Abstract
Pyrrolysine (Pyl), the 22nd natural amino acid, is genetically encoded by UAG and inserted into proteins by the unique suppressor tRNAPyl1. The Methanosarcinaceae produce Pyl and express Pyl-containing methyltransferases that allow growth on methylamines2. Homologous methyltransferases and the Pyl biosynthetic and coding machinery are also found in two bacterial species1,3. Pyl coding is maintained by pyrrolysyl-tRNA synthetase (PylRS), which catalyzes the formation of Pyl-tRNAPyl4,5. Pyl is not a recent addition to the genetic code. PylRS was already present in the last universal common ancestor6; it then persisted in organisms that utilize methylamines as energy sources. Recent protein engineering efforts added non-canonical amino acids to the genetic code7,8. This technology relies on the directed evolution of an ‘orthogonal’ tRNA synthetase:tRNA pair in which an engineered aminoacyl-tRNA synthetase (aaRS) specifically and exclusively acylates the orthogonal tRNA with a non-canonical amino acid. For Pyl the natural evolutionary process developed such a system some 3 billion years ago. When transformed into Escherichia coli, Methanosarcina barkeri PylRS and tRNAPyl function as an orthogonal pair in vivo5,9. Here we demonstrate that Desulfitobacterium hafniense PylRS:tRNAPyl is an orthogonal pair in vitro and in vivo, and present the crystal structure of this orthogonal pair. The ancient emergence of PylRS:tRNAPyl allowed for the evolution of unique structural features in both the protein and the tRNA. These structural elements manifest an intricate, specialized aaRS:tRNA interaction surface highly distinct from those observed in any other known aaRS:tRNA complex; it is this general property that underlies the molecular basis of orthogonality.
Unlike the archaeal PylRS sequences, the bacterial versions are encoded in two separate genes. The pylS gene encodes the D. hafniense PylRS (DhPylRS) presented here, which includes a tRNA recognition domain (the tRNA binding domain 1), the conserved tRNA synthetase class II catalytic domain, the bulge domain, and a C-terminal tail, which is also involved in tRNA recognition (Fig. 1a). In Pyl-decoding bacteria, a second gene (pylSn) encodes a 110 residue polypeptide that is homologous (20% identity) to the N-terminal domain of archaeal PylRSs (Supplementary Fig. 1). The hydrophobic nature of this domain reduces the solubility of PylRS9 and encumbers crystallography6,10. While both the DhPylRS and truncated versions of M. barkeri PylRS are active in aminoacylating tRNAPyl in vitro, only the full length archaeal PylRSs, with a KD ~10 times lower than the bacterial enzyme, displayed sufficient activity to support in vivo protein synthesis9.
Figure 1.
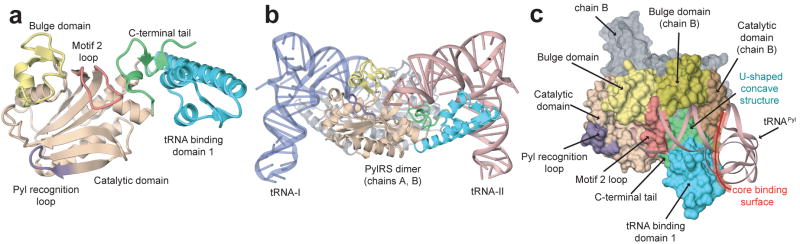
Overall structures. (a) DhPylRS subunit A, shown as a ribbon model, consisting of the tRNA binding domain 1 (1.68, blue), the catalytic domain (69.96 and 128.266, beige), the bulge domain (97.127, yellow), and the C-terminal tail (267.288, light green), with the motif 2 loop (160–170) colored red and the Pyl recognition loop (212–218) colored purple. (b) The dimeric DhPylRS:tRNAPyl complex structure, shown as a ribbon model. The asymmetric unit contains one PylRS dimer and two tRNAPyl molecules; PylRS-A (colored as in panel a), PylS-B (gray), tRNA-I (blue), and tRNA-II (pink). (c) Binding of tRNAPyl (ribbon representation) to the surface model of DhPylRS, structural domains are colored as in (a).
When M. barkeri PylRS and tRNAPyl were transformed into Escherichia coli, they function as an orthogonal pair in the heterologous environment5,9. We now show both in vitro (Supplementary Fig. 2) and in vivo (Supplementary Fig. 3) that DhPylRS:tRNAPyl is an orthogonal pair with reduced enzyme activity compared to its full-length archaeal counterpart, and that addition of PylSn did not significantly enhance DHPylRS activity. Aminoacylation was performed with N-ε-cyclopentyloxycarbonyl-L-lysine (Cyc) because of the difficulty in chemically synthesizing Pyl11. DhPylRS acylates 80% of tRNAPyl transcript with Cyc, which is greater than that of the homologous M. mazei PylRS (MmPylRS) fragment (53%), and comparable to the 85% level reached by the full length archaeal PylRS (Supplementary Fig. 2). Unfractionated E. coli tRNA is not a substrate for DhPylRS, and Cyc-tRNAPyl formation is not perturbed by competition with total E. coli tRNA. Similar results for in vitro aminoacylation by full length and truncated MmPylRSs were reported recently10. In attempting to suppress a lacZ amber mutant, D. hafniense pylS did not make enough Cyc-tRNAPyl to yield detectable β-galactosidase activity9. Therefore we applied a strong selection12,13 in which Cyc-tRNAPyl was required to suppress an E. coli trpA amber mutation and thus convert the test strain from Trp auxotrophy to prototrophic growth (Supplementary Fig. 3). We observed no growth on solid media in the negative controls, and growth was only observed when PylRS, tRNAPyl and Cyc were present (Supplementary Fig. 3a–c). While addition of Trp to minimal liquid medium results in wild type growth rate (2.3 h doubling time), the M. barkeri PylRS:tRNAPyl (4 h) and the DhPylRS:tRNAPyl (5.8 h) display slower but significant growth with Cyc supplementation in the absence of Trp (Supplementary Fig. 3d).
To understand the molecular details of the PylRS:tRNAPyl interaction, we determined the crystal structures of the apo enzyme and of DhPylRS complexed to D. hafniense tRNAPyl at 2.5 A and 3.1 A resolution, respectively (Fig. 1, Supplementary Table 1 and 2). The protein forms a dimer in the crystal and in solution (data not shown). The final model of DhPylRS:tRNAPyl includes residues 10-288 of DhPylRS and tRNAPyl. The asymmetric unit of the complex crystal contains a DhPylRS dimer and two tRNAPyl molecules. Each tRNAPyl interacts predominantly with one subunit, but also makes specific contacts to the other protomer (Fig. 1b). The tRNA binding domain 1 and C-terminal tail are unique to PylRSs. The α1 helix of the tRNA binding domain 1, the C terminal tail, and the bulge domain of the opposite subunit form a U-shaped concave structure that is shape complementary to the acceptor helix and directs the 3′-terminus of tRNAPyl to the motif 2 loop (Arg160-Asn170) in the catalytic site (Fig. 1c, Supplementary discussion). In addition to the core binding surface (Fig. 1c), these unique protein structural elements contribute to the orthogonality of PylRS:tRNAPyl.
The aminoacyl-tRNA synthetases are found in two protein families that are distinguished by their evolutionarily unrelated catalytic core domains and by how they bind opposing sides of the tRNA (reviewed in14,15). The class I aaRSs share a conserved Rossmann fold aminoacylation domain and (with the exception of TyrRS and TrpRS) approach from the minor groove side of the tRNA acceptor stem. PylRS includes the conserved class II catalytic domain fold (only also observed in biotin synthetase and lipoyltransferase), and like all other class II aaRSs, PylRS approaches its tRNA from the major groove side of the acceptor stem (Fig 2). Similarities between the DhPylRS:tRNAPyl and other class II aaRS:tRNA complexes do not extend much beyond these general features.
Figure 2.
Class I (a) and class II (b) tRNA synthetase:tRNA complexes are structurally aligned. Only a single monomer of the catalytic core domains are displayed, color coded according to structural similarity. Viewed from the major groove side of the acceptor stem, a phosphate backbone outline of the tRNAs is shown (tan), and tRNAPyl is shown in purple. In space filling representation, a glutamyl-adenylate (a) and a pyrrolysyl-adenylate (b) highlight the class I and class II active site pockets, respectively. The small substrates are partially obscured by the protein backbone due to the need to show both aaRS families in the same orientation relative to the tRNA.
There are 31 protein residues in contact with the tRNA (annotated in Supplementary Figs. 1 and 4). As in other aaRS:tRNA complexes that also lack the ATP or aminoacyl-adenylate substrate16,17, the terminal adenosine (A76) occupies the ATP binding pocket. In subsequent aaRS:tRNA complexes with ATP or aminoacyl-adenylate substrates the terminal adenosine flips out of the ATP binding pocket; this is accompanied by a slight conformational shift of the terminal CCA bases without affecting most of the interactions between the protein and tRNA18,19. Comparison with the MmPylRS:ATP complex6 shows that only three residues (Arg160, Leu169 and Phe172) are incompatible with simultaneous binding to ATP and the A76 adenylate.
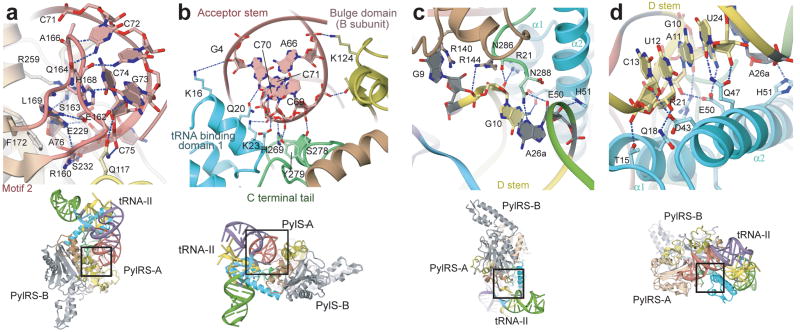
Unique interactions between PylRS and tRNAPyl contribute to orthogonality. Half of the 28 remaining residues, many of which participate in the core binding surface, emerge from PylRS specific domains. Twelve residues from the class II catalytic domain and two residues from the bulge domain complete the tRNA binding surface (Supplementary Fig. 4a). A comparison with the available class II aaRS co-crystal structures with completely docked tRNAs18–23 indicates that PylRS uses typical tRNA binding residues in unusual ways and also uses inserted residues, i.e., those without homologous counterparts in other class II aaRSs, to make novel contacts to the tRNA. The imidazole ring of His168, which is located in the motif 2 loop, participates in base stacking with C74 (Fig 3a). In other class II aaRSs, this position is occupied by His or Arg residues that invariably interact with C74 via hydrogen bonding. Gln164 establishes three sequence specific hydrogen bonds to C71, C72, and G73 (Fig. 3a). Only the yeast AspRS also places a tRNA binding residue (Ser329) at the homologous location, which establishes a single hydrogen bond to C74. Ser163, an insertion in the motif 2 loop of PylRS, forms a hydrogen bond to the 6-amino group of A76 (Fig. 3a). Four contacts (Lys124, Arg140, Arg144, and Glu245) emerge from the B-chain of DhPylRS to contact the tRNA bound to the A-chain. Another inserted residue, Lys124, interacts with the phosphate backbone at A66 (Fig. 3b). The side chains of Arg140, Arg144 and Glu245 interact with G9 (Fig. 3c), which, particular to tRNAPyl, is flipped outside of the main tRNA body, so these interactions also are absent from other aaRS:tRNA complexes.
Figure 3.
DhPylRS:tRNAPyl interface. (a) View showing the recognition of the CCA terminus by the motif-2 loop of DhPylRS. (b) View showing the recognition of the tRNAPyl acceptor helix by DhPylRS. (c,d) Views showing the recognition of the tRNAPyl minimal core by the core-binding surface of DhPylRS.
The tertiary core of a tRNA molecule is the location of sequence-distant interactions that are responsible for the canonical L-shape tertiary structure of the tRNA. The core of tRNAPyl is recognized by the core binding surface which is composed of tRNA binding domain 1, the C-terminal tail, and the α6 helix from the opposing protomer (Figs. 1c and 3b–d). In tRNAPyl, the deletion of the otherwise invariant U8 base, and an atypically short variable region and D-loop contribute to the compact core. Other unusual features of the tRNAPyl sequence and secondary structure have been detailed elsewhere1,24,25. Deletion of U8 disrupts one of the most highly conserved tertiary base pairs (U8:A14) in tRNAs26, leaving the non-standard guanosine at position 14 to base pair with C59 from the T-loop (Supplementary Fig. 5b,c). The U8 deletion also allows G9 to flip away from the tRNA body where it is specifically recognized as minor identity element27 principally via a conserved cation-π interaction from Arg140 (Fig. 3c). Due to the absence of canonical position 48, a typical base pair between the T and D-loops is also missing (Supplementary Fig. 5b,c). In summary, these deletions in tRNAPyl lead to a structurally re-arranged and tightly packed tertiary core (Supplementary discussion). PylRS evolved to form specific contacts with the compact core of tRNAPyl including four strictly conserved contacts to the identity element base pairs G10:C25 and A11:U2427 (Fig. 3c,d). Interactions with the tertiary core, largely provided by tRNA binding domain 1, make PylRS sterically incompatible with other canonical tRNAs due to their bulkier tertiary core.
Most of the interaction between PylRS and tRNAPyl is captured by the D. hafniense pylS gene product. The genetic code is maintained in all cellular life on earth by the accurate aminoacylation of tRNAs with their cognate amino acids. In part, the fidelity of this interaction is due to tRNA identity elements, i.e., those bases, base pairs and structural features of a tRNA that are crucial for aminoacylation by its cognate aaRS. The DhPylRS:tRNAPyl structure allows a more complete interpretation of tRNAPyl identity elements, elucidated by previous biochemical work. Identity elements for DhPylRS27 include the discriminator base (G73), the first base pair in the acceptor stem (G1:C72), the D.stem base pairs G10:C25 and A11:U24, and G9, all of which are in direct contact with DhPylRS (Fig. 3a,c,d). While the full-length PylRS does not specifically recognize the anticodon, the two bases adjacent to the anticodon (U33 and A37) are identity elements for MmPylRS25. These two bases are possibly recognized in a sequence-specific manner by residues from the N-terminal domain (PylSn).
The tRNA binding surface in DhPylRS shows a high degree of evolutionary conservation among PylRSs. Of the 28 tRNA binding residues identified here, 15 (54%) are strictly conserved among all PylRS sequences, which compares to only 39% sequence identity between DhPylRS and its archaeal counterparts (Supplementary Fig. 1 and 6). Five additional residues in the interface have generically conserved hydrogen bonding potential, either through similar side chain, e.g., Asp versus Asn at position 264, or the hydrogen bond is via the protein backbone and thus sequence independent, e.g., at Ala166. Two residues are highly conserved, although not strictly (Lys16, Ser278). The hydrogen bonds presented at these positions likely represent somewhat less important interactions. Only four residues provide hydrogen bonds to the tRNA, but are not conserved among PylRS sequences. Taken together, among PylRS sequences 81% of the residues in the DhPylRS:tRNAPyl interface are identical, highly conserved, or have conservation of an amino acid property that is important for the interaction such as charge or hydrogen bonding potential. One residue (Glu245), which is involved in a hydrogen bond network that contacts G9, is conserved only in the bacterial PylRSs. Archaeal tRNAs have a U at this position and the smaller pyrimidine supports fewer contacts to the protein, which may contribute to the more robust aminoacylation by DhPylRS compared to a similar fragment of MmPylRS (Supplementary Fig. 2).
A recent attempt to identify the PylRS:tRNAPyl interaction using homology modeling suggested 10 protein residues that might play a role in the interface10. Alanine scanning mutations showed that 7 of these residues affected aminoacylation yields moderately to severely. The mutational analysis could not distinguish between mutants that alter protein stability and those that affect stability of the protein-nucleic acid interface. Of the 31 amino acid residues in direct contact with tRNAPyl, only six residues were correctly predicted. That homology modeling could not capture most of the PylRS:tRNAPyl interface highlights the distinctiveness of this complex compared to other aaRS:tRNA complexes.
In addition to the DhPylRS structures presented here, other structures are available of the homologous MmPylRS fragment6,10. Although MmPylRS and DhPylRS share only 39% sequence identity in this fragment of the molecule the two proteins are highly structurally similar, displaying an RMSD of 2.4 A and a structural similarity of 63% according to the metric QH14. The only significant difference in the active site of the DhPylRS enzyme is the replacement of a bulky Trp139 residue with the diminutive Leu209 residue in MmPylRS, also recently observed in a DHPylRS apoenzyme structure28. Homology modeling shows that Pyl fits into the smaller active site of DhPylRS (Supplementary Fig. 7).
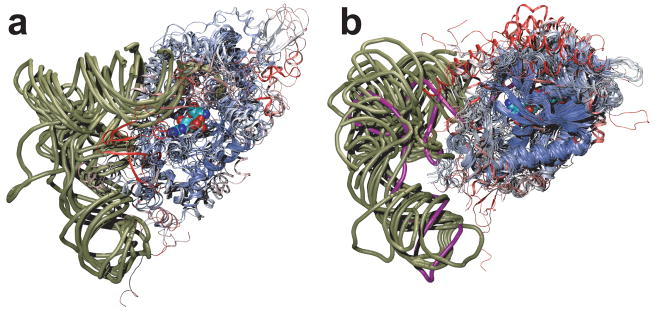
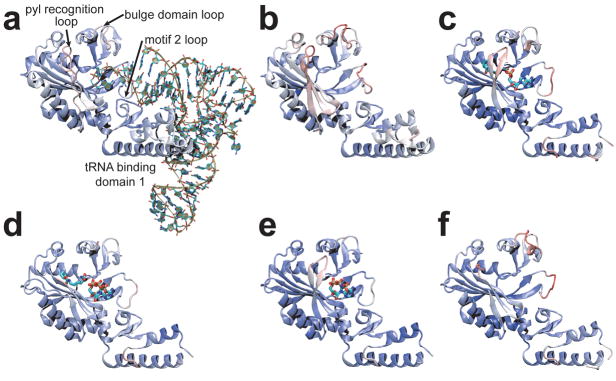
There are four regions (i–iv) displaying significant differences between the solved structures of PylRS (Fig. 4). (i) tRNA binding domain 1 is well ordered in the DhPylRS structures, but not in the MmPylRS structures. This region appears to be more stable in general in the DhPylRS context, possibly explaining why the DhPylRS shows higher aminoacylation yield than the homologous fragment of the M. mazei enzyme (Supplementary Fig. 2). Comparison of the DhPylRS structures shows that tRNA binding induces side chain order due to specific interaction between the tRNA and tRNA binding domain 1. (ii) A conserved Tyr (DhPylRS Tyr217, Supplementary Fig. 7) in the Pyl recognition loop (Fig. 1, Supplementary Fig. 1) forms a hydrogen bond to the pyrrole ring nitrogen in the MmPylRS pyrrolysyl-adenylate complex6. As in other PylRS structures that lack the substrate Pyl, the Pyl recognition loop is not well ordered in our structures. The flexible nature of this loop was captured here, since one of the three asymmetric molecules in the C2 crystal was observed in the closed form while in the other molecules the loop is bent away from the empty active site. (iii) Upon tRNA binding, the motif 2 loop drastically alters its conformation and becomes intercalated into the major groove of the acceptor end (Fig. 3a), providing base-specific recognition of the G1:C72 base pair as well as the discriminator G73 (Fig. 3a). (iv) The bulge domain loop (residues 110–117) is resolved and well ordered only in the tRNA complex. Loop ordering is induced directly via a hydrogen bond between Gln117 and the backbone phosphate oxygen of C75 and also indirectly due to interactions (e.g., a hydrogen bond between Glu162 and Gln117) with motif 2, which orders upon tRNA binding (Fig. 3a).
Figure 4.
Comparison of PylRS structures: (a) DhPylRS:tRNAPyl complex, (b) DhPylRS apo, (c) MmPylRS:Pyl-AMP complex (PDB code 2zim)6, (d) MmPylRS:ATP, Cyc complex (2q7g)6, (e) MmPylRS:ATP analog complex (2q7e)6, (f) MmPylRS apo (2e3c)10. The structures are colored according to B-factor (indicating more [red] or less [blue] structurally dynamic regions), and four regions that show conformational changes in the different structures are labeled in panel a. Only one subunit of the dimer is shown for clarity.
The class II aminoacyl-tRNA synthetases can be divided into three subclasses. Except for SerRS the members of subclass IIa (HisRS, GlyRS α2, ThrRS, ProRS, and SerRS) and subclass IIb (AspRS, AsnRS, and LysRS) include subclass-specific anticodon binding domains. The homologous domain architecture results in a more similar mode of tRNA binding among members of the subclass than between members of different subclasses. The amino acids genetically encoded by subclass IIb members are chemically similar (large polar or charged amino acids Asn, Asp and Lys). The subclass IIa aaRSs are responsible for the small amino acids (Gly, Pro) and small polar amino acids (His, Thr, Ser). These trends are also observed among the subclasses of class I aaRSs14. Subclass IIc, including PylRS, PheRS, O-phosphoseryl-tRNA synthetase (SepRS), GlyRS (αβ)2, and AlaRS, represent a different kind of evolutionary phenomenon. The amino acid substrates are chemically more diverse than those in any other subclass, and they do not share homologous anticodon binding domains. Comparison of the DhPylRS:tRNAPyl complex with tRNA complexes of other subclass IIc synthetases further shows that the detailed interactions between protein and tRNA are not conserved among subclass IIc members. The genesis of the class II aaRS family involved the initial radiation of three molecular lineages, i.e., the ancestral molecules from which the three subclasses evolved. Two of the three aaRS progenitors were restricted in their evolution, being selected to recognize large polar (subclass IIb) or small polar amino acid substrates (subclass IIa). The subclass IIc ancestor, from which PylRS evolved, was more adaptable than the other two, and perhaps its selective value is due to a greater inherent evolutionary plasticity.
Methods Summary
DHPylRS was overproduced and purified by published methods9,27. D. hafniense tRNAPyl was transcribed with T7 RNA polymerase, and purified under denaturing conditions by polyacrylamide gel electrophoresis. For crystallization DhPylRS was mixed with tRNAPyl in a molar ratio of 2:2.2, at a final protein concentration of 5 mg/ml. The complex crystals grew at 20°C by hanging-drop vapor diffusion against reservoir solution of 90 mM MES-NaOH buffer (pH 6.0) containing 5.4% 2-propanol, 180 mM calcium acetate, 2% ethanol, 10 mM Tris-HCl (pH 8.5).
Additional details are presented in Supplementary Methods; the crystallographic, data-collection and refinement statistics are in Supplementary Tables 1 and 2.
Supplementary Material
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank the beam-line staff at BL41XU of SPring-8 (Harima, Japan) and NW12 of PF-AR (Tsukuba, Japan) for technical help during data collection. P.O. holds a National Science Foundation postdoctoral fellowship in Biological Informatics. This work was supported by a grants from the Japan Science and Technology Agency (to O.N.), from the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology (to O.N.), from the Ministry of Education, Culture, Sports, Science and Technology (to R.I. and O.N.), from the Mitsubishi Foundation (to O.N.), from the Kurata Memorial Hitachi Science and Technology Foundation (to O.N.), from National Institute of General Medical Sciences (to D.S.), from the Department of Energy (to D.S.), and from the National Science Foundation (to D.S.).
Footnotes
Author Contributions
K.N. did purification, crystallization and structure determination. S.G. and T.U. did biochemical analyses. R.I. performed molecular dynamics. Y.A., R.I, and O.N. assisted the structure determination. P.O’D. analyzed the data and performed bioinformatic analysis. P.O’D., K.N., O.N. and D.S. wrote the paper. O.N. and D.S. conceived and supervised the work.
Coordinates and structure factors and deposited in the Protein Data Bank under accession codes 2ZNI (DhPylRS:tRNAPyl complex) and 2ZNJ (DhPylRS apo).
References
- 1.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 2.Krzycki JA. The direct genetic encoding of pyrrolysine. Curr Opin Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Zhang Y, Gladyshev VN. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res. 2007;35:4952–4963. doi: 10.1093/nar/gkm514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polycarpo C, et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci USA. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight SK, et al. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 6.Kavran JM, et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci USA. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, Xie J, Schultz PG. Expanding the genetic code. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 8.Neumann H, Peak-Chew SY, Chin JW. Genetically encoding Nε-acetyllysine in recombinant proteins. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]
- 9.Herring S, et al. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007;581:3197–3203. doi: 10.1016/j.febslet.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanagisawa T, et al. Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase. J Mol Biol. 2008;378:634–652. doi: 10.1016/j.jmb.2008.02.045. [DOI] [PubMed] [Google Scholar]
- 11.Polycarpo CR, et al. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanofsky C, Horn V. Tryptophan synthetase chain positions affected by mutations near the ends of the genetic map of trpA of Escherichia coli. J Biol Chem. 1972;247:4494–4498. [PubMed] [Google Scholar]
- 13.Murgola EJ. tRNA, suppression, and the code. Annu Rev Genet. 1985;19:57–80. doi: 10.1146/annurev.ge.19.120185.000421. [DOI] [PubMed] [Google Scholar]
- 14.O’Donoghue P, Luthey-Schulten Z. On the evolution of structure in aminoacyl-tRNA synthetases. Microbiol Mol Biol Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasil’eva IA, Moor NA. Interaction of aminoacyl-tRNA synthetases with tRNA: general principles and distinguishing characteristics of the high-molecular-weight substrate recognition. Biochemistry (Mosc) 2007;72:247–263. doi: 10.1134/s0006297907030029. [DOI] [PubMed] [Google Scholar]
- 16.Ruff M, et al. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp. Science. 1991;252:1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- 17.Goldgur Y, et al. The crystal structure of phenylalanyl-tRNA synthetase from Thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 18.Cavarelli J, et al. The active site of yeast aspartyl-tRNA synthetase: structural and functional aspects of the aminoacylation reaction. EMBO J. 1994;13:327–337. doi: 10.1002/j.1460-2075.1994.tb06265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moor N, Kotik-Kogan O, Tworowski D, Sukhanova M, Safro M. The crystal structure of the ternary complex of phenylalanyl-tRNA synthetase with tRNAPhe and a phenylalanyl-adenylate analogue reveals a conformational switch of the CCA end. Biochemistry. 2006;45:10572–10583. doi: 10.1021/bi060491l. [DOI] [PubMed] [Google Scholar]
- 20.Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 21.Eiler S, Dock-Bregeon A, Moulinier L, Thierry JC, Moras D. Synthesis of aspartyl-tRNAAsp in Escherichia coli–a snapshot of the second step. EMBO J. 1999;18:6532–6541. doi: 10.1093/emboj/18.22.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sankaranarayanan R, et al. The structure of threonyl-tRNA synthetase-tRNAThr complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 23.Briand C, et al. An intermediate step in the recognition of tRNAAsp by aspartyl-tRNA synthetase. J Mol Biol. 2000;299:1051–1060. doi: 10.1006/jmbi.2000.3819. [DOI] [PubMed] [Google Scholar]
- 24.Théobald-Dietrich A, Frugier M, Giegé R, Rudinger-Thirion J. Atypical archaeal tRNA pyrrolysine transcript behaves towards EF-Tu as a typical elongator tRNA. Nucleic Acids Res. 2004;32:1091–1096. doi: 10.1093/nar/gkh266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambrogelly A, et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc Natl Acad Sci USA. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herring S, Ambrogelly A, Polycarpo CR, Söll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35:1270– 1278. doi: 10.1093/nar/gkl1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MM, et al. Structure of Desulfitobacterium hafniense PylSc, a pyrrolysyl-tRNA synthetase. Biochem Biophys Res Commun. 2008;374:470–474. doi: 10.1016/j.bbrc.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905– 921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Herring S, Ambrogelly A, Polycarpo CR, Söll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35:1270–1278. doi: 10.1093/nar/gkl1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polycarpo CR, et al. Pyrrolysine analogues as substrates for pyrrolysyl-tRNA synthetase. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yanofsky C, Horn V. Tryptophan synthetase chain positions affected by mutations near the ends of the genetic map of trpA of Escherichia coli. J Biol Chem. 1972;247:4494–4498. [PubMed] [Google Scholar]
- 36.Murgola EJ. tRNA, suppression, and the code. Annu Rev Genet. 1985;19:57–80. doi: 10.1146/annurev.ge.19.120185.000421. [DOI] [PubMed] [Google Scholar]
- 37.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: Unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz VM, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36:D534–538. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is linked to the online version of the paper at www.nature.com/nature.