 4 hours (n=2).
Table 1.
Effect of TRAIL on C16-ceramide and sphingosine in SW480 and SW620 cells
| Treatment | SW480* | SW620 | ||
|---|---|---|---|---|
|
|
||||
| C16-cer | Sph | C16-cer | Sph | |
| none | 2050 ± 148 | 225 ± 24 | 727 ± 89 | 30 ± 7 |
| 4h TRAIL | 3478 ± 248 | 149 ± 11 | 835 ± 65 | 25 ± 4 |
| 14h TRAIL | 3955 ± 224 | 108 ± 2 | 550 ± 49 | 29 ± 1 |
| 22h TRAIL | 4368 ± 216 | 70 ± 6 | 758 ± 36 | 25 ± 0 |
All values of TRAIL treated samples are significantly different from the control (p<0.05). Units are pmol sphingolipid/nmol Pi
SW480 and SW620 differ in expression of CerS6
In humans, six CerS homologues have been identified (Mizutani et al., 2005; Pewzner-Jung et al., 2006). These gene products synthesize ceramide with a high degree of fatty acid specificity. For example, overexpression of CerS4/LASS4/TRH-1 corresponded to increased C18-, C20- and C22-ceramide synthesis, whereas overexpression of CerS5/LASS5/TRH-4 resulted in increased generation of C16-ceramide (Riebeling et al., 2003; Venkataraman et al., 2002). The CerS homologues 5 and 6 were of particular interest for this study, since both preferentially generate C16-ceramide and we previously observed a selective increase in this ceramide species during TRAIL induced apoptosis (Mizutani et al., 2005; Riebeling et al., 2003; Voelkel-Johnson et al., 2005). In addition, CerS6 is expressed at relatively high levels in intestinal tissue (pers. comm., Dr. Anthony Futerman, Weizman Institute, Israel). In order to characterize and compare CerS5 and CerS6 expression, we performed real-time PCR and expressed mRNA levels in SW620 cells relative to levels in SW480 cells (Figure 2A). Little difference was observed (less than 2-fold), which may be explained by a recent finding that SW620 cells have global alterations in their ability to translate mRNA and suggests that mRNA levels may not properly reflect protein expression in these cells (Provenzani et al., 2006). CerS6 protein expression was evaluated by western blotting using a commercially available antibody (Supplementary Fig. 1). We found that CerS6 expression in SW620 cells was much lower than in SW480 cells, which is consistent with a lack of correlation between CerS6 mRNA and protein in SW620 cells (Figure 2B). Endogenous levels of CerS5 could not be evaluated due to lack of commercially available antibodies against this protein and subsequent studies focused on CerS6.
Figure 2.
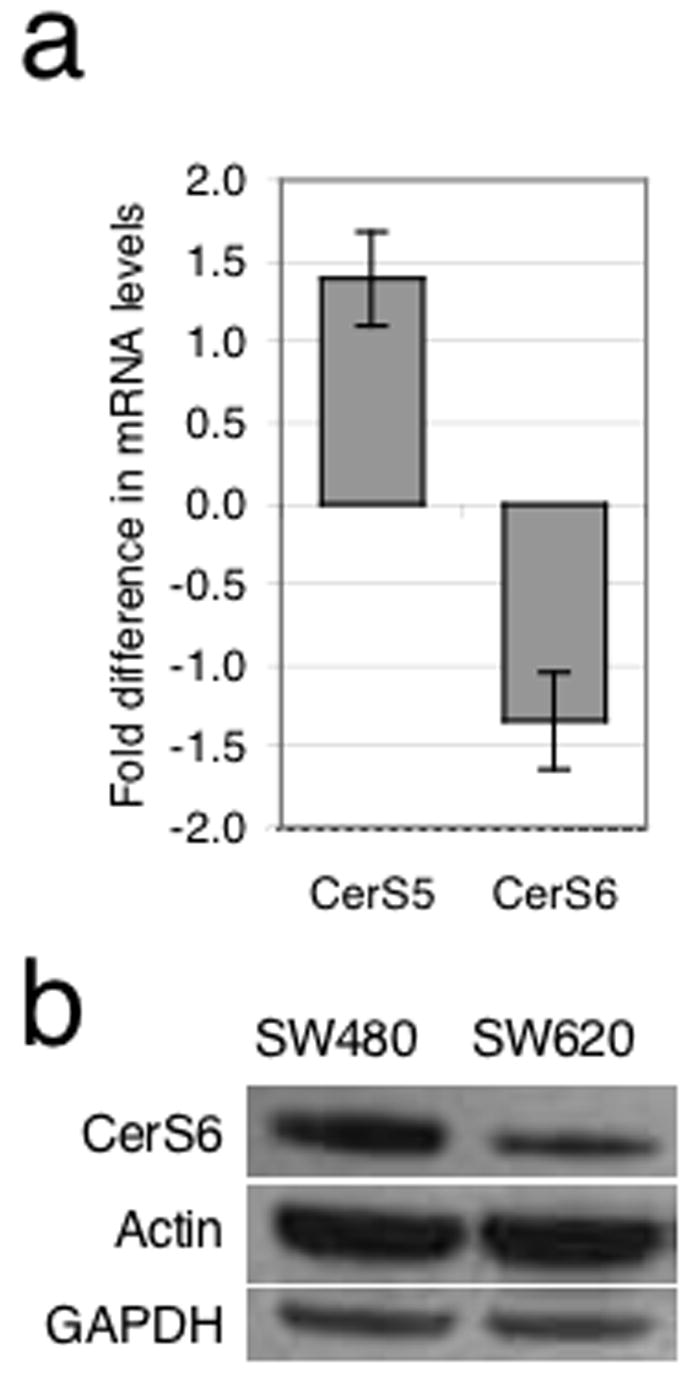
SW480 cells express more CerS6 than SW620 cells. (A) Measurement by real-time PCR showed slightly more (<2 fold more) expression of CerS5 mRNA in SW620 and slightly less (<2 fold less) CerS6 mRNA in comparison to SW480 cells, n=7. (B) Western blot analysis of CerS6 protein levels revealed a pronounced decline in SW620 cells compared to SW480 cells. Actin and GAPDH are included as loading controls.
RNAi against CerS6 results in a specific reduction in C16-ceramide
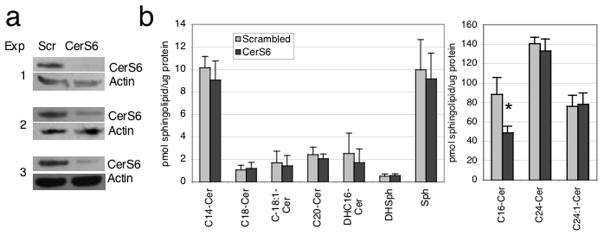
Next we investigated the effect of specifically reducing expression of CerS6 in SW480 cells. We transfected SW480 cells with scrambled and CerS6 siRNAs and determined CerS6 expression using western blotting and sphingolipid profile using LC/MS. CerS6 protein was decreased but not completely abrogated in each of the three experiments performed (Fig 3A). Since the siRNA efficiently decreases CerS6 mRNA (data not shown), residual protein expression is indicative of a long protein half-life. The sphingolipid profile did not show differences in minor ceramide species (C14, dhC16, C18, C18:1, C20) or sphingosines (Fig 3B). Among the major ceramide species, a specific decrease was detected only in C16-ceramide but not C24-ceramides (Figure 3B). These results demonstrate that downregulation of CerS6 results in a specific decrease in C16-ceramide without significantly affecting other ceramide species.
Figure 3.
Downregulation of CerS6 in SW480 cells by siRNA results in a specific decrease of C16-ceramide. SW480 cells were transfected with scrambled siRNA or siRNA against CerS6 and harvested 72 hours later for protein and LC/MS analysis. (A) Western blot analysis of samples used for LC/MS analysis. (B) Sphingolipid profile. The only significant change detected by LC/MS was a nearly 50% reduction in C16-ceramide (* p<0.05).
Decreased CerS6 expression results in resistance to TRAIL-induced apoptosis
Having confirmed that CerS6 siRNA results in a specific reduction of C16-ceramide, we investigated the effect of CerS6 knockdown on TRAIL-induced apoptosis. Cleavage of PARP, a caspase target that is frequently used as a marker of apoptosis, was diminished when CerS6 expression was reduced (Figure 4A). Analysis of chromatin condensation and nuclear degradation by Hoechst staining revealed that TRAIL treatment of SW480 cells transfected with the control siRNA resulted in chromatin condensation and fragmentation of the nucleus into a grape-like appearance, which is consistent with apoptotic morphology (Fig. 4B, supplementary Fig. 2). In contrast, SW480 cells transfected with CerS6 siRNA maintained a normal nuclear morphology upon exposure to TRAIL. Taken together, these data suggest that reduced CerS6 expression is sufficient to impair TRAIL-mediated apoptosis in SW480 cells.
Figure 4.
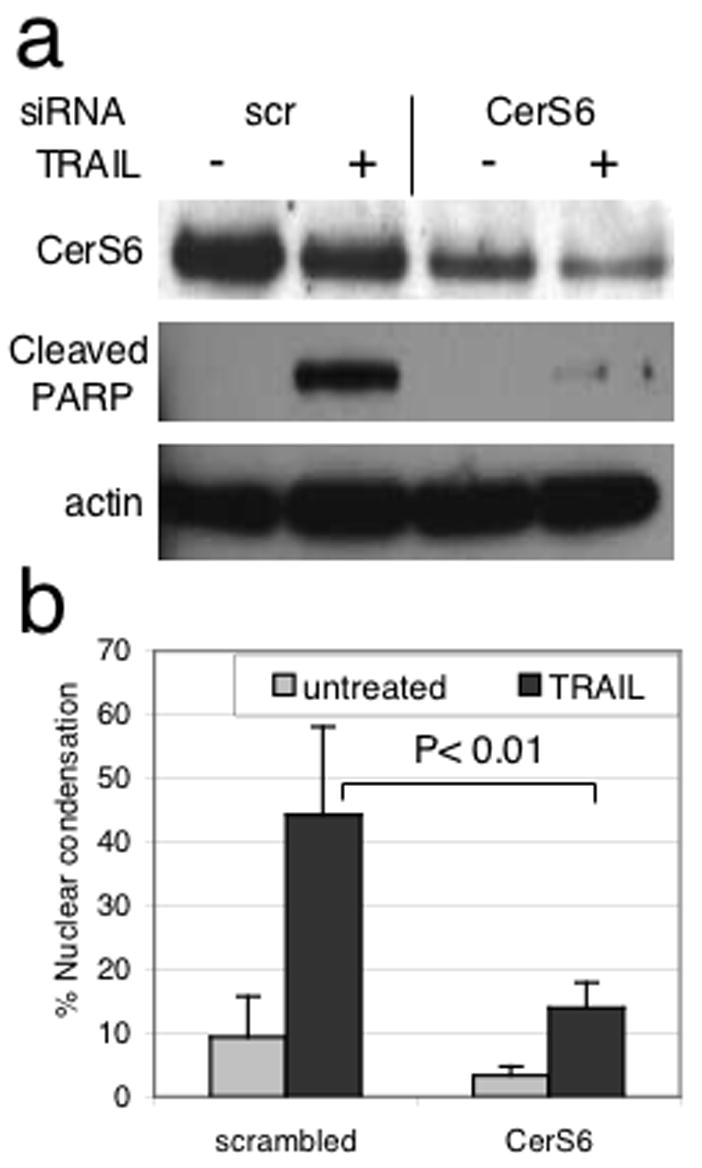
Downregulation of CerS6 in SW480 cells by siRNA results in a TRAIL-resistant phenotype. SW480 cells were transfected with siRNA and exposed to 100 ng/ml TRAIL. (A) Analysis of CerS6 expression and PARP cleavage by western blot. Actin serves as a loading control. Similar results were obtained in at least 3 independent experiments. (B) Quantitation of nuclear condensation by Hoechst stain. Condensed nuclei in three randomly chosen fields/well were counted and apoptotic nuclei expressed as a percentage of total nuclei (n=3).
CerS6 inhibits the apoptotic signal downstream of caspase-3 activation
To pinpoint the lesion in the TRAIL apoptotic pathway resulting from CerS6 downregulation, we analyzed caspase cleavage. Surprisingly, we failed to find significant differences in cleavage of caspases-8, -2, -9, or -3 regardless of CerS6 expression (Figure 5A). A decrease in CerS6 resulted only in inhibition of PARP cleavage. PARP is a known target of caspase-3 and therefore we wished to verify that the cleaved form of caspase-3 detected by Western blot was functionally active. However, downregulation of CerS6 did not result in significant differences in caspase-3/7 activity following TRAIL treatment (Figure 5B). To confirm these results, we generated SW480 cells that stably expressed a CerS6 shRNA against a different target sequence than the siRNA. An shRNA against GFP served as a control in these experiments. Consistent with data obtained with siRNA, shRNA against CerS6 also inhibited nuclear condensation and PARP cleavage but not cleavage of caspase-3 (Fig. 6, supplementary Fig. 3). Similar data were obtained with mass and different individual clones at both 8 and 22 hours after TRAIL treatment (data not shown).
Figure 5.
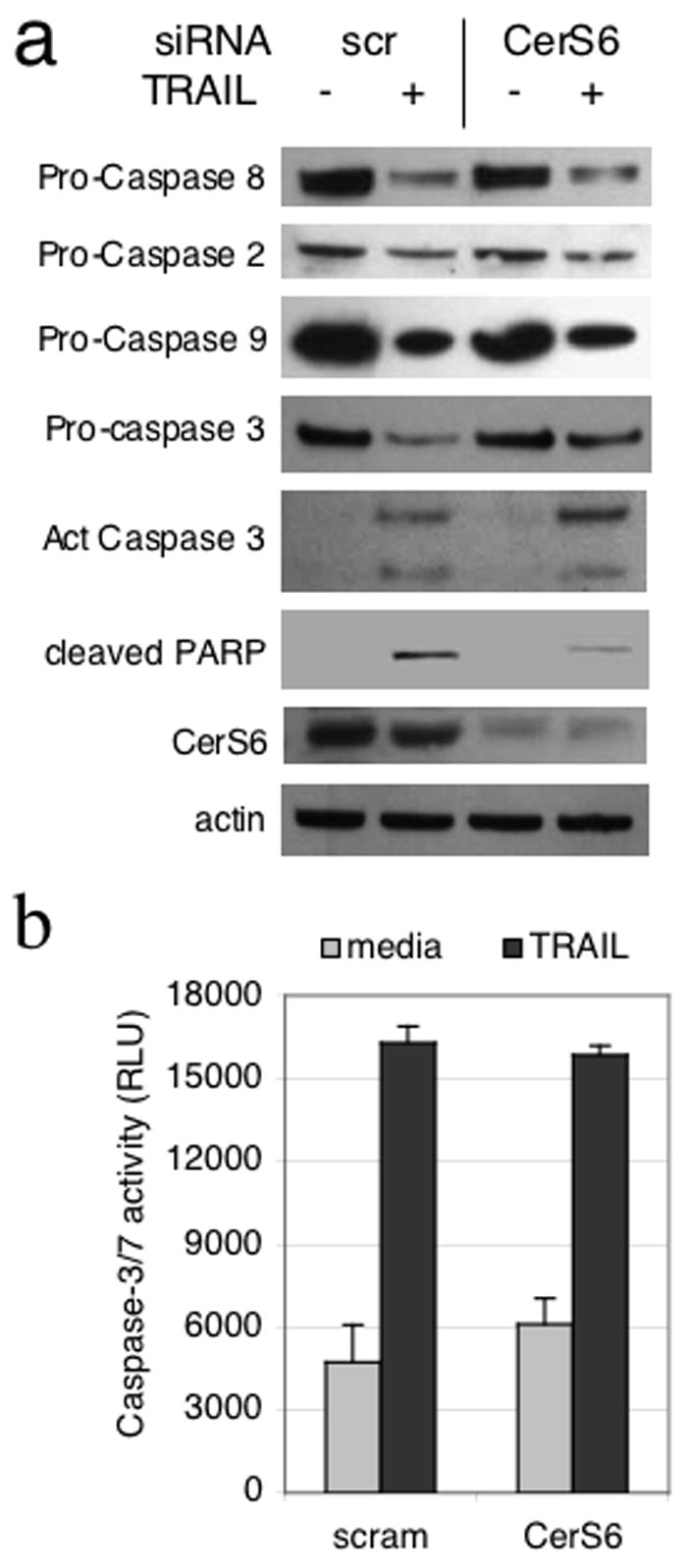
CerS6 deficiency inhibits TRAIL-induced apoptosis downstream of caspase-3 activation. (A) Western blot analysis of caspase and PARP cleavage. A representative experiment is shown. Similar results were obtained in a least two additional experiments. Actin is included as a loading control (B) Caspase-3/7 activity assay, n=2.
Figure 6.
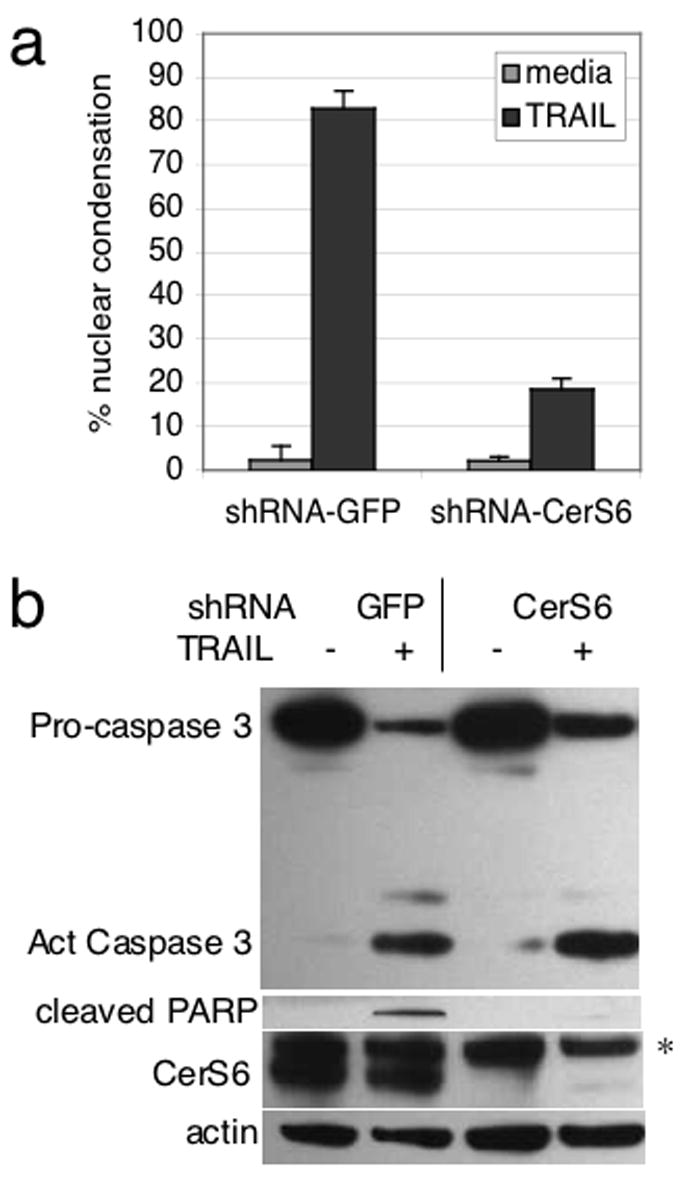
Apoptosis resistance despite caspase-3 cleavage in SW480 cells stably transfected with CerS6 shRNA. Cells were treated with 100 ng/ml TRAIL for 22 hrs. (A) Quantitation of nuclear condensation by Hoechst stain. Condensed nuclei in three randomly chosen fields/well were counted and apoptotic nuclei expressed as a percentage of total nuclei (n=2). (B) Western blot analysis. *The actin membrane was reprobed with CerS6. The upper band in the CerS6 blot is the actin signal.
Nuclear translocation of active caspase 3 is inhibited upon CerS6 downregulation
Based on our observation that cells with reduced CerS6 expression were able to maintain a normal morphology despite caspase-3 activity, we hypothesized that active caspase-3 was unable to translocate into the nucleus in these cells. To test this hypothesis we used the FLICA-DEVD assay, in which a fluorochrome caspase inhibitor binds to active caspases-3/7. In TRAIL treated cells transfected with scrambled siRNA, evidence of active caspase-3/7 coincided with visible nuclear condensation (Fig 7A and suppl.Fig 4). In contrast, in TRAIL treated cells transfected with CerS6 siRNA we observed cells in which the rhodamine signal was not accompanied by nuclear condensation. To quantitate the amount of active caspase-3 translocating into the nucleus, isolated nuclei were incubated with a FITC-conjugated antibody specific for active caspase-3 followed by flow cytometry analysis. As shown in Figure 7B, the amount of active caspase-3 in nuclei of cells transfected with CerS6 siRNA was near baseline after TRAIL treatment and significantly less than in nuclei isolated from TRAIL-treated cells transfected with control siRNA. Taken together, these results suggest that deficiency in CerS6 expression inhibits the ability of active caspase-3 to translocate into the nucleus.
Figure 7.
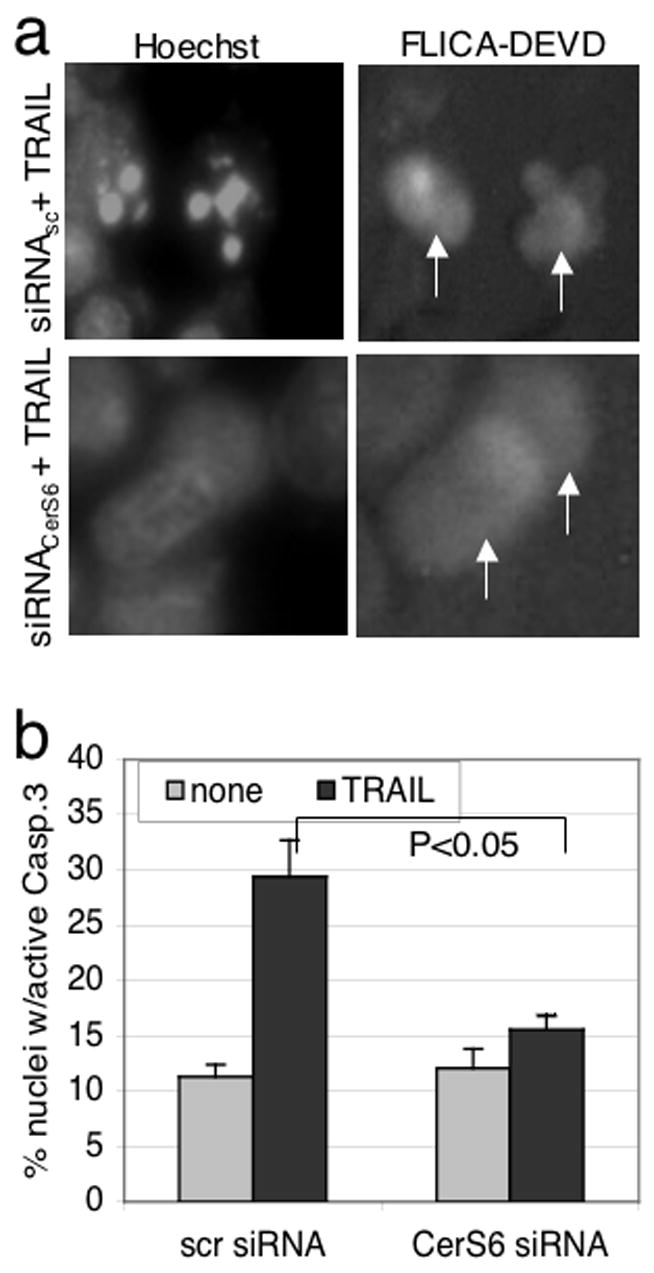
CerS6 deficiency inhibits translocation of active caspase into the nucleus. (A) Analysis of active caspase-3/7 (FLICA-DEVD) and nuclear morphology (Hoechst). Similar results were obtained in a least two additional experiments. (B) Quantitative analysis of activated caspase-3 by flow cytometry of isolated nuclei, n=2.
Exogenous CerS6 localizes primarily to the perinuclear region
Ceramide synthases have been identified as proteins that localize to the endoplasmic reticulum (Kitatani et al., 2006; Mizutani et al., 2005; Riebeling et al., 2003; Senkal et al., 2007; Venkataraman et al., 2002). The membranes of the endoplasmic reticulum and nucleus are continuous and since our results suggested a role for CerS6 in mediating caspase-3 translocation through the nuclear membrane, we were interested in determining the subcellular location of CerS6 in SW480 cells. Detection of endogenous CerS6 was hampered by non-specific signals from the LASS6 (CerS6) antibody. Thus, we transiently transfected SW480 cells with a plasmid that expresses CerS6 with an N-terminal FLAG-tag. We probed for the FLAG-tag as well as Lamin B, calreticulin, and the nuclear stain DRAQ5. In SW480 cells, Lamin B however did not localize to the nuclear membrane (suppl. Fig 5). Calreticulin, a protein found in the endoplasmic reticulum did co-localize with FLAG-tagged CerS6 but the pattern did not completely overlap. In agreement with a previous study investigating the intracellular location of CerS6, we found that the majority of the protein was detected in the perinuclear region (Fig. 8) (Mizutani et al., 2005).
Figure 8.
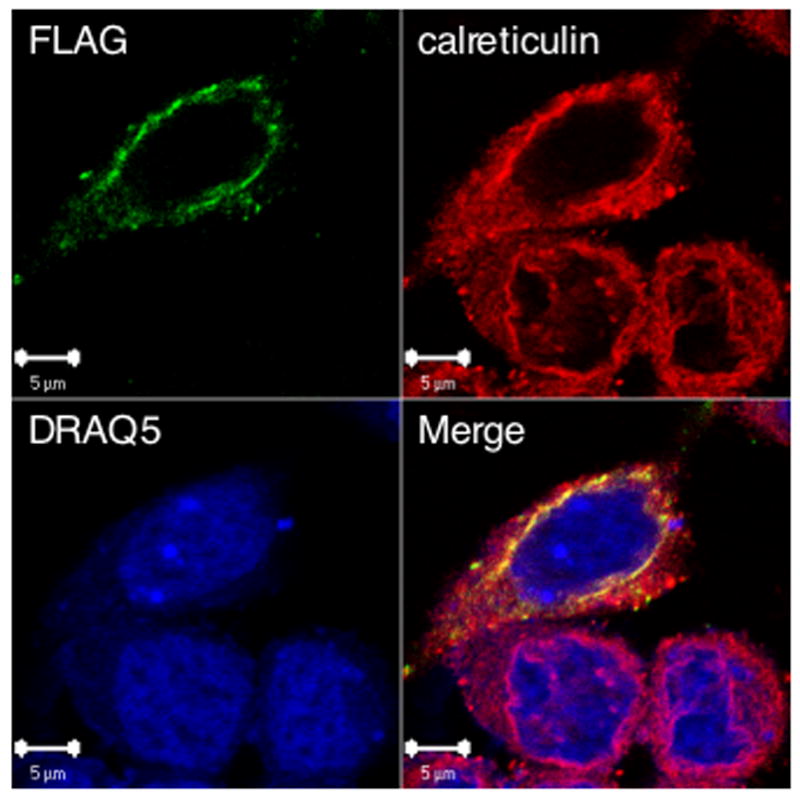
Localization of transfected CerS6 in SW480 cells. FLAG-tagged CerS6 was transiently transfected into SW480 cells and visualized by confocal microscopy using an antibody against the FLAG tag. Endoplasmic reticulum and the nucleus were visualized by staining with calreticulin or DRAQ5, respectively.
Upregulation of CerS6 in SW620 cells results in TRAIL sensitivity
The difference in TRAIL sensitivity between SW480 and SW620 cells does not appear to be mediated by significant alterations to levels of pro- or anti-apoptotic proteins with the possible exception of XIAP (Huerta et al., 2007; Ndozangue-Touriguine et al., 2008). However, in our cells we did not observe elevated expression of anti-apoptotic proteins such as cFLIP, Bcl-2 or XIAP that would explain the TRAIL resistant phenotype of SW620 cells (suppl. Fig. 6). Having observed that the downregulation of CerS6 generates a TRAIL-resistant phenotype in TRAIL sensitive SW480 cells, we investigated whether SW620 cells, which are TRAIL resistant and have reduced CerS6 expression, could be sensitized by increasing levels of CerS6. We generated a plasmid encoding CerS6 and EGFP separated by an IRES sequence, which allows the expression of two proteins from one mRNA, so that GFP serves as an indicator for transfected cells. SW620 cells were transfected with a control plasmid (pIRES2-EGFP) or pCerS6-IRES2-EGFP. Untreated and TRAIL treated cells were then analyzed morphologically. CerS6 over-expression alone did not induce morphological changes associated with apoptosis (Figure 9A). However, when cells transfected with pCerS6-IRES2-EGFP were treated with TRAIL, GFP positive cells appeared to be undergoing apoptosis (Figure 9A, arrows). Some GFP-negative cells also appeared apoptotic, indicating a possible bystander effect that may be mediated by a high local ceramide concentration.
Figure 9.
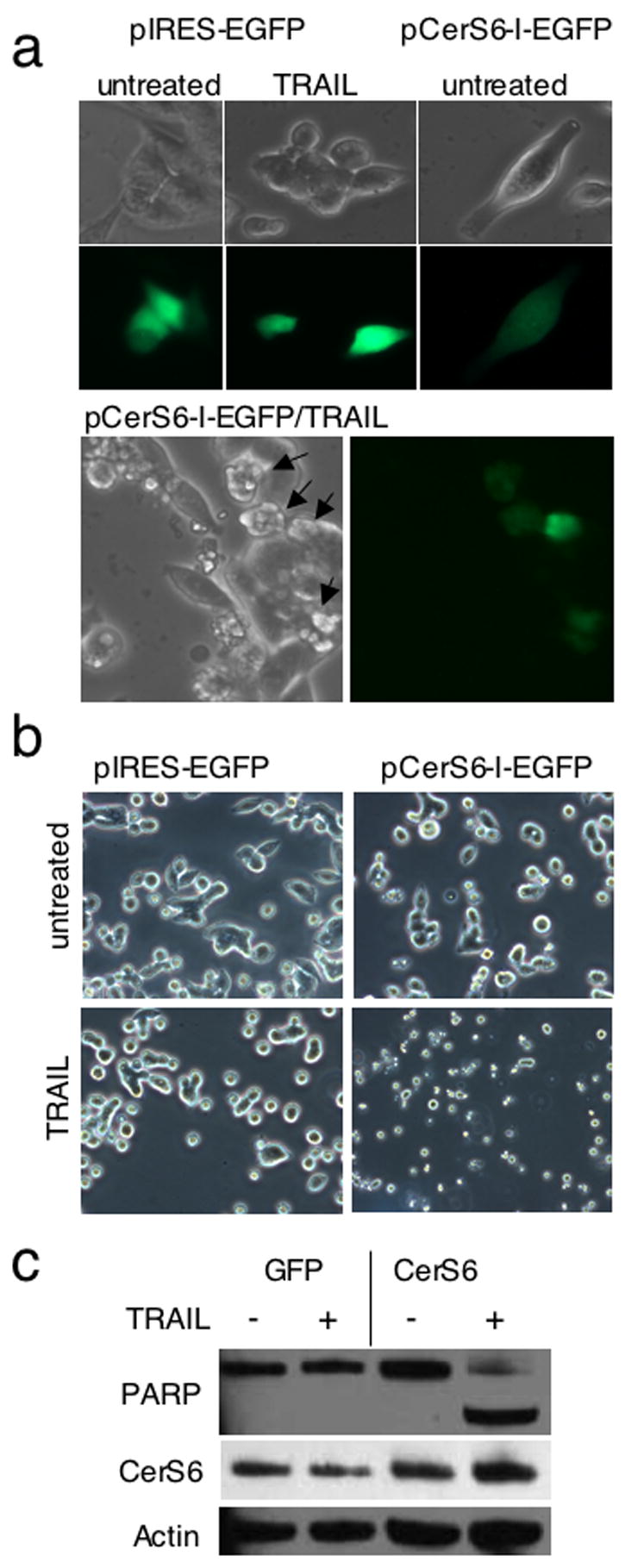
Overexpression of CerS6 in SW620 cells results in sensitization to TRAIL. (A) SW620 cells were transiently transfected with pIRES-GFP as a negative control or pCerS6-IRES-GFP. Cells were visualized at a magnification of 200X 24 hours after treatment with 100 ng/ml TRAIL was initiated. Representative images from one of three experiments are shown. (B/C) SW620 mass clones stably transfected with pIRES-GFP or pCerS6-IRES-GFP were treated with 100 ng/ml TRAIL for 4 hours and examined for morphological evidence of apoptosis (B) or PARP cleavage by western blot analysis (C). A representative experiment is shown. Similar results have been obtained at least three times.
SW620 cells transfect poorly and in order to perform a more detailed analysis of an entire cell population, we generated stable transfectants. The amount of CerS6 expression was moderately elevated in these cells, indicating that high levels of CerS6 expression may result in a survival disadvantage (Figure 9C). Consistent with the morphological observation following transient transfection, cells stably transfected with CerS6 but not with the empty plasmid acquired a TRAIL sensitive phenotype (Fig. 9B). The TRAIL sensitive phenotype was confirmed by PARP cleavage (Fig 9C).
DISCUSSION
The goal of this study was to extend our original observation that isogenic TRAIL sensitive and resistant cells differed in basal and induced generation of ceramide, which had led to our hypothesis that TRAIL resistant cells have defects in sphingolipid metabolism. In this study we found that the TRAIL-induced increase in intracellular C16-ceramide was accompanied by a decrease in sphingosine, leading us to investigate the potential involvement of ceramide synthases. We were specifically interested in further studying CerS5 and CerS6, which are the two CerS family member that preferentially generate C16-ceramide but found that mRNA levels of these enzymes did not differ significantly between the cell lines. We were unable to study endogenous CerS5 due to lack of antibodies but found that CerS6 protein expression was reduced in TRAIL resistant SW620 cells. We demonstrated that RNAi against CerS6 results in a specific decrease in C16-ceramide and that this decrease was sufficient to result in TRAIL resistance.
Ceramide has been shown to play a role in multiple subcellular compartments during apoptosis. At the plasma membrane, ceramide generation is involved in aggregation of surface receptors into “ceramide-enriched platforms”, which results in more efficient transduction of death receptor signals (Cremesti et al., 2001; Gulbins & Kolesnick, 2003). At the mitochondria, ceramide plays an important role in Bax oligomer formation (Birbes et al., 2005). Thus we were surprised to find that CerS6 downregulation did not appear to interfere with caspase-3 activation, which functions downstream of events at the plasma membrane and mitochondria. The only other protein reported to interfere with apoptosis downstream of caspase-3/7 activation is Hsp70 (Jaattela et al., 1998).
Our data suggested that reduced levels of CerS6 interfere with translocation of active caspase-3 into the nucleus. The nuclear envelope consists of a double membrane containing nuclear pore complexes responsible for selectively exchanging material between the cytoplasm and the nucleus (Tran & Wente, 2006). An earlier study revealed that inhibition of nuclear transport by microinjection of wheat-germ agglutinin inhibits Fas-induced apoptosis and caspase-3-induced nuclear changes (Yasuhara et al., 1997). It is known that active caspase-3 enters the nucleus during apoptosis but the mechanism by which this occurs is not well understood. One study suggested that the diffusion limits of nuclear pores are increased by activated caspase-9 during apoptosis to allow passive import of caspase-3 fragments (Faleiro & Lazebnik, 2000). The hypothesis that active caspase-3 enters the nucleus by passive diffusion was supported by Beckman et al. who found that active caspase-3 degrades POM, a protein which anchors nuclear pores, and a number of nuclear porins such as Nup153, Nup93, and Nup96, thereby destabilizing the pores and creating leakiness in the nuclear membrane (Beckman et al., 2004). Another group found that active caspase-3 is transported into the nucleus by a specific mechanism and later identified A-kinase-anchoring protein 95 (AKAP95) as a potential carrier of active caspase-3 (Kamada et al., 2005a; Kamada et al., 2005b). CerS6-generated C16-ceramide may influence the mechanisms responsible for active transport of caspase-3 into the nucleus or could directly influences the permeability of the nuclear membrane by increasing pore size. It has been demonstrated that nucleocytoplasmic transport is altered early during apoptosis as evidenced by increased permeability to larger size dextrans (Ferrando-May et al., 2001) but whether ceramide plays a role in this process is unknown. The ratio of ceramide to dihydroceramide, its precursor in the de novo pathway, can control the formation of pores in micelles and organelles, thus it is possible that cells with reduced basal C16-ceramide or the inability to generate C16-ceramide in response to apoptotic stimuli have impaired pore formation (Siskind & Colombini, 2000; Stiban et al., 2008). The observation that transfected CerS6 primarily localizes to the perinuclear region (this study and (Mizutani et al., 2005)) and that downregulation of CerS6 results in a lesion downstream of caspase-3 but upstream of nuclear condensation and PARP cleavage warrants further investigation of a possible role of CerS6 and/or its products in regulation of nucleocytoplasmic exchange. The development of antibodies that detect endogenous CerS6 by immunostaining will be useful to further explore the role of CerS6 in the perinuclear region. The general idea that membrane permeability plays a role in TRAIL-induced apoptosis has been examined recently in the context of leukemic T-cells, in which heat shock and alcohol treatments were used as methods to increase membrane fluidity. Both treatments resulted in sensitization to TRAIL, which could be completely abrogated by inhibiting the ceramide generation resulting from heat shock and alcohol treatments (Moulin et al., 2007). Although the sensitization methods used are not entirely specific to membrane fluidity, the results support the idea that the TRAIL apoptotic signal is strengthened by ceramide and that membrane changes are part of the enhancement.
Inhibitor studies have shown that ceramide synthases are required for apoptosis induced by androgen ablation, oxidative stress, taxol, daunorubicin, and radiation (Basnakian et al., 2005; Bose et al., 1995; Charles et al., 2001; Eto et al., 2003; Garzotto et al., 1999; Ueda et al., 2001). However, since cloning of CerS, only LASS1/CerS1 has been studied with regard to apoptosis in cancer. Alterations in CerS1 and its product C18-ceramide have previously been reported to play a role in the regulation of HNSCC pathogenesis and therapy (Karahatay et al., 2007; Koybasi et al., 2004; Senkal et al., 2007), suggesting the possibility that other members of the CerS family play similar, yet specific roles in other cancer types or tissues. For example, LASS5/CerS5 is important in the lung (Xu et al., 2005) and CerS6 is highly expressed in intestinal tissue (pers. comm., Dr. Anthony Futerman, Weizman Institute, Israel). Since ceramide levels are significantly lower in colon tumors compared to the surrounding normal mucosa (Selzner et al., 2001), it will be interesting to determine whether this decrease is associated with altered expression of ceramide synthases. We have shown that a moderate increase in CerS6 expression is sufficient for reversing TRAIL resistance. Therefore, our data suggest that restoring CerS6 expression may constitute a new therapeutic strategy for enhancing TRAIL sensitivity.
Supplementary Material
Acknowledgments
The authors would like to thank Rick Peppler and Kylie Martin for assistance with flow cytometry experiments, Dr. Jacek Bielawski of the Lipidomics Core Facility for assistance with the mass spectrophotometic characterization of ceramide profiles, and Dr. Kaushal Rege for providing the EDGE3’3 polymer, and Tejas Tirodkar for technical assistance. In addition, we would like to express our thanks to Dr. Yusuf Hannun for reviewing our data and making insightful suggestions.
This work was supported by 1P20-RR17698 NIH COBRE award to CVJ; R01 NIH/NIA AG016583 and NIH/NCI P01CA097132 awards to LMO; CA-088932, DE016572, and CA-097132 awards to BO; NIH/NGA: 1T32ES012878 "Training Program in Environmental Stress Signaling" to TM; and by a scholarship from the Abney Foundation to SWG. The MUSC Lipidomics Core Facility (NIH C06 RR018823) and the Flow Cytometry & Cell Sorting Shared Resource Facility was in part supported by MUSC and the Hollings Cancer Center.
References
- Ashkenazi A, Herbst RS. To kill a tumor cell: the potential of proapoptotic receptor agonists. J Clin Invest. 2008;118:1979–90. doi: 10.1172/JCI34359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua S, Joshi A, Banerjee A, Matthews D, Sharfstein ST, Cramer SM, et al. Parallel Synthesis and Screening of Polymers for Non-Viral Gene Delivery. Molecular Pharmaceutics. 2008 doi: 10.1021/mp800151j. Accepted pending revisions. [DOI] [PubMed] [Google Scholar]
- Basnakian AG, Ueda N, Hong X, Galitovsky VE, Yin X, Shah SV. Ceramide synthase is essential for endonuclease-mediated death of renal tubular epithelial cells induced by hypoxia-reoxygenation. Am J Physiol Renal Physiol. 2005;288:F308–14. doi: 10.1152/ajprenal.00204.2004. [DOI] [PubMed] [Google Scholar]
- Beckman M, Kihlmark M, Iverfeldt K, Hallberg E. Degradation of GFP-labelled POM121, a non-invasive sensor of nuclear apoptosis, precedes clustering of nuclear pores and externalisation of phosphatidylserine. Apoptosis. 2004;9:363–8. doi: 10.1023/b:appt.0000025813.75258.b5. [DOI] [PubMed] [Google Scholar]
- Birbes H, Luberto C, Hsu Y, El Bawab S, Hannun Y, Obeid L. A mitochondrial pool of sphingomyelin is involved in TNFalpha-induced Bax translocation to mitochondria. Biochem J. 2005;386:445–51. doi: 10.1042/BJ20041627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose R, Verheij M, Haimovitz-Friedman A, Scotto K, Fuks Z, Kolesnick R. Ceramide synthase mediates daunorubicin-induced apoptosis: an alternative mechanism for generating death signals. Cell. 1995;82:405–14. doi: 10.1016/0092-8674(95)90429-8. [DOI] [PubMed] [Google Scholar]
- Charles AG, Han TY, Liu YY, Hansen N, Giuliano AE, Cabot MC. Taxol-induced ceramide generation and apoptosis in human breast cancer cells. Cancer Chemother Pharmacol. 2001;47:444–50. doi: 10.1007/s002800000265. [DOI] [PubMed] [Google Scholar]
- Cremesti A, Paris F, Grassme H, Holler N, Tschopp J, Fuks Z, et al. Ceramide enables fas to cap and kill. Journal of Biological Chemistry. 2001;276:23954–61. doi: 10.1074/jbc.M101866200. [DOI] [PubMed] [Google Scholar]
- de Jong S, Timmer T, Heijenbrok FJ, de Vries EG. Death receptor ligands, in particular TRAIL, to overcome drug resistance. Cancer Metastasis Rev. 2001;20:51–6. doi: 10.1023/a:1013112624971. [DOI] [PubMed] [Google Scholar]
- Dohi T, Okada K, Xia F, Wilford CE, Samuel T, Welsh K, et al. An IAP-IAP complex inhibits apoptosis. J Biol Chem. 2004;279:34087–90. doi: 10.1074/jbc.C400236200. [DOI] [PubMed] [Google Scholar]
- Eto M, Bennouna J, Hunter OC, Hershberger PA, Kanto T, Johnson CS, et al. C16 ceramide accumulates following androgen ablation in LNCaP prostate cancer cells. Prostate. 2003;57:66–79. doi: 10.1002/pros.10275. [DOI] [PubMed] [Google Scholar]
- Faleiro L, Lazebnik Y. Caspases disrupt the nuclearcytoplasmic barrier. J Cell Biol. 2000;151:951–9. doi: 10.1083/jcb.151.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando-May E, Cordes V, Biller-Ckovric I, Mirkovic J, Gorlich D, Nicotera P. Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 2001;8:495–505. doi: 10.1038/sj.cdd.4400837. [DOI] [PubMed] [Google Scholar]
- Garzotto M, Haimovitz-Friedman A, Liao WC, White-Jones M, Huryk R, Heston WD, et al. Reversal of radiation resistance in LNCaP cells by targeting apoptosis through ceramide synthase. Cancer Res. 1999;59:5194–201. [PubMed] [Google Scholar]
- Gulbins E, Kolesnick R. Raft ceramide in molecular medicine. Oncogene. 2003;22:7070–7. doi: 10.1038/sj.onc.1207146. [DOI] [PubMed] [Google Scholar]
- Huerta S, Heinzerling JH, Anguiano-Hernandez YM, Huerta-Yepez S, Lin J, Chen D, et al. Modification of gene products involved in resistance to apoptosis in metastatic colon cancer cells: roles of Fas, Apaf-1, NFkappaB, IAPs, Smac/DIABLO, and AIF. J Surg Res. 2007;142:184–94. doi: 10.1016/j.jss.2006.12.551. [DOI] [PubMed] [Google Scholar]
- Jaattela M, Wissing D, Kokholm K, Kallunki T, Egeblad M. Hsp70 exerts its anti-apoptotic function downstream of caspase-3-like proteases. Embo J. 1998;17:6124–34. doi: 10.1093/emboj/17.21.6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada S, Kikkawa U, Tsujimoto Y, Hunter T. A-kinase-anchoring protein 95 functions as a potential carrier for the nuclear translocation of active caspase 3 through an enzyme-substrate-like association. Mol Cell Biol. 2005a;25:9469–77. doi: 10.1128/MCB.25.21.9469-9477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada S, Kikkawa U, Tsujimoto Y, Hunter T. Nuclear translocation of caspase-3 is dependent on its proteolytic activation and recognition of a substrate-like protein(s) J Biol Chem. 2005b;280:857–60. doi: 10.1074/jbc.C400538200. [DOI] [PubMed] [Google Scholar]
- Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, et al. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 2007;256:101–11. doi: 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley SK, Harris LA, Xie D, Deforge L, Totpal K, Bussiere J, et al. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: Characterization of in vivo efficacy, pharmacokinetics, and safety. Journal of Pharmacology & Experimental Therapeutics. 2001;299:31–38. [PubMed] [Google Scholar]
- Kitatani K, Idkowiak-Baldys J, Bielawski J, Taha TA, Jenkins RW, Senkal CE, et al. Protein kinase C-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 MAPK. J Biol Chem. 2006;281:36793–802. doi: 10.1074/jbc.M608137200. [DOI] [PubMed] [Google Scholar]
- Koschny R, Walczak H, Ganten TM. The promise of TRAIL--potential and risks of a novel anticancer therapy. J Mol Med. 2007;85:923–35. doi: 10.1007/s00109-007-0194-1. [DOI] [PubMed] [Google Scholar]
- Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, et al. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. Journal of Biological Chemistry. 2004;279:44311–9. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- Kroesen BJ, Jacobs S, Pettus BJ, Sietsma H, Kok JW, Hannun YA, et al. BcR-induced apoptosis involves differential regulation of C16 and C24-ceramide formation and sphingolipid-dependent activation of the proteasome. Journal of Biological Chemistry. 2003;278:14723–31. doi: 10.1074/jbc.M210756200. [DOI] [PubMed] [Google Scholar]
- MacFarlane M. TRAIL-induced signalling and apoptosis. Toxicology Letters. 2003;139:89–97. doi: 10.1016/s0378-4274(02)00422-8. [DOI] [PubMed] [Google Scholar]
- Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–71. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, Carpentier S, Levade T, Arrigo AP. Potential roles of membrane fluidity and ceramide in hyperthermia and alcohol stimulation of TRAIL apoptosis. Apoptosis. 2007;12:1703–20. doi: 10.1007/s10495-007-0096-2. [DOI] [PubMed] [Google Scholar]
- Ndozangue-Touriguine O, Sebbagh M, Merino D, Micheau O, Bertoglio J, Breard J. A mitochondrial block and expression of XIAP lead to resistance to TRAIL-induced apoptosis during progression to metastasis of a colon carcinoma. Oncogene. 2008 doi: 10.1038/onc.2008.197. [DOI] [PubMed] [Google Scholar]
- Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–16. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- Ogretmen B, Pettus BJ, Rossi MJ, Wood R, Usta J, Szulc Z, et al. Biochemical mechanisms of the generation of endogenous long chain ceramide in response to exogenous short chain ceramide in the A549 human lung adenocarcinoma cell line. Role for endogenous ceramide in mediating the action of exogenous ceramide. J Biol Chem. 2002;277:12960–9. doi: 10.1074/jbc.M110699200. [DOI] [PubMed] [Google Scholar]
- Peter ME. The flip side of FLIP. Biochem J. 2004;382:e1–3. doi: 10.1042/BJ20041143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–5. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006;27:1323–33. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- Riebeling C, Allegood JC, Wang E, Merrill AH, Jr, Futerman AH. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. Journal of Biological Chemistry. 2003;278:43452–9. doi: 10.1074/jbc.M307104200. [DOI] [PubMed] [Google Scholar]
- Selzner M, Bielawska A, Morse MA, Rudiger HA, Sindram D, Hannun YA, et al. Induction of apoptotic cell death and prevention of tumor growth by ceramide analogues in metastatic human colon cancer. Cancer Res. 2001;61:1233–40. [PubMed] [Google Scholar]
- Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, et al. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol Cancer Ther. 2007;6:712–22. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- Sharpe J, Arnoult D, Youle R. Control of mitochondrial permeability by Bcl-2 family members. Biochimica et Biophysica Acta. 2004;1644:107–113. doi: 10.1016/j.bbamcr.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Siskind LJ, Colombini M. The lipids C2- and C16-ceramide form large stable channels. Implications for apoptosis. J Biol Chem. 2000;275:38640–4. doi: 10.1074/jbc.C000587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiban J, Caputo L, Colombini M. Ceramide synthesis in the endoplasmic reticulum can permeabilize mitochondria to proapoptotic proteins. J Lipid Res. 2008;49:625–34. doi: 10.1194/jlr.M700480-JLR200. [DOI] [PubMed] [Google Scholar]
- Thomas RL, Jr, Matsko CM, Lotze MT, Amoscato AA. Mass spectrometric identification of increased C16 ceramide levels during apoptosis. J Biol Chem. 1999;274:30580–8. doi: 10.1074/jbc.274.43.30580. [DOI] [PubMed] [Google Scholar]
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–53. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Ueda N, Camargo SM, Hong X, Basnakian AG, Walker PD, Shah SV. Role of ceramide synthase in oxidant injury to renal tubular epithelial cells. J Am Soc Nephrol. 2001;12:2384–91. doi: 10.1681/ASN.V12112384. [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, et al. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. Journal of Biological Chemistry. 2002;277:35642–9. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- Voelkel-Johnson C, Hannun YA, El-Zawahry A. Resistance to TRAIL is associated with defects in ceramide signaling that can be overcome by exogenous C6-ceramide without requiring down-regulation of cellular FLICE inhibitory protein. Mol Cancer Ther. 2005;4:1320–7. doi: 10.1158/1535-7163.MCT-05-0086. [DOI] [PubMed] [Google Scholar]
- Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem. 2005;94:1078–87. doi: 10.1002/jcb.20409. [DOI] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity. 1995;3:673–82. doi: 10.1016/1074-7613(95)90057-8. [DOI] [PubMed] [Google Scholar]
- Wilson MR. Apoptosis: unmasking the executioner. Cell Death Differ. 1998;5:646–52. doi: 10.1038/sj.cdd.4400394. [DOI] [PubMed] [Google Scholar]
- Xu Z, Zhou J, McCoy DM, Mallampalli RK. LASS5 is the predominant ceramide synthase isoform involved in de novo sphingolipid synthesis in lung epithelia. J Lipid Res. 2005;46:1229–38. doi: 10.1194/jlr.M500001-JLR200. [DOI] [PubMed] [Google Scholar]
- Yasuhara N, Eguchi Y, Tachibana T, Imamoto N, Yoneda Y, Tsujimoto Y. Essential role of active nuclear transport in apoptosis. Genes Cells. 1997;2:55–64. doi: 10.1046/j.1365-2443.1997.1010302.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.