Surface-enhanced Raman scattering (SERS) is under active investigation in biomedical diagnostics due to its high sensitivity, increased levels of multiplexing, robustness, and ability to perform detection in blood and other biological matrices. SERS has been successfully applied for labeling cells1 and tissues,2 for multiplexed biomarker labeling to monitor apoptotic processes,3 and for real-time monitoring of single live cell signaling processes.4
We have developed SERS tags (Nanoplex biotags, a trademark of Oxonica Inc.), comprised of one or more SERS-active metal nanoparticles (Au) and a sub-monolayer of reporter molecules adsorbed to the metal surface, all encapsulated in a protective and functionalized silica coat (Figure 1A).5 Nanoplex biotags have been successfully used in biological applications.6,7 Here we describe a novel application of Nanoplex biotags for the direct detection of rare cancer cells in whole blood.
Figure 1.
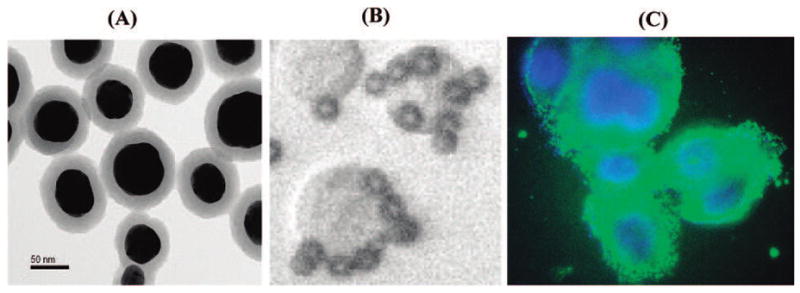
(A) TEM image of Nanoplex biotags. (B) Small magnetic beads binding to SKBR3 cells. (C) her2 antibody-conjugated SERS tag (green) labeling of the SKBR3 membrane (Hoechst dye-labeled nuclei are in blue).
Malignant cells are shed and circulate in the bloodstream of patients with solid tumors.8 Since the “seed and soil” theory for circulating tumor cells (CTCs) was hypothesized9 and confirmed,10 two major approaches, based on polymerase chain reaction or cytometric methods (such as the CellSearch system), have been established for CTC detection.8,11 However, high instrument cost and labor-intensive and time-consuming procedures remain a major concern and hamper their use in clinical diagnostics. Taking advantage of the intrinsic properties of the SERS tags, we have developed a novel, homogeneous, no-wash assay platform that overcomes the current assay limitations. We use magnetic beads for CTC capture and Nanoplex biotags for rapid and sensitive detection directly in human whole blood. Scheme 1 illustrates the concept in which magnetic beads, conjugated to an epithelial cell-specific antibody (epithelial cell adhesion molecule, anti-EpCAM), and the SERS tags, conjugated to an anti-her2 antibody (human epidermal growth factor receptor-2), bind to a tumor cell. Since the breast cancer cell is of epithelial origin, the magnetic bead–EpCAM antibody will specifically bind to this tumor cell but not regular circulating blood cells. Since the her2 receptor is highly expressed on the breast cancer cell membrane, the anti-her2–SERS tag will specifically recognize these tumor cells. By adding the magnetic bead–EpCAM and SERS tag–her2 conjugates to a patient’s blood sample, circulating breast cancer cells (CTCs) can be detected rapidly and with good sensitivity in the presence of whole blood.
Scheme 1.
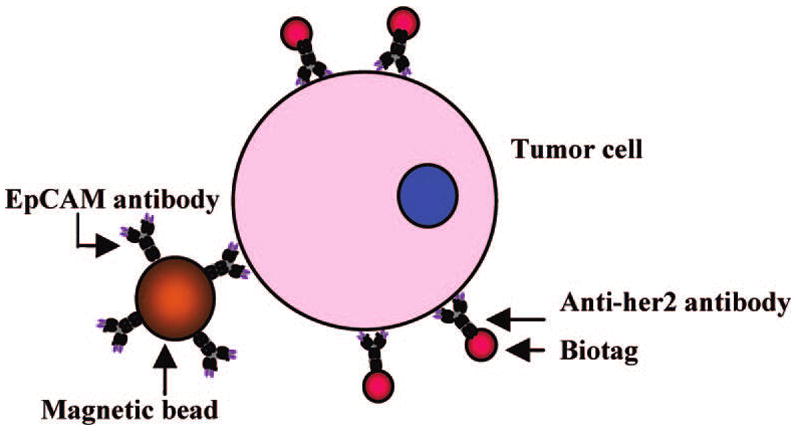
Schematic Illustration of the Ternary Immuno-Complex Formed by Nanoplex Biotags and Magnetic Bead Conjugates Binding to the Model Tumor Cell
In a proof-of-concept experiment, the breast cancer cell line SKBR3, expressing high levels of her2 receptor on the cell surface,12 was used as a model target. After a 30 min incubation of SKBR3 cells with magnetic bead–EpCAM and SERS–her2 conjugates, small volumes of the reaction mixtures were loaded on a glass slide. Bright-field microscopy imaging showed specific binding of the magnetic beads–EpCAM to the tumor cells (Figure 1B), and anti-her2 antibody-conjugated SERS tag binding to the tumor cell membrane was confirmed via immunostaining (Figure 1C). In a typical experiment, the reaction tube is placed by a magnet to concentrate the magnetic particles along with captured cells and biotags to a specific location on the side of the tube where a Raman spectrum is acquired. To determine assay sensitivity, SKBR3 cells were serially diluted with buffer. The titration curve (Figure 2) demonstrates a high correlation between Raman signal intensity and cell concentration (linear correlation shown in the inset in Figure 2). The low Raman signal for the negative controls (absence of SKBR3 cells) indicates negligible nonspecific binding of the SERS tags to the magnetic particles. The calculated limit of detection (LOD) is less than 10 cells/mL, with 99.7% confidence in the buffer system. Control experiments using a nonrelevant anti-cTnI (cardiovascular biomarker) antibody conjugated to SERS tags, replacing the her2–SERS tags, showed no detectable Raman signal after incubation with the tumor cells and demonstrated specificity of SERS–her2 conjugate.
Figure 2.
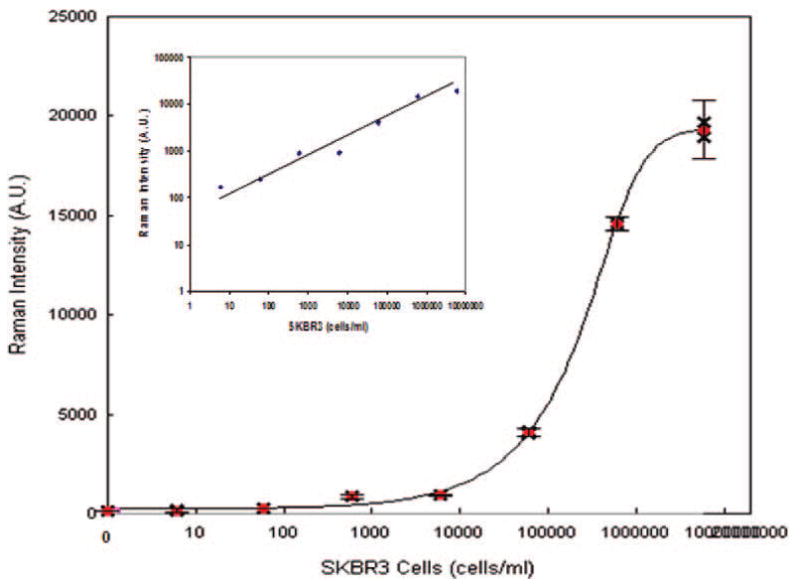
Detection of circulating tumor cell in buffer. Dose–response curve of SKBR3 cells spiked into buffer. The LOD is the cell concentration corresponding to the average signal of three replicate negative controls plus three standard deviations.
In contrast to other optical detection methods, near-IR excitation permits the use of Nanoplex biotags in biological matrices such as whole blood. Thus, tumor cells were spiked directly into whole blood prior to a short incubation of 30 min with magnetic bead–EpCAM and SERS tag–her2 conjugates. The results show no detectable Raman signal from whole blood (Figure 3A, red trace). In the absence of SKBR3 cells, a low background signal was observed when magnetic beads and SERS tags were added to whole blood (Figure 3A, blue trace). However, a strong Raman signal was detected when SKBR3 cells were spiked into whole blood (Figure 3A, green trace), indicating that a signal can be directly acquired in whole blood without washing or additional handling steps. To our knowledge, this is the first demonstration of SERS-based detection of tumor cells in a homogeneous, no-wash whole blood assay.
Figure 3.
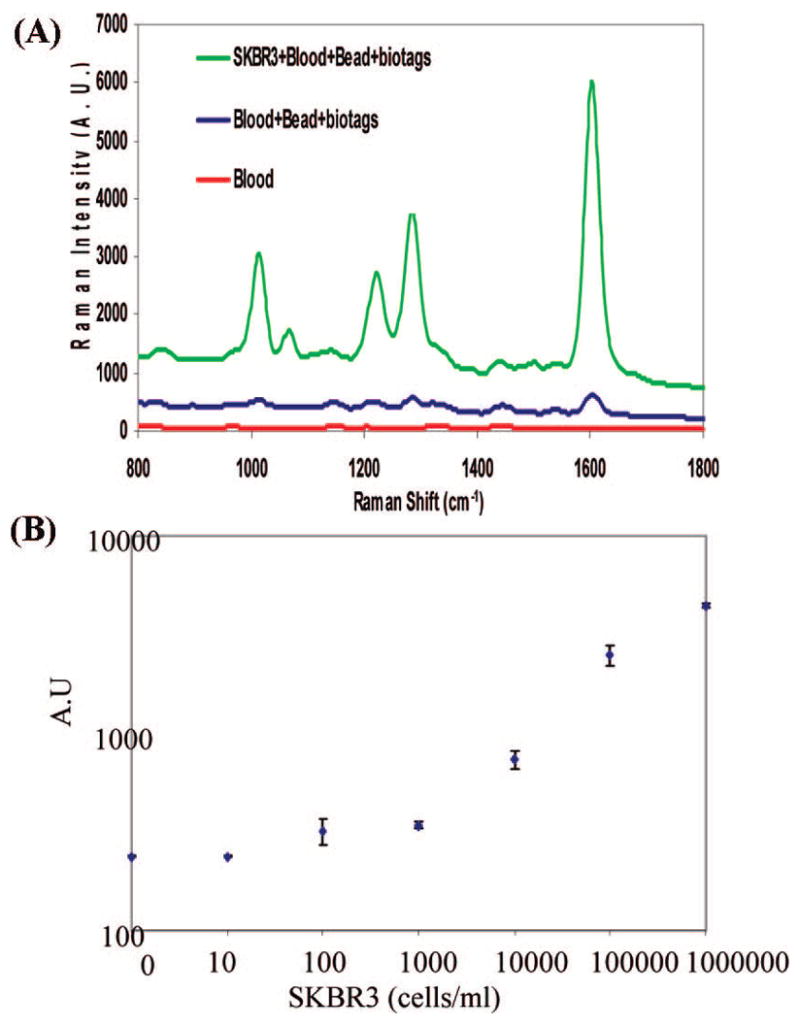
(A) Detection of SKBR3 spiked into whole blood. Raman spectra of whole blood (red) and of beads and biotag reagents in blood without a cell spike (blue) and with SKBR3 cells spike (green). (B) Dose–response curve of SKBR3 cells spiked into whole blood. Blood with no cell spike was used as a negative control.
To determine assay sensitivity, SKBR3 cells were spiked into whole blood and serially diluted with unspiked blood. Figure 3B shows a dose–response curve of SKBR3 cells spiked into blood. The calculated LOD is about 50 cells/mL, with 99.7% confidence. This illustrates that whole blood only marginally affects assay performance, with a small suppression of signal due to biological matrix interference. It should be noted that a small blood sample (100 μL) was used in this test. We expect further improvements in assay accuracy and sensitivity with larger sample volumes.11
In conclusion, using a combination of magnetic particles and Nanoplex biotags, we have demonstrated a novel homogeneous, no-wash assay for the direct detection of circulating tumor cells in whole blood. Multiplexed labeling of biomarkers with available multiplex SERS tags will further improve assay accuracy and specificity. This sensitive, simple, and rapid assay platform, requiring no sample preparation, could provide enormous benefit to clinical diagnostics, real-time monitoring of therapy progress, and drug development.
Supplementary Material
Supporting Information Available: Experimental details and complete ref 2. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
Thanks to Drs. Ed Holland, Glenn Davis, Gabriela Chakarova, and James Leung. Partial support of this work was provided by NIH CCNE program (U54CA119338-01, sub-contract 5-40255-G6).
References
- 1.Kim JH, Kim JS, Choi H, Lee SM, Jun BH, Yu KN, Kuk E, Kim YK, Jeong DH, Cho MH, Lee YS. Anal Chem. 2006;78:6967–6973. doi: 10.1021/ac0607663. [DOI] [PubMed] [Google Scholar]
- 2.Sun L, et al. Nano Lett. 2007;7:351–356. doi: 10.1021/nl062453t. [DOI] [PubMed] [Google Scholar]
- 3.Yu KN, Lee SM, Han JY, Park H, Woo MA, Noh MS, Hwang SK, Kwon JT, Jin H, Kim YK, Hergenrother PJ, Jeong DH, Lee YS, Cho MH. Bioconjugate Chem. 2007;18:1155–1162. doi: 10.1021/bc070011i. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Yin H, Cooper JM, Haswell SJ. Anal Bioanal Chem. 2008;390:833–840. doi: 10.1007/s00216-007-1564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mulvaney SP, Musick MD, Keating CD, Natan MJ. Langmuir. 2003;19:4784–4790. [Google Scholar]
- 6.Doering WE, Piotti M, Natan MJ, Freeman RG. Adv Mater. 2007;19:3100–3108. [Google Scholar]
- 7.Sha MY, Xu H, Penn SG, Cromer R. Nanomedicine. 2007;2:725–734. doi: 10.2217/17435889.2.5.725. [DOI] [PubMed] [Google Scholar]
- 8.Mocellin S, Keilholz U, Rossi CR, Nitti D. Trends Mol Med. 2006;12:130–139. doi: 10.1016/j.molmed.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Fidler IJ. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 10.Ulmer A, Schmidt-Kittler O, Fischer J, Ellwanger U, Rassner G, Riethmuller G, Fierlbeck G, Klein C. Clin Cancer Res. 2004;10:531–537. doi: 10.1158/1078-0432.ccr-0424-03. [DOI] [PubMed] [Google Scholar]
- 11.Shaffer DR, Leversha MA, Danila DC, Lin O, Gonzalez-Espinoza R, Gu B, Anand A, Smith K, Maslak P, Doyle GV, Terstappen LW, Lilja H, Heller G, Fleisher M, Scher HI. Clin Cancer Res. 2007;13:2023–2029. doi: 10.1158/1078-0432.CCR-06-2701. [DOI] [PubMed] [Google Scholar]
- 12.Hayes DF, Walker TM, Singh B, Vitetta ES, Uhr JW, Gross S, Rao C, Doyle GV, Terstappen LW. Int J Oncol. 2002;21:1111–1117. doi: 10.3892/ijo.21.5.1111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Available: Experimental details and complete ref 2. This material is available free of charge via the Internet at http://pubs.acs.org.


