Abstract
Evolutionary biologists often wish to explore the impact of a particular historical event (e.g., the origin of a novel morphological trait, an episode of biogeographic dispersal, or the onset of an ecological association) on rates of diversification (speciation minus extinction). We describe a Bayesian approach for evaluating the correlation between such events and differential rates of diversification that relies on cross-validation predictive densities. This approach exploits estimates of the marginal posterior probability for the rate of diversification (in the unaffected part of the tree) and the marginal probability for the timing of the event to generate a predictive distribution of species diversity that would be expected had the event not occurred. The realized species diversity can then be compared to this predictive diversity distribution to assess whether rates of diversification associated with the event are significantly higher or lower than expected. Although simple, this Bayesian approach provides a robust inference framework that accommodates various sources of uncertainty, including error associated with estimates of divergence times, diversification-rate parameters, and event history. Furthermore, the proposed approach is relatively flexible, allowing exploration of various types of events (including changes in discrete morphological traits, episodes of biogeographic movement, etc.) under both hypothesis-testing and data-exploration inference scenarios. Importantly, the cross-validation predictive densities approach facilitates evaluation of both replicated and unique historical events. We demonstrate this approach with empirical examples concerning the impact of morphological and biogeographic events on rates of diversification in Adoxaceae and Lupinus, respectively.
Keywords: Adoxaceae, key innovations, Lupinus, speciation, extinction
Documenting the patterns and understanding the causes of variation in diversification rates is a central objective of evolutionary biology. Rates of diversification may be influenced both by the origin of intrinsic traits—“key innovations,” such as morphological, behavioral, or physiological novelties—and the incidence of extrinsic events—“key opportunities,” such as episodes of biogeographic or climatic change. Accordingly, a comprehensive understanding of the causes of differential diversification requires the ability to explore the impact of a diverse array of both intrinsic and extrinsic factors.
Several recent phylogeny-based methods have greatly enhanced our ability to test key innovation hypotheses regarding the influence of intrinsic factors, principally discrete binary traits, on rates of diversification (1–3). Despite remarkable progress in this area, we perceive the need to extend the phylogenetic study of diversification-rate correlates in 3 ways: to more fully accommodate inherent sources of uncertainty (associated with estimated divergence times, diversification-rate parameters, event histories, etc.), to address a wider range of historical events (associated with episodes of change in morphology, biogeography, and ecology), and to expand the fundamental mode of inference (to enable both hypothesis testing and data exploration).
With these considerations in mind, we describe a Bayesian approach for identifying correlates of differential diversification rates that relies on cross-validation predictive densities, which effectively asks “How diverse would the effected lineage be if the inferred event had no impact on rates of diversification?” The cross-validation predictive densities approach provides a versatile inference framework (under both hypothesis-testing and data-exploration scenarios) for investigating the influence of diverse types of historical events (whether unique or replicated), while simultaneously accommodating various sources of uncertainty. We illustrate this approach with two empirical analyses, first exploring the impact of fruit evolution on rates of diversification in the plant group Adoxaceae and then investigating the influence of biogeographic dispersal into South America on rates of diversification in lupines.
Cross-Validation Predictive Diversity Densities
Cross-validation predictive densities (4, 5) are related to the more familiar technique known as posterior predictive densities simulation, which has been applied to a number of problems in evolutionary biology, such as detecting positive selection on amino acid sites (6), evaluating the adequacy of nucleotide substitution models (7), and mapping mutations (8) and traits on phylogenies (9). Both are sampling-based approaches that provide a means of evaluating the adequacy of a given model. Posterior predictive simulation involves drawing model parameter values from their respective marginal posterior probability distributions (previously estimated from the original data under the candidate model) to generate a distribution of “future” (predictive) observations. If the model provides an adequate description of the original data, relevant aspects of the predictive and realized observations should be similar.
Cross-validation predictive density simulation is similar, but includes the additional step of parsing the data into two complementary subsets, referred to as the “training” and “testing” partitions (10). This entails estimating the marginal posterior densities for parameters of the model from a subset of the data (the training partition), which are then sampled to generate a posterior predictive distribution. This predictive distribution is compared to the complementary subset of the data (the testing partition), and the adequacy of the model is then assessed by its ability to predict relevant aspects of the excluded data. Importantly, by integrating over their respective marginal posterior probability densities, this approach accommodates uncertainty associated with those parameter estimates.
More formally, suppose that X is a set of observations { xi; i = 1, 2, …, n}. The cross-validation predictive densities compose the set {f(xi|X(i)); i = 1, 2, …, n}, where X(i) denotes all elements except xi. Conveniently, the density f(xi|X(i)) predicts what values of xi are likely when the model is fitted to all of the observations except xi. The actual value xi,obs can then be compared with this predictive density in various ways to evaluate whether xi,obs is likely under the model (4). These cross-validation predictive densities are calculated as
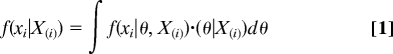 |
where the integrand is evaluated over the marginal density of the model parameters, θ.
The application of cross-validation predictive densities to the current problem is straightforward. Suppose that the history of our study group includes an event k that is inferred to occur along an internal branch vk of the phylogeny at some time in the past tk, which subtends a subclade comprising nk species. We may suspect that event k is a key innovation/opportunity (hypothesis-testing scenario), or we may wish to investigate whether this event coincides with a shift in diversification rate (data-exploration scenario). Under either mode of inference, we are essentially attempting to address the same question: “If k did not influence rates of diversification, how many species would have descended from vk?”
Given point estimates for tk and λ(k) (the rate of diversification in lineages not descended from vk, for the moment assuming a Yule stochastic branching process), the expected diversity of the group associated with event k is simply
(11). However, this conditional expectation assumes that the age and the diversification rate are known without error. Because these parameter values are estimates from data, they are associated with uncertainty. Accordingly, it would be more appropriate if the predictive diversity distribution E(nk) accommodated uncertainty in estimates of tk and λ(k). This could be achieved by integrating over the marginal posterior densities for these parameters Pr(tk|X) and Pr(λ(k)|X(k)) estimated from the joint posterior probabilities of the entire data set and the training data set, respectively. This cross-validation predictive diversity density is calculated as
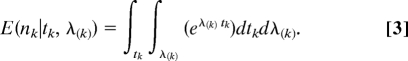 |
Although possible to solve analytically, Eq. 3 is conveniently evaluated using Monte Carlo integration. This merely involves repeatedly sampling pairs of event times and diversification-rate estimates from their respective marginal posterior densities, calculating the expected diversity, and storing each result to a list. The resulting array of expected species diversities is then summarized as the predictive diversity density, E(nk).
By comparing the predictive diversity density to the realized diversity nk,obs we can calculate the probability of realizing the observed species diversity under the estimated background diversification rate. This posterior predictive P-value is calculated as the proportion of predictive diversities that exceeds the realized diversity,
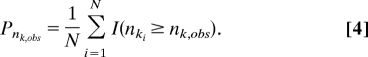 |
(5, 12, 13), where I(·) is the indicator function that takes the value 1 when its argument is true and the value 0 otherwise, and nki is the predicted species diversity based on the ith sample from the marginal probabilities of tk and λ(k). Probabilities more extreme than a specified value (conventionally α = 0.05) suggest that the observed species diversity nk,obs differs significantly from that predicted under the background diversification rate λ(k), which therefore suggests that event k is correlated with a significantly increased rate of diversification. Of course, we can also evaluate both tails of the cross-validation predictive diversity density if the hypothesis predicts that the event may be correlated with either significant increases or decreases in diversification rate or if we are adopting an exploratory data analysis (EDA) perspective in which the direction of the effect is unspecified.
We have so far implicitly assumed the availability of marginal posterior densities for the parameters required to generate the cross-validation predictive diversity density. Our approach leverages dedicated implementations of Bayesian MCMC methods to estimate marginal posterior densities for these parameters. A number of existing programs can be used to approximate the joint posterior probability density of phylogeny and absolute or relative divergence times (under a strict or a relaxed molecular clock) and, importantly, to allow estimation of the marginal posterior densities of the diversification-rate parameters [i.e., μ and/or λ, depending on whether a birth–death or a Yule prior is used to model the branching process, respectively (14–17)]. Similarly, estimation of event history relies on methods deemed most appropriate to the particular problem at hand. For instance, we can infer evolutionary changes in discrete morphological and molecular traits (8, 9, 18–22), the evolution of ecological associations (23, 24), and events in biogeographic history (25, 26).
The methods described above have been implemented in the freely available program, tRate. This command-line R package can be run on multiple platforms (Windows, Macintosh OSX, and Unix versions) to perform cross-validation predictive density simulation and to calculate posterior predictive P-values. tRate works in conjunction with other applications, which provide estimates of the marginal posterior probability distributions of divergence times and diversification-rate parameter and inferred event histories (e.g., using programs such as BEAST, BayesTraits, and AReA, as illustrated in the empirical examples below). The tRate distribution bundle may be obtained by contacting B.R.M.
Two Worked Examples
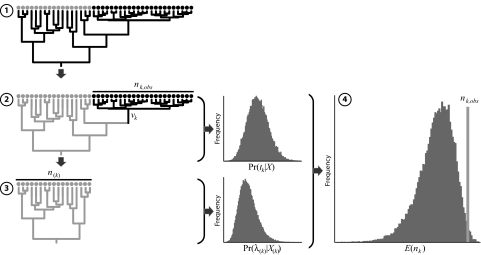
In practice, evaluating correlates of differential diversification by means of cross-validation predictive densities entails a 4-step protocol (Fig. 1). Step 1 involves estimating the joint posterior probability density of the phylogeny and divergence times from the complete set of nucleotide sequences. In step 2, the history of the event is estimated from the resulting posterior probability distribution of trees. The training data partition is defined in step 3 by excluding the subset of species associated with the event and then subjecting the remaining data set to a second round of phylogeny/divergence-time estimation. Finally, we draw from the marginal posterior densities of tk and λ(k) (estimated in steps 2 and 3, respectively) to generate the cross-validation predictive diversity density (using Eq. 3), which allows us to assess whether the realized species diversity correlated with the event is significantly higher or lower than predicted (using Eq. 4). Below we illustrate this procedure with two examples: the first investigates the influence of morphological events on rates of diversification under an EDA perspective, and the second case evaluates the impact of biogeographic events under a hypothesis-testing inference scenario.
Fig. 1.
Evaluating correlates of differential diversification rates using cross-validation predictive densities. Step 1 involves estimating the joint posterior probability density of the phylogeny and divergence times from the entire data set of nucleotide sequences. In step 2, the history of the event is estimated from the resulting posterior probability distribution of trees. Step 3 entails constructing the training data partition by removing the subset of species associated with the event and then subjecting this reduced data set to a second round of phylogeny/divergence-time estimation. Finally, we draw from the marginal posterior densities of tk and λ(k) (estimated in steps 2 and 3, respectively) to generate the cross-validation predictive diversity density (using Eq. 3), which permits us to assess whether the realized species diversity is significantly higher or lower than predicted (using Eq. 4).
Is Fruit Type Correlated with Differential Rates of Diversification in Adoxaceae?
In a recent exploration of biogeographic and morphological correlates of diversification rate in the plant clade Dipsacales (27), we inferred several instances in which trait changes occurred in close proximity to significant shifts in diversification rate [independently inferred using a maximum-likelihood approach (refs. 28 and 29)]. However, a number of these apparent correlations were nonsignificant under the key-innovation test of Ree (2). One such case involved the evolution of fruits with a single seed in Adoxaceae, which we revisit using the cross-validation predictive densities approach.
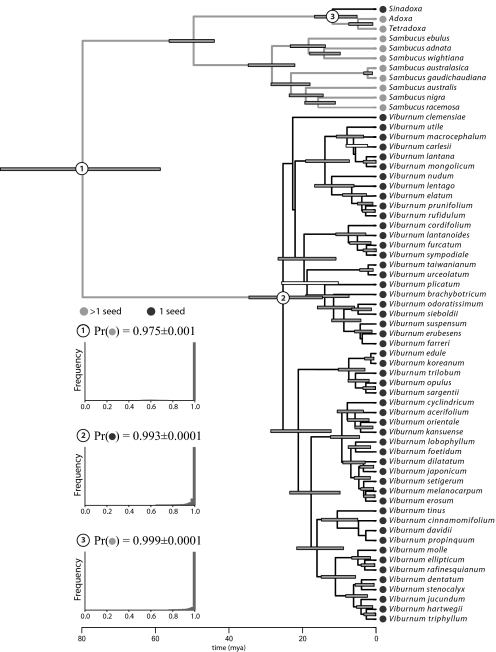
We estimated the posterior probability density of phylogeny and divergence times for Adoxaceae using BEAST (16), assuming that substitution rates evolved under the uncorrelated-lognormal model and that the branching process conformed to a Yule prior, and the evolutionary history of fruit morphology was then inferred using a Bayesian model-averaging approach (20, 22) [see supporting information (SI Text, Figs. S1 and S2, Tables S1–S3, and Dataset S1, and Table S4) for details]. These findings suggest that fruits with a single seed evolved independently in the most recent common ancestor of Viburnum and separately in Sinadoxa corydalifolia (Fig. 2).
Fig. 2.
Dated phylogeny for Adoxaceae based on a Bayesian analysis of the combined data set. Uncertainty in the tree topology and divergence times are indicated by bars on internal nodes: their length corresponds to the 95% highest posterior density (HPD) of node ages, and their shading reflects the posterior probabilities of nodes (shaded bars, nodal posterior probabilities ≥0.90; open bars, those <0.90). Circles adjacent to the tips are shaded to indicate the observed fruit type in the respective species, and branches of the tree have been shaded to reflect the posterior probability estimates for ancestral states. Numbered internal nodes correspond to the fossil constraints used to estimate divergence times (Table S2) and/or to internal nodes for which the posterior probabilities of the ancestral fruit morphologies were inferred. The inset histograms depict the estimated posterior probabilities of the ancestral trait values for the corresponding labeled nodes, which indicate that single-seeded fruits arose once along the branch subtending Sinadoxa corydalifolia and once along the branch subtending node 2.
This example illustrates several features of the cross-validation predictive densities approach. Credible intervals for divergence times suggest that these estimates are associated with considerable uncertainty. As detailed above, the current approach accommodates this uncertainty by integrating estimates over the marginal posterior density of divergence times for the node associated with the event. We are also free to adopt either a punctuated model of evolution, in which trait change is assumed to occur only at speciation [as required by some methods (ref. 1)], or a more gradualist model, in which change may occur along branches [as assumed by other methods (refs. 2 and 3)]. Under the punctuated scenario, tk corresponds only to the posterior density of divergence times for the event node (e.g., the crown node of Viburnum: node 2, Fig. 2). By contrast, under the gradualist scenario, tk comprises the composite posterior density of the two nodes bracketing the branch along which the event is inferred (e.g., nodes 1 and 2 bounding the stem of Viburnum: Fig. 2). More generally, tk can comprise the composite posterior density of divergence times for a series of ancestor-descendant branches over which the event is plausible, providing a flexible means for accommodating uncertainty in the inferred event history.
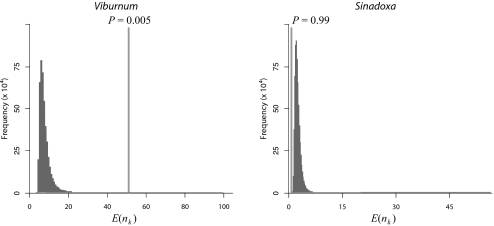
Results of the cross-validation predictive densities approach indicate that origin of single-seeded fruits in Viburnum is correlated with significantly higher species diversity than predicted under the background rate of diversification in Adoxaceae, whereas the independent origin in S. corydalifolia evidently had little impact on diversification rates (Fig. 3). By contrast, no overall correlation between fruit form and diversification rate is detected when these 2 events are evaluated collectively—either by using the key innovation test of Ree (2) or by explicitly combining the 2 posterior predictive P-values estimated for each event using the cross-validation predictive densities approach (see SI Text for details). Considering the 2 independent events in the same analysis fails to indentify the correlation of diversification rate and single-seeded fruits in the case of Viburnum.
Fig. 3.
Estimated correlation of fruit morphology with diversification rates in Adoxaceae. The graphs summarize the inferred correlation between diversification rate and the two instances of single-seeded fruits: each histogram depicts the predictive distribution of single-seeded species diversity, and the vertical line indicates the observed species diversity, with the corresponding posterior-predictive probability. These tests indicate that single-seeded fruits are correlated with significantly increased rates of diversification in Viburnum relative to background rates of diversification in Adoxaceae, but not in Sinadoxa corydalifolia.
The ability to dissect the partial correlations in these data motivates reevaluation of the problem—although both Viburnum and S. corydalifolia possess fruits with a single seed, they appear to be dispersed in different ways. The drupes of Viburnum are dispersed by birds, whereas the fruits of S. corydalifolia may be dispersed by water (30). Accordingly, we may wish to revise our original question (the influence of single-seeded fruits) to focus on the role of bird-dispersed fruits, which could subsequently be explored in other groups. This example illustrates how data exploration—and specifically the evaluation of individual events—may guide the formulation of hypotheses regarding correlates of diversification rate.
Did Dispersal to South America Promote Rates of Diversification in Lupinus?
Hughes and Eastwood (31) recently documented the adaptive radiation of Lupinus, in which the movement of this North American plant clade into South America apparently promoted extremely rapid rates of diversification via geographically driven speciation across the newly forming Andean mountain range. Although this study demonstrated high absolute rates of diversification in the Andean lineage, the lack of appropriate methods precluded statistical evaluation of the predicted correlation between elevated rates of diversification and dispersal of Lupinus into the Andes. Because biogeographic range and morphological traits are inherited in fundamentally different ways (25, 32), existing trait-based tests are inappropriate for evaluating the impact of biogeographic history on rates of diversification. Under the cross-validation predictive densities approach, however, biogeographic dispersal is merely another type of historical event.
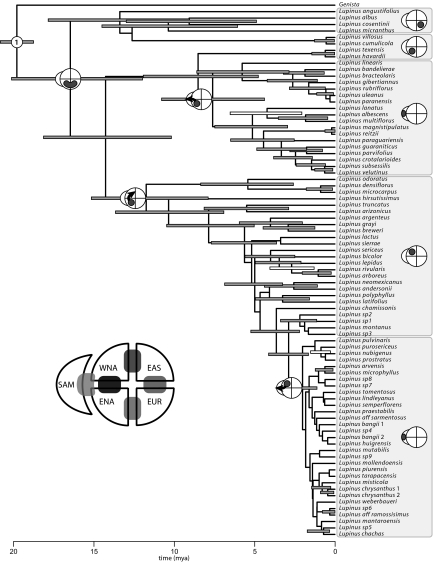
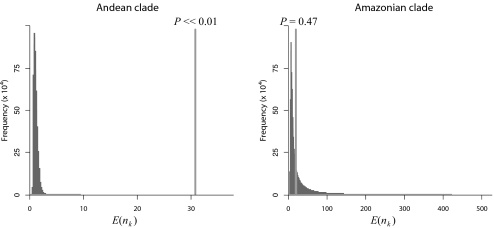
The posterior probability density of chronograms for Lupinus was estimated as in the previous example, and the biogeographic history of the group was inferred using a maximum-likelihood approach (25, 26) [see SI Text, Fig. S1, Tables S1 and S2, Dataset S2, and Table S5]. Two episodes of dispersal into South America were inferred: one lineage dispersed from eastern North America into lowland habitats of South America and the second from Central America into the Andean region (Fig. 4). Results of the cross-validation predictive densities approach suggest that movement into the Andes of South America is correlated with significantly higher species diversity in Lupinus [as predicted by Hughes and Eastwood (ref. 31)], whereas the dispersal into the lowland habitats apparently had little impact on diversification rate (Fig. 5).
Fig. 4.
Dated phylogeny for Lupinus based on a Bayesian analysis of the combined data set. Uncertainty in the tree topology and divergence times are indicated by bars on internal nodes: their length corresponds to the 95% highest posterior density (HPD) of node ages, and their shading reflects the posterior probabilities of nodes (shaded bars, nodal posterior probabilities ≥0.90; open bars, those <0.90). The numbered internal node corresponds to the fossil constraint used to estimate divergence times (Table S2). Observed species ranges are indicated across the tips of the tree, and the diagrams adjacent to internal nodes indicate the inferred locations of biogeographic dispersal events, including 2 into South America.
Fig. 5.
Estimated correlation between biogeographic dispersal into South America and rates of diversification in Lupinus. Each histogram depicts the predictive distribution of South American species diversity, and the vertical line indicates the observed species diversity, with the corresponding posterior-predictive probability. These results indicate that the Andean clade experienced significantly increased rates of diversification relative to background rates of diversification in Lupinus, whereas the other South American lineage did not.
Importantly, combination of the posterior predictive P-values for the 2 episodes of South American dispersal in Lupinus does not support an overall correlation between dispersal into South America and increased diversification rate (see SI). Again, the analysis combining both cases obscures the strong correlation between diversification and Andean dispersal. The hypothesis that dispersal of Northern Hemisphere groups into the Andes promotes rates of diversification can be tested by evaluating other groups.
Discussion
The versatility of the current approach is attractive, as it expands the scope of evolutionary events that can be considered (to include essentially any estimable historical variable) and also extends the mode of inference from a strictly hypothesis-testing realm to enable an exploratory data analysis mode of inquiry. Perhaps most importantly, this approach permits investigation of both replicated and unique historical events. As demonstrated by the empirical examples, the ability to evaluate the influence of unique events may permit new insights into factors influencing rates of diversification. Furthermore, the impact of a number of similar historical events may be considered collectively to explore the broader context and generality of a putative correlation. Moreover, the investigator is free to combine probabilities associated with individual events in a manner that is most appropriate to the predicted effect. That is, different omnibus statistics capture different aspects of what we mean by a “significant correlation” (33, 34). Our hypothesis may lead us to predict that the study variable will exert a consistent (but moderate) influence on diversification rates or may lead us to expect large (but variable) differences in diversification rates, etc. The diversity of available omnibus statistics should engender a more statistically nuanced understanding of the relationships between replicated events and diversification rates.
In addition to providing a measure of flexibility, the cross-validation predictive densities approach is also appealing for its ability to accommodate various sources of uncertainty. As an inference problem, evaluating diversification-rate correlates relies critically on parameters that are inherently associated with considerable uncertainty (estimates of divergence times, diversification-rate parameters, and event history). Accordingly, our inferences are apt to be more robust if we endeavor to incorporate these sources of error (35, 36). Significantly, the current approach is also robust to a common concern associated with key-innovation tests. Significance under these tests is often assessed by reference to expectations specified by a Yule stochastic branching process in which the probability of speciation in all lineages is equal and independent at any moment in time (37). However, empirical data typically exhibit a poor fit to this model (owing to ubiquitous fluctuation in rates across lineages), which is apt to increase type I error rates (38). By contrast, the current approach estimates the diversification-rate parameter from all of the lineages that are not associated with the event λ(k), which effectively captures the distribution of diversification rates associated with stochastic fluctuations and/or the variation stemming from other deterministic factors. Accordingly, the current approach provides a more appropriate, empirically based null expectation with which to identify events that are correlated with significant departures in diversification rate.
The cross-validation predictive densities approach will be ill suited to various inference scenarios. For instance, candidate traits that evolve at relatively high rates are likely to give rise to complex histories (with multiple reversals, etc.), which will complicate evaluation under the approach described here. Fortunately, methods designed to accommodate complex trait histories are available (e.g., ref. 2). Furthermore, as noted by Maddison (39), a preponderance of species may exhibit a given character state either because that trait has promoted rates of diversification or because of asymmetric transition probabilities between states (e.g., state 1 may be more prevalent than state 0 in the study group if q01 ≫ q10). Accordingly, when we have reason to suspect such a bias (e.g., on the basis of estimated rate coefficients or from knowledge of the underlying genetic mechanisms of character change, e.g., ref. 40), it will be most appropriate to use methods developed to address this problem (e.g., ref. 3).
As a practical point, application of the current method should carefully consider the issue of taxon sampling. Ideally, taxa should be sampled so as to provide a proportional representation of species that are and are not associated with the event of interest [nk and N(k), respectively], as this will reduce associated biases. Moreover, the taxonomic scope of an analysis will circumscribe the scope of possible conclusions. For example, the conclusion that single-seeded fruits are associated with increased rates of diversification in Viburnum is a relative statement made with reference to Adoxaceae (which is the group that we used to estimate the background diversification rate). Investigators should caution against overstating the scope of such conclusions. Finally, it is important to include an adequate sample from N(k), as this will reduce the error variance in estimates of λ(k). If too few nonevent lineages are sampled, the estimated marginal posterior probability density for the background diversification rate is likely to become vague, which will artificially broaden the posterior predictive distribution and decrease the ability to detect the effect of an event on diversification rate.
As described above, the approach assumes a pure-birth, Yule stochastic branching model with a single rate parameter, λ. In principle, this approach could readily be generalized to more complex stochastic branching processes. For instance, divergence-time estimators that employ birth–death priors (14, 15, 17) could be used to approximate the posterior densities for the corresponding birth and death rate parameters, λ and μ, which could then be used to generate the predictive diversity density [e.g., using estimators similar to those proposed by Magallón and Sanderson (ref. 41)]. In practice, however, it may prove difficult to reliably estimate the rate parameters of a birth–death process (1, 42).
The search for correlates of diversification typically involves a single (usually morphological) variable. For several reasons, however, the evaluation of a particular event in isolation may not be ideal. Imagine, for example, that the probability of diversification has been significantly increased by the origin of a given trait and that the phylogenetic distribution of this trait is nested within a second, more widely distributed trait. In this situation, the bona fide causal correlation between diversification rate and the first trait may well create an illusory correlation between diversification rate and the second trait. Accordingly, this “piggy-backing” effect may lead to the inference of spurious correlations (27), particularly when we ignore the potential influence of other events. Moreover, the evaluation of events in isolation precludes the discovery of interactions in which the evolution of one trait mediates or facilitates the effects of subsequently evolved features (43). Such considerations emphasize the need for methods that can simultaneously evaluate the influence of multiple variables on rates of diversification. In theory, the cross-validation predictive density approach could be extended in this direction by comparing the predictive “influence” of various events (4).
Concluding Thoughts.
Existing methods have been developed in the context of testing key innovation hypotheses, which (appropriately) evaluate the overall association between a given character state and diversification rate. Ideally, such hypotheses draw upon robust evolutionary theories of the general mechanisms that influence probabilities of speciation and/or extinction, from which specific instances can be identified and tested. Unfortunately, theory on the factors influencing rates of diversification is currently rather thin, such that the need to approach all inference problems from a hypothesis-testing perspective is somewhat problematic. For instance, the virtually exclusive reliance on this inference mode increases the incidence of type III error (i.e., “testing” hypotheses that were initially suggested by the data) and limits the ability of empirical data to directly inform theory. Accordingly, it is often desirable (and entirely appropriate) to alternate between hypothesis testing and data exploration; it is reasonable to explore empirical data, learn something in the process, and then (re)formulate a hypothesis that can subsequently be evaluated using additional data.
Moreover, some inference problems naturally pertain to the influence of particular historical events on rates of diversification. That is, rather than asking “Are rates of diversification correlated with a particular morphological/biogeographic/ecological state?” we may instead wish to ask “Are rates of diversification correlated with a particular morphological/biogeographic/ecological event?” We believe that it is both useful and valid for evolutionary biologists to search for the cause(s) of individual evolutionary events, even when broader comparative tests may not be possible. Our hope is that the cross-validation predictive densities approach will facilitate the study of both unique and replicated historical events and thereby will foster the reciprocal illumination between empirical patterns and theoretical explanations that is critical to understanding of the causes of diversification.
Supplementary Material
Acknowledgments.
We thank Mary Moore, John Huelsenbeck, Tom Near, Stephen Smith, and David Tank for stimulating discussion. We are indebted to Paul Fine and Mark Pagel for constructive and insightful reviews. Our work on this problem has been supported by the NSF through the Phytogeography Working Group at the National Evolutionary Synthesis Center (NESCent), Cyberinfrastructure for Phylogenetic Research (CIPRES), and two Assembling the Tree of Life (ATOL) awards.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807230106/DCSupplemental.
References
- 1.Paradis E. Statistical analysis of diversification with species traits. Evolution. 2005;59:1–12. [PubMed] [Google Scholar]
- 2.Ree RH. Detecting the historical signature of key innovations using stochastic models of character evolution and cladogenesis. Evolution. 2005;59:257–265. [PubMed] [Google Scholar]
- 3.Maddison WP, Midford PE, Otto SP. Evaluating a binary character's effect on speciation and extinction. Syst Biol. 2007;56:701–710. doi: 10.1080/10635150701607033. [DOI] [PubMed] [Google Scholar]
- 4.Gelfand AE. In: Markov Chain Monte Carlo in Practice. Gilks WR, Richardson S, Spiegelhalter D, editors. New York: Chapman & Hall; 1996. pp. 145–161. [Google Scholar]
- 5.Gelman AE, Dey DK, Chang H. In: Bayesian Statistics. Bernardo JM, Berger JO, Dawid AP, Smith AFM, editors. Vol 4. New York: Oxford Univ Press; 1992. pp. 147–167. [Google Scholar]
- 6.Nielsen R, Huelsenbeck JP. In: Altman RB, Dunker AK, Hunter L, Lauderdale K, Klein TE, editors. Pacific Symposium on Biocomputing, Proceedings; Singapore: World Scientific; 2002. pp. 576–588. [Google Scholar]
- 7.Bollback JP. Bayesian model adequacy and choice in phylogenetics. Mol Biol Evol. 2002;19:1171–1180. doi: 10.1093/oxfordjournals.molbev.a004175. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen R. Mapping mutations on phylogenies. Syst Biol. 2002;51:729–739. doi: 10.1080/10635150290102393. [DOI] [PubMed] [Google Scholar]
- 9.Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Syst Biol. 2003;52:131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- 10.Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. Proceedings of the Fourteenth International Joint Conference on Artificial Intelligence; 1995. pp. 1137–1143. [Google Scholar]
- 11.Harris TE. The Theory of Branching Processes. Berlin: Springer; 1964. [Google Scholar]
- 12.Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian Data Analysis. London: Chapman & Hall; 1996. [Google Scholar]
- 13.Meng X-L. Posterior predictive P-values. Ann Stat. 1994;22:1142–1160. [Google Scholar]
- 14.Aris-Brosou S, Yang Z. The effects of models of rate evolution on estimation of divergence dates with a special reference to the metazoan 18S rRNA phylogeny. Syst Biol. 2002;51:703–714. doi: 10.1080/10635150290102375. [DOI] [PubMed] [Google Scholar]
- 15.Aris-Brosou S, Yang Z. Bayesian models of episodic evolution support a late Precambrian explosive diversification of the Metazoa. Mol Biol Evol. 2003;20:1947–1954. doi: 10.1093/molbev/msg226. [DOI] [PubMed] [Google Scholar]
- 16.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst Biol. 2007;56:453–466. doi: 10.1080/10635150701420643. [DOI] [PubMed] [Google Scholar]
- 18.Huelsenbeck JP, Bollback JP. Empirical and hierarchical Bayesian estimation of ancestral states. Syst Biol. 2001;50:351–366. [PubMed] [Google Scholar]
- 19.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 20.Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am Nat. 2006;167:808–825. doi: 10.1086/503444. [DOI] [PubMed] [Google Scholar]
- 21.Bollback JP. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics. 2006;7:88. doi: 10.1186/1471-2105-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 23.Huelsenbeck JP, Rannala B, Larget B. A Bayesian framework for the analysis of cospeciation. Evolution. 2000;54:352–364. doi: 10.1111/j.0014-3820.2000.tb00039.x. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck JP, Rannala B, Larget B. In: Tangled Trees: Phylogenies, Cospeciation, and Coevolution. Page RDM, editor. Chicago: Chicago Univ Press; 2002. pp. 93–119. [Google Scholar]
- 25.Ree RH, Moore BR, Donoghue MJ. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59:2299–2311. [PubMed] [Google Scholar]
- 26.Moore BR, Smith SA, Donoghue MJ. A likelihood model for incorporating fossil data in the inference of historical biogeography. Evolution. 2009 in press. [Google Scholar]
- 27.Moore BR, Donoghue MJ. Correlates of diversification in the plant clade Dipsacales: geographic movement and evolutionary innovations. Am Nat. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- 28.Moore BR, Chan KMA, Donoghue MJ. In: Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life. Bininda-Emonds ORP, editor. Dordrecht, The Netherlands: Kluwer Academic; 2004. pp. 487–533. [Google Scholar]
- 29.Chan KMA, Moore BR. SymmeTREE: performing whole-tree tests of diversification rate variation. Bioinformatics. 2005;21:1709–1710. doi: 10.1093/bioinformatics/bti175. [DOI] [PubMed] [Google Scholar]
- 30.Donoghue MJ, Bell CD, Winkworth RC. The evolution of reproductive characters in Dipsacales. Int J Plant Sci. 2003;164:S453–S464. [Google Scholar]
- 31.Hughes C, Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc Natl Acad Sci USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronquist F. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst Biol. 1997;46:195–203. [Google Scholar]
- 33.Nee S, Barraclough TG, Harvey PH. In: Biodiversity: A Biology of Numbers and Differences. Gaston KJ, editor. Oxford: Blackwell Science; 1996. pp. 230–252. [Google Scholar]
- 34.Chan KMA, Moore BR. Whole-tree methods for detecting differential diversification rates. Syst Biol. 2002;51:855–865. doi: 10.1080/10635150290102555. [DOI] [PubMed] [Google Scholar]
- 35.Huelsenbeck JP, Rannala B, Masly JP. Accommodating phylogenetic uncertainty in evolutionary studies. Science. 2000;288:2349–2350. doi: 10.1126/science.288.5475.2349. [DOI] [PubMed] [Google Scholar]
- 36.Lutzoni F, Pagel M. Accelerated molecular evolution as a consequence of transitions to mutualism. Proc Natl Acad Sci USA. 1997;94:11422–11427. doi: 10.1073/pnas.94.21.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yule GU. A mathematical theory of evolution, based on the conclusions of Dr. J. C. Willis. Philos Trans R Soc Lond B. 1924;213:21–87. [Google Scholar]
- 38.de Queiroz A. Interpreting sister-group tests of key innovation hypotheses. Syst Biol. 1998;47:710–718. doi: 10.1080/106351598260699. [DOI] [PubMed] [Google Scholar]
- 39.Maddison WP. Confounding asymmetries in evolutionary diversification and character change. Evolution. 2006;60:1743–1746. [PubMed] [Google Scholar]
- 40.Donoghue MJ, Ree RH, Baum DA. Phylogeny and the evolution of flower symmetry in the Asteridae. Trends Plants Sci. 1998;3:311–317. [Google Scholar]
- 41.Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 42.Kubo T, Iwasa Y. Inferring rates of branching and extinction from molecular phylogenies. Evolution. 1995;49:694–704. doi: 10.1111/j.1558-5646.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 43.Donoghue MJ. Key innovations, convergence, and success: macroevolutionary lessons from plant phylogeny. Paleobiology. 2005;31:77–93. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.