Abstract
RNA amplification is a series of molecular manipulations designed to amplify genetic signals from small quantities of starting materials (including single cells and homogeneous populations of individual cell types) for microarray analysis and other downstream genetic methodologies. A novel methodology named terminal continuation (TC) RNA amplification has been developed in this laboratory to amplify RNA from minute amounts of starting material. Briefly, an RNA synthesis promoter is attached to the 3′ and/or 5′ region of cDNA utilizing the TC mechanism. The orientation of amplified RNAs is “antisense” or a novel “sense” orientation. TC RNA amplification is utilized for many downstream applications, including gene expression profiling, microarray analysis, and cDNA library/subtraction library construction. Input sources of RNA can originate from a myriad of in vivo and in vitro tissue sources. Moreover, a variety of fixations can be employed, and tissues can be processed for histochemistry or immunocytochemistry prior to microdissection for TC RNA amplification, allowing for tremendous cell type and tissue specificity of downstream genetic applications.
Keywords: expression profiling, functional genomics, IVT (in vitro transcription), microarray, postmortem human brain, RNA amplification
1 Introduction
Conventional molecular biology techniques allow for gene expression analysis in a wide variety of paradigms and experimental systems. Methods include in situ hybridization (RNA detection), Northern analysis (RNA detection), polymerase chain reaction (PCR; DNA detection), reverse transcription-PCR (RT-PCR; RNA detection), ribonuclease (RNase) protection assay, and Southern analysis (DNA detection), among others. Most of these methods evaluate the abundance of individual elements one at a time (or a few at a time). Advancements in high-throughput genomic methodologies now enable the assessment of dozens to hundreds to thousands of genes simultaneously in a coordinated manner [1].
RNA can originate from a variety of in vivo and in vitro sources. Fresh, frozen, and fixed tissues are useful to varying degrees, depending on the paradigm and tissue quality. Many laboratories isolate genomic DNA and total RNA from paraffin-embedded fixed tissues as well as fresh and frozen tissues [2–6]. In addition, genetic material preserved by fixation is a superior resource because fresh or frozen tissues frequently are not available, whereas archived fixed human and animal model tissues exist in many tissue banks and individual laboratories. With mRNA as starting material, it cannot be emphasized enough the importance of preservation of RNA integrity in tissues and cells. RNA species are particularly sensitive to degradation by RNases. Because they play an important role in nucleic acid metabolism, RNases are found in virtually every cell type [7]. RNases degrade RNA species through endonuclease and exonuclease activity. RNases are quite stable and retain their activity over a broad pH range [7, 8]. Thus, RNase-free precautions need to be taken prior to and during all RNA amplification procedures. All biological samples require prompt handling, either through rapid RNA extraction, flash freezing, or fixation to minimize the degradation of intracellular RNAs by RNases.
Significant discourse centers on devising optimal methods to prepare brain tissues for downstream genetic analyses. Consensus on which protocol should be utilized has not yet been achieved. RNAs have been mined from tissue samples using cross-linking fixatives, including 10% neutral buffered formalin and 4% paraformaldehyde, as well as precipitating fixatives, such as 70% ethanol buffered with 150 mM sodium chloride [3, 4, 9, 10]. A useful method for assessing RNA quality in tissue sections prior to performing expression profiling studies is acridine orange (AO) histofluorescence. AO is a fluorescent dye that intercalates selectively into nucleic acids and has been used to detect RNA and DNA in brain tissues [11–14]. AO can be employed in combination with immunocytochemistry to identify cytoplasmic RNAs and specific antigens of interest and is compatible with confocal microscopy [15, 16]. In brain tissue sections, AO-positive neurons are in contrast to the pale background of white matter tracts that lack abundant nucleic acids. Nonneuronal cells tend to have less AO histofluorescence than neurons and brain tumor cells [17]. Importantly, individual RNA species (e.g., mRNA, rRNA, and tRNA) cannot be delineated by AO histofluorescence. Rather, this method provides a diagnostic test employed on adjacent tissue sections to ensure the likelihood that an individual case has abundant RNA prior to performing expensive expression profiling studies. A more thorough examination of RNA quality can be obtained via bioanalysis (e.g., 2100 Bioanalyzer, Agilent Technologies), which utilizes capillary gel electrophoresis to detect RNA quality and quantitate relative abundance [18–20]. Bioanalysis displays the analytical assessment of RNAs in an electropherogram or digital gel format, with relatively high sensitivity.
Microarray analysis has emerged as a useful and relatively cost-effective tool to assess transcript levels in a myriad of systems and paradigms. A disadvantage to these high-throughput technologies is a requirement for significant amounts of high-quality input sources of RNA for increased sensitivity and reproducibility. Whole organism and regional studies can generate significant input amounts of RNA species without any amplification procedures [21, 22]. Unfortunately, the quantity of RNA harvested from a single cell, estimated to be approximately 0.1–1.0 picograms, is not sufficient for standard RNA extraction procedures [23–25]. As a result, molecular-based methods have been developed to increase the amount of input RNA for downstream genetic analyses, including exponential PCR-based analyses and linear RNA amplification procedures. This protocol describes one RNA amplification procedure, terminal continuation (TC) RNA amplification.
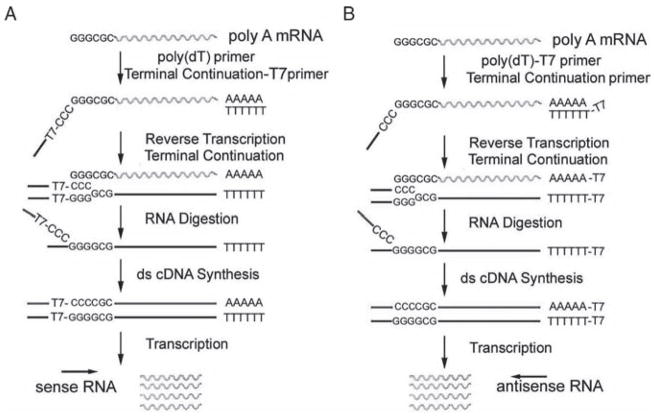
A new procedure was developed in our laboratory, TC RNA amplification (Fig. 10.1) [18, 19, 26]. TC RNA amplification of genetic signals includes synthesizing the first-strand cDNA complementary to the mRNA template, subsequently generating second-strand cDNA complementary to the first-strand cDNA, and finally in vitro transcription (IVT) using the double-stranded cDNA as a template [18, 19, 26]. First-strand cDNA synthesis complementary to the template mRNA entails the use of two oligonucleotide primers, a first-strand poly d(T) primer and a TC primer. The poly d(T) primer is similar to conventional primers that exploit the poly(A) sequence present on most mRNAs. The TC primer contains a span of three cytidine triphosphates (CTPs) or guanosine triphosphates (GTPs) at the 3′ terminus [18]. Adenosine triphosphates (ATPs) or thymidine triphosphates (TTPs) do not perform well as constituents of the TC primer [20]. In this configuration, second-strand cDNA synthesis can be initiated by annealing a second oligonucleotide primer complementary to the attached oligonucleotide [18], and can be performed with robust DNA polymerases, such as Taq polymerase. One round of amplification is sufficient for downstream genetic analyses [18, 26]. Additionally, TC RNA transcription can be performed using a promoter sequence (e.g., T7, T3, or SP6) attached to either the 3′ or 5′ oligonucleotide primers. Therefore, transcript orientation can be in an antisense orientation (similar to conventional amplified antisense RNA methods) when the bacteriophage promoter sequence is placed on the first-strand poly d(T) primer, or in a sense orientation when the promoter sequence is attached to the TC primer, depending on the design of the experimental paradigm (Fig. 10.1) [19, 27]. Regional and single cell gene expression studies within the brains of animal models and human postmortem brain tissues have been performed via microarray analysis coupled with TC RNA amplification (Fig. 10.2) [1, 14, 27–31].
Fig. 10.1.
Overview and analysis of the TC RNA amplification method. (A) A TC primer (containing a bacteriophage promoter sequence for sense orientation) and a poly d(T) primer are added to the mRNA population to be amplified. First-strand synthesis occurs as an mRNA-cDNA hybrid is formed after reverse transcription and terminal continuation of the oligonucleotide primers. Following RNase H digestion to remove the original mRNA template strand, second-strand synthesis is performed using Taq polymerase. The resultant double-stranded product is utilized as template for IVT, yielding high-fidelity, linear RNA amplification of sense orientation (rippled lines). (B) Schematic similar to A, illustrating the TC RNA amplification procedure amplifying RNA in the antisense orientation (rippled lines). Adapted from Ginsberg 2005 [26]
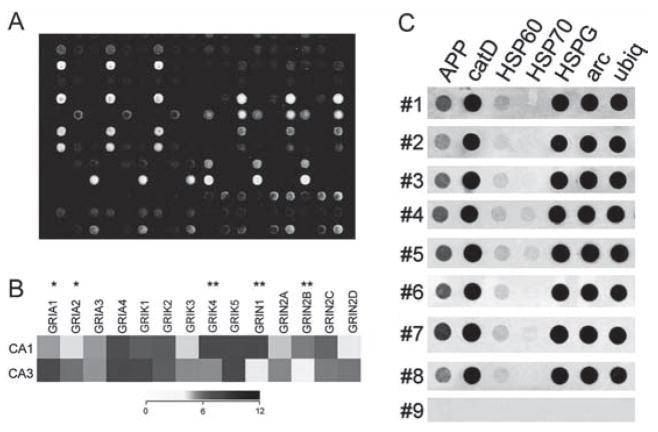
Fig. 10.2.
Representative array platforms illustrating utility of RNA amplification procedures for single cell and population cell analysis. (A) Spotted cDNA array platform using RNA amplified from individual hippocampal CA1 pyramidal neurons from normal control brains and from neurofibrillary tangle (NFT) bearing CA1 neurons from Alzheimer’s disease (AD) patients.
(B) Dendrogram demonstrating relative expression levels of representative genes in CA1 and CA3 pyramidal neurons microaspirated from human hippocampus. A heatmap matrix plot illustrates relative expression levels for individual glutamate receptor transcripts in CA1 and CA3 neurons. A single asterisk indicates a significant increase in expression in CA3 neurons as compared to CA1 neurons and a double asterisk denotes a significant increase in expression in CA1 neurons as compared to CA3 neurons. Key: GRIA 1, alpha-amino-3-hydroxy-5-methyl-4-isoxa-zolepropionate receptor 1 (AMPA1); GRIA2, AMPA2, GRIA3, AMPA3, GRIA4, AMPA4; GRIK1, kainate (KA) receptor GluR5; GRIK2, KA GluR6; GRIK3, KA GluR7; GRIK4, KA receptor KA1; GRIK5, KA receptor KA2; GRIN1, N-methyl D-aspartate receptor 1 (NR1); GRIN2A, NR2A; GRIN2B, NR2B; GRIN2C, NR2C; GRIN2D, NR2D. Adapted from Ginsberg and Che 2005 [32].
(C) Single-cell array analysis of human CA1 pyramidal neurons using custom-designed cDNA arrays and TC RNA amplification. Representative arrays illustrate a wide dynamic range of hybridization signal intensities for eight human postmortem cases (numbers 1–8). The negative control (number 9) is a single CA1 pyramidal neuron from the first case (number 1) that does not have the primers necessary for TC RNA amplification. In addition, a moderate variation of gene level expression across the eight human cases is observed, indicating the utility of using postmortem human samples for normative and neuropathological investigations. Key: APP, amyloid-β precursor protein; catD, cathepsin D; HSP60, heat shock protein 60; HSP70, heat shock protein 70; arc, activity regulated cytoskeletal-associated protein; ubiq, ubiquitin thiolesterase. Adapted from Ginsberg and Che 2002 [10]
2 Materials
The methodology described here is a step-by-step protocol for TC RNA amplification as described by Che and Ginsberg [18, 19]. For clarity, the protocol begins at the point where cells are captured via laser-capture microdissection (LCM) or a microaspiration strategy and follows through IVT using biotinylated, fluorescent, or radioactive methods to label the TC RNA amplified products (see Notes 1 and 2).
2.1 Isolation of RNA
Trizol reagent (Invitrogen, Carlsbad, CA), stored at 4 °C.
25:24:1 (v/v) Phenol:chloroform:isoamyl alcohol (Invitrogen), stored at 4 °C.
5 mg/mL linear acrylamide (Ambion, Austin, TX), stored at −20 °C.
2.2 First-Strand cDNA Synthesis
First strand synthesis primers: 10 ng/μL poly d(T) and 10 ng/μL TC primer (Table 10.1).
-
Freshly prepared (on wet ice) reverse transcription (RT) master mix (see Note 2), comprising (for each RNA sample)
4 μL of 5 × first-strand buffer (Invitrogen).
1 μL of 4 × 10 mM dNTPs (Invitrogen).
1 μL of 0.1 M dithiothreitol (DTT).
0.5 μL of 20 U/μL Superase-in RNase inhibitor (Ambion).
5.5 μL of 18.2 MΩ RNase-free water.
200 U/μL Superscript III (Invitrogen); stored at −20 °C.
Table 10.1.
Representative oligonucleotide sequences utilized for the poly d(T) and TC primers for the TC RNA amplification method
| Antisense RNA orientation |
| First-strand synthesis primer (66 bp): 3′-AAA CGA CGG CCA GTG AAT TGT AAT ACG ACT CAC TAT AGG CGC TTT TTT TTT TTT TTT TTT TTT TTT -5′ |
| TC primer (17 bp): 5′-TAT CAA CGC AGA GTC CC -3′ |
| Sense RNA orientation |
| First-strand synthesis primer (18 bp): 3′-TTT TTT TTT TTT TTT TTT -5′ |
| TC-T7 primer (51 bp): 5′-AAA CGA CGG CCA GTG AAT TGT AAT ACG ACT CAC TAT AGG CGC GAG AGC CCC -3′ |
2.3 Second-Strand cDNA Synthesis
-
Freshly prepared (on wet ice) second-strand master mix, comprising (for each first-strand cDNA sample)
10 μL of 10 × PCR buffer, containing 15 mM MgCl2 (PE Biosystems, Foster City, CA).
0.5 μL of 1 U/μL RNase H (Invitrogen).
69 μL of 18.2 MΩ RNase-free water.
5 U/μL Taq polymerase (PE Biosystems), stored at −20 °C.
2.4 Double-Stranded cDNA Preparation
25:24:1 (v/v/v) Phenol:chloroform:isoamyl alcohol, stored at 4 °C.
50 mL conical tubes (Falcon) filled with 18.2 MΩ RNase-free water.
0.025 μm membrane filters for drop dialysis (VSWP01300, Millipore, Billerica, MA).
2.5 IVT for TC RNA Amplification: Biotinylation/Fluorescent Probe Labeling
Freshly prepared (on wet ice) IVT MASTER Mix, comprising (for each double-stranded cDNA sample)
4 μL of 10 × RNA amplification buffer (Ambion; see Note 3).
4 μL of 10 × Biotin-labeled ribonucleotides (Enzo Life Sciences, Farmingdale, NY; see Note 4).
4 μL of 10 × DTT (Invitrogen).
4 μL of 10 × RNase inhibitor mix (Enzo).
12 μL of 18.2 MΩ RNase-free water.
2 μL of 1,000 U/μL T7 RNA polymerase (Epicentre, Madison, WI).
2.6 IVT for TC RNA Amplification: Radioactive Probe Labeling
-
Freshly prepared (on wet ice) IVT master mix, comprising (for each double-stranded cDNA sample)
2 μL of 10 × RNA amplification buffer (Ambion).
1 μL of 0.1 M DTT (Invitrogen).
2 μL of 3 × 2.5 mM rATP, rCTP, and rGTP (Invitrogen).
2 μL of 100 μM UTP (Invitrogen).
0.5 μL of 20 U/μL Superase-in RNase inhibitor (Ambion).
20 mCi/mL 33P-UTP (GE Healthcare, Piscataway, NJ).
1,000 U/μL T7 RNA polymerase (Epicentre, Madison, WI).
3 Methods
3.1 Isolation of RNA
Add 250 μL of Trizol reagent to the 500 μL thin-wall PCR tube that will receive the microdissected regions or cells, acquired via LCM, and keep on wet ice.
Invert the tube so that the Trizol reagent bathes the microdissected material, and keep on wet ice (samples can be stored at −80 °C at this juncture for future use).
Add 50 μL of phenol:chloroform:isoamyl alcohol, vortex vigorously for 15 s and centrifuge at 12,000 g for 15 min at 4 °C.
Collect the clear upper aqueous phase, containing RNA, by aspirating with a pipette.
Add 125 μL of 100% (v/v) 2-propanol and 5 μL of linear acrylamide to precipitate the RNA from the aqueous phase (store at −80 °C to precipitate RNA if desired).
Vortex vigorously for 15 s and centrifuge at 12,000 g for 15 min at 4 °C.
Decant the supernatant, by inverting the tube, being careful not to dislodge the pellet.
Add 250 μL of 75% (v/v) ethanol to wash RNA pellet. Vortex vigorously for 15 s and centrifuge at 7,500 g for 5 min at 4 °C.
Decant the supernatant, by inverting the tube, being careful not to dislodge the pellet.
Air dry the sample by inverting the tube for 5 min in a fume hood (see Note 5).
Resuspend the pellet in 5 μL of 18.2 MΩ RNase-free water.
3.2 First-Strand cDNA Synthesis
Add 1 μL of first-strand synthesis primer (Table 10.1) to a 5 μL RNA sample. Pulse centrifuge for 10 s.
Heat the mixture for 2 min at 75 °C. Pulse centrifuge for 10 s and place on ice.
Prepare the RT master mix (on wet ice; see Note 2).
Warm the RT master mix (without primers or Superscript III) for 2 min at 50 °C.
Heat denature an aliquot (1 μL for each RNA sample) of 10 ng/μL TC primer (Table 10.1) for 2 min at 70 °C, place on ice for several min and add to the RT master mix (see Note 6).
Add an aliquot of Superscript III (1 μL for each RNA sample) to the RT master mix (see Note 7). The RT master mix should constitute 14 μL per RT reaction.
Add the RT master mix to the 6 μL of sample, pipette vigorously, and centrifuge briefly. Incubate the mixture for 60 min at 50 °C.
Inactivate the Superscript III by heating the reaction mixture at 65 °C for 15 min.
Centrifuge briefly and cool immediately on wet ice. Samples can be stored at −20 °C for a short time, or at −80 °C for the long term.
3.3 Second-Strand cDNA Synthesis
Prepare the second-strand master mix (on wet ice).
Dispense 79.5 μL (for each second-strand synthesis reaction) of second-strand master mix into a 0.5-mL thin-wall PCR tube.
Add 20 μL of first-strand cDNA to the second-strand master mix and mix thoroughly with a pipette tip.
Place in a thermal cycler (see Note 8), programmed with the following second-strand cDNA synthesis protocol: 37 °C, 10 min (RNA degradation); 95 °C, 3 min (hot-start denaturation); 60 °C, 3 min (annealing); 75 °C, 30 min (elongation).
Start the thermocycle program. As soon as the block temperature reaches 95 °C, pause the reaction (this is the hot-start) and (using a dedicated PCR pipette) add 0.5 μL (for each reaction) of Taq polymerase.
Mix thoroughly with a pipette tip (see Note 9).
Continue the second-strand synthesis program by pushing the Continue function.
Once the program is complete, centrifuge briefly and store at −20 °C for the short term or at −80 °C for the long term.
3.4 Double-Stranded cDNA Preparation
To each sample, add 100 μL of 5 M ammonium acetate and 170 μL of phenol: chloroform:isoamyl alcohol.
Vortex vigorously for 30 s, centrifuge at 14,000 g for 5 min at 22 °C, and collect the upper aqueous phase in a fresh 1.7 mL microcentrifuge tube.
To precipitate each sample, add 1mL of 100% (v/v) ice-cold ethanol, and centrifuge at 14,000 g for 30 min at 4 °C.
Carefully discard the supernatant by inverting the tube and blotting on a piece of lint-free tissue. Air dry the pellet, in an inverted position, for 5 min in a fume hood.
Resuspend the double-stranded cDNA pellet in 20 μL of 18.2 MΩ RNase-free water.
Fill a 50-mL conical tube with 18.2 MΩ RNase-free water and float one 0.025-μm millipore membrane filter on the surface of the water (see Note 10). Load a cDNA sample onto the center of the floating membrane and carefully place the cap back on the conical tube. Allow it to stand undisturbed for 4 h at 22 °C.
Carefully remove the drop dialyzed sample into a new microfuge tube using a pipette tip.
Centrifuge briefly and store at −20 °C for the short term or at −80 °C for the long term.
3.5 IVT for TC RNA Amplification: Biotinylation/Fluorescent Probe Labeling
Prepare the IVT master mix (on wet ice).
Add 10 μL of double-stranded cDNA sample to the IVT master mix (30 μL for each cDNA sample).
Mix thoroughly (very important). Centrifuge briefly and incubate for 5 h at 37 °C.
The TC RNA amplified products are now ready for purification, fragmentation, and application to cDNA or oligonucleotide microarrays (see Note 11, Fig. 10.2A).
3.6 IVT for TC RNA Amplification: Radioactive Probe Labeling
Prepare the IVT master mix (on wet ice).
Centrifuge briefly and (on wet ice) add 7.5 μL of double-stranded cDNA sample to the IVT master mix (7.5 μL for each cDNA sample).
Add 4 μL of 33P-UTP and 1 μL of T7 RNA polymerase.
Mix thoroughly (very important), centrifuge briefly, and incubate for 4 h at 37 °C.
The TC RNA amplified products now are ready to be hybridized to membrane-based arrays (see Note 12, Fig. 10.2C).
Acknowledgments
I thank Shaoli Che, MD, PhD; Melissa Alldred, PhD; Irina Elarova; Shaona Fang; and Krisztina M. Kovacs for expert technical assistance Support for this project comes from the NINDS (NS43939, NS48447) and NIA (AG10668, AG14449, AG17617, AG09466) and Alzheimer’s Association.
Footnotes
All solutions used throughout the entire protocol should be made in 18.2 MΩ RNase-free water (e.g., Nanopure Diamond, Barnstead, Dubuque, IA), which is referred to 18.2 MΩ RNase-free water in the text.
Prepare sufficient master mix for each reaction step using this basic formula: Tabulate the total number of samples plus two extra (i.e.. calculate the volumes of the mix components for χ number of samples + control + one to control for any volume loss or experimenter error).
An alternative receipe for 10 × transcription buffer (store in 1ml aliquots) is 400 mM Tris-HCl, pH 7.5, 70 mM MgCl2, 100 mM NaCl, 20 mM spermidine.
This protocol is based on using the BioArray RNA Transcript Labeling Kit (Enzo), although any biotinylation or fluorescent labeling protocols are suitable (minor modifications may apply).
Do not air dry RNA pellets longer than 5 min, as the pellets become difficult to resuspend.
Each RNA sample to which the RT master mix is added receives 1 μL of TC RNA primer; therefore, if there are 8 RNA samples, the RT master mix must be sufficient for ten RNA samples and 10 μL of the TC primer must be added to the RT master mix prior to adding it to the RNA samples.
When removing aliquots of Superscript III from the tube, be extremely careful to not contaminate the vial of enzyme with primers or RNA samples.
For the PCR cycling of the second-strand synthesis, it is a good idea to create a specific stepwise program using a final volume of 100 μL.
A crucial mistake that is commonly made is failure to mix the hot-start second-strand synthesis reaction when the Taq polymerase is added. Thorough mixing with a pipette tip ensures a suitable reaction environment.
This step requires a little practice. Float the membrane filter shiny side up, on the surface of the water using a pair of dedicated RNase-free tissue forceps. Take care not to sink the membrane. Do not overfill the 50-mL conical tube. Conversely, underfilling (<40 mL) the conical tube makes placement of the filter difficult.
There are numerous Internet sites and published protocols for the hybridization of labeled probes to microarray platforms, subsequent washing, and imaging protocols (e.g., www.affymetrix.com, www.enzo.com,. [26, 27, 33]).
References
- 1.Ginsberg SD, Mirnics K. Functional genomic methodologies. Prog Brain Res. 2006;158:15–40. doi: 10.1016/S0079-6123(06)58002-1. [DOI] [PubMed] [Google Scholar]
- 2.Kabbarah O, et al. Expression profiling of mouse endometrial cancers microdissected from ethanol-fixed, paraffin-embedded tissues. Am J Pathol. 2003;162:755–762. doi: 10.1016/S0002-9440(10)63872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su JM, et al. Comparison of ethanol versus formalin fixation on preservation of histology and RNA in laser capture microdissected brain tissues. Brain Pathol. 2004;14:175–182. doi: 10.1111/j.1750-3639.2004.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanji N, et al. Effect of tissue processing on the ability to recover nucleic acid from specific renal tissue compartments by laser capture microdissection. Exp Nephrol. 2001;9:229–234. doi: 10.1159/000052616. [DOI] [PubMed] [Google Scholar]
- 5.Fend F, et al. Immuno-LCM: Laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61–66. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinnecom K, Pachter JS. Selective capture of endothelial and perivascular cells from brain microvessels using laser capture microdissection. Brain Res Protoc. 2005;16:1–9. doi: 10.1016/j.brainresprot.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Farrell RE., Jr . RNA methodologies. 2. Academic Press; San Diego: 1998. [Google Scholar]
- 8.Blumberg DD. Creating a ribonuclease-free environment. Methods Enzymol. 1987;152:20–24. doi: 10.1016/0076-6879(87)52005-5. [DOI] [PubMed] [Google Scholar]
- 9.Goldsworthy SM, et al. Effects of fixation on RNA extraction and amplification from laser capture microdissected tissue. Mol Carcinog. 1999;25:86–91. [PubMed] [Google Scholar]
- 10.Ginsberg SD, Che S. RNA amplification in brain tissues. Neurochem Res. 2002;27:981–992. doi: 10.1023/a:1020944502581. [DOI] [PubMed] [Google Scholar]
- 11.Mai JK, Schmidt-Kastner R, Tefett H-B. Use of acridine orange for histologic analysis of the central nervous system. J Histochem Cytochem. 1984;32:97–104. doi: 10.1177/32.1.6197440. [DOI] [PubMed] [Google Scholar]
- 12.Vincent VA, et al. Analysis of neuronal gene expression with laser capture microdissection. J Neurosci Res. 2002;69:578–586. doi: 10.1002/jnr.10329. [DOI] [PubMed] [Google Scholar]
- 13.Ginsberg SD, et al. Sequestration of RNA in Alzheimer’s disease neurofibrillary tangles and senile plaques. Ann Neurol. 1997;41:200–209. doi: 10.1002/ana.410410211. [DOI] [PubMed] [Google Scholar]
- 14.Mufson EJ, Counts SE, Ginsberg SD. Single cell gene expression profiles of nucleus basalis cholinergic neurons in Alzheimer’s disease. Neurochem Res. 2002;27:1035–1048. doi: 10.1023/a:1020952704398. [DOI] [PubMed] [Google Scholar]
- 15.Ginsberg SD, et al. RNA sequestration to pathological lesions of neurodegenerative disorders. Acta Neuropathol. 1998;96:487–494. doi: 10.1007/s004010050923. [DOI] [PubMed] [Google Scholar]
- 16.Tatton NA, Kish SJ. In situ detection of apoptotic nuclei in the substantia nigra compacta of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-treated mice using terminal deoxynucleotidyl transferase labelling and acridine orange staining. Neuroscience. 1997;77:1037–1048. doi: 10.1016/s0306-4522(96)00545-3. [DOI] [PubMed] [Google Scholar]
- 17.Sarnat HB, et al. Gliosis and glioma distinguished by acridine orange. Can J Neurol Sci. 1987;14:31–35. doi: 10.1017/s0317167100026135. [DOI] [PubMed] [Google Scholar]
- 18.Che S, Ginsberg SD. Amplification of transcripts using terminal continuation. Lab Invest. 2004;84:131–137. doi: 10.1038/labinvest.3700005. [DOI] [PubMed] [Google Scholar]
- 19.Che S, Ginsberg SD. RNA amplification methodologies. In: McNamara PA, editor. Trends in RNA Research. Nova Science Publishing; Hauppauge. NY: 2006. pp. 277–301. [Google Scholar]
- 20.Ginsberg SD, Che S. Combined histochemical staining, RNA amplification, regional, and single cell analysis within the hippocampus. Lab Invest. 2004;84:952–962. doi: 10.1038/labinvest.3700110. [DOI] [PubMed] [Google Scholar]
- 21.Shaulsky G, Loomis WF. Gene expression patterns in Dictyostelium using microarrays. Protist. 2002;153:93–98. doi: 10.1078/1434-4610-00087. [DOI] [PubMed] [Google Scholar]
- 22.Alter O, Brown PO, Botstein D. Generalized singular value decomposition for comparative analysis of genome-scale expression data sets of two different organisms. Proc Natl Acad Sci USA. 2003;100:3351–3356. doi: 10.1073/pnas.0530258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kacharmina JE, Crino PB, Eberwine J. Preparation of cDNA from single cells and subcellular regions. Methods Enzymol. 1999;303:3–18. doi: 10.1016/s0076-6879(99)03003-7. [DOI] [PubMed] [Google Scholar]
- 24.Phillips J, Eberwine JH. Antisense RNA amplification: A linear amplification method for analyzing the mRNA population from single living cells. Methods Enzymol Suppl. 1996;10:283–288. doi: 10.1006/meth.1996.0104. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 26.Ginsberg SD. RNA amplification strategies for small sample populations. Methods. 2005;37:229–237. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Ginsberg SD, et al. Cell and tissue microdissection in combination with genomic and proteomic applications. In: Zaborszky L, Wouterlood FG, Lanciego JL, editors. Neuroanatomical tract tracing 3: Molecules, neurons, and systems. Springer; New York: 2006. pp. 109–141. [Google Scholar]
- 28.Ginsberg SD, et al. Shift in the ratio of three-repeat tau and four-repeat tau mRNAs in individual cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Neurochem. 2006;96:1401–1408. doi: 10.1111/j.1471-4159.2005.03641.x. [DOI] [PubMed] [Google Scholar]
- 29.Ginsberg SD, et al. Down regulation of trk but not p75 gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem. 2006;97:475–487. doi: 10.1111/j.1471-4159.2006.03764.x. [DOI] [PubMed] [Google Scholar]
- 30.Ginsberg SD, et al. Single cell gene expression profiling in Alzheimer’s disease. NeuroRx. 2006;3:302–318. doi: 10.1016/j.nurx.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Counts SE, et al. Galanin fiber hypertrophy within the cholinergic nucleus basalis during the progression of Alzheimer’s disease. Dement Geriatr Cogn Disord. 2006;21:205–214. doi: 10.1159/000090906. [DOI] [PubMed] [Google Scholar]
- 32.Ginsberg SD, Che S. Expression profile analysis within the human hippocampus: Comparison of CA1 and CA3 pyramidal neurons. J Comp Neurol. 2005;487:107–118. doi: 10.1002/cne.20535. [DOI] [PubMed] [Google Scholar]
- 33.Cheung VG, et al. Making and reading microarrays. Nat Genet. 1999;21:15–19. doi: 10.1038/4439. [DOI] [PubMed] [Google Scholar]