Abstract
Protein glycosylation is an important element of biologic systems because of its significant effects on protein properties and functions. Although prominent within all domains of life, O-linked glycosylation systems modifying serine and threonine residues within bacteria and eukaryotes differ substantially in target protein selectivity. In particular, well-characterized bacterial systems have been invariably dedicated to modification of individual proteins or related subsets thereof. Here we characterize a general O-linked glycosylation system that targets structurally and functionally diverse groups of membrane-associated proteins in the Gram-negative bacterium Neisseria gonorrhoeae, the etiologic agent of the human disease gonorrhea. The 11 glycoproteins identified here are implicated in activities as varied as protein folding, disulfide bond formation, and solute uptake, as well as both aerobic and anaerobic respiration. Along with their common trafficking within the periplasmic compartment, the protein substrates share quasi-related domains bearing signatures of low complexity that were demonstrated to encompass sites of glycan occupancy. Thus, as in eukaryotes, the broad scope of this system is dictated by the relaxed specificity of the glycan transferase as well as the bulk properties and context of the protein-targeting signal rather than by a strict amino acid consensus sequence. Together, these findings reveal previously unrecognized commonalities linking O-linked protein glycosylation in distantly related life forms.
Keywords: bacteria, glycoprotein, pilin, post-translational modification, pgl
Targeting of a broad array of heterogeneous protein substrates is a fundamental feature of O-linked glycosylation in eukaryotes and N-linked glycosylation systems in all domains of life (1). The covalent addition of uniform glycan tags enables diverse proteins to be recognized and modified by conserved core processes influencing protein trafficking, folding, and turnover. Glycosylation substrate selection involves not only colocalization with the glycosylation machinery but also the presence and availability of occupancy sites. In bacterial, archaeal, and eukaryotic N-linked systems, glycosylation sites are characterized by the N-X-S/T tripeptide sequon that is necessary but not sufficient for modification (2). In contrast, structural determinants intrinsic to O-linked protein glycosylation have been more difficult to characterize (3). In higher eukaryotes, the seemingly most prevalent (and arguably the most thoroughly studied) form of O-linked glycosylation is termed mucin-type glycosylation, in which cell-surface proteins are modified with N-acetylgalactosamine through the action of polypeptide N- α-acetylgalactosaminyltransferases (4). Glycosylation acceptor sites in this system are associated with domains rich in serine, threonine, and proline that are localized to coiled or turn regions and that are often tandemly repeated (5). In the case of the eukaryotic O-linked N-acetylglucosamine modification mediated by the O-GlcNAc transferase, few clear patterns have emerged although systematic peptide-based assays have begun to reveal some sequence preferences (6, 7).
Bacterial protein glycosylation systems have come under enhanced scrutiny because of the increasing frequencies with which they are seen in pathogenic and symbiotic species, as well as their potential for exploitation in recombinant glycoprotein engineering strategies. Studies of these systems also have the potential to provide insight into the evolution of glycan biosynthesis, glycosyltransferases, and structural determinants of substrate recognition. Indeed, the Gram-negative species Campylobacter jejuni targets multiple proteins at asparagine residues like its eukaryotic counterparts and uses an oligosaccharyltransferase (OTase) originally recognized on the basis of its sequence identity with the STT3, the catalytic subunit of eukaryotic OTases (8). An STT3-like OTase also acts in N-linked glycosylation in the thermophilic archaeaon Pyrococcus furiosus (9). In contrast, no clear relationships have been delineated for conventional O-linked systems that predominate in eukaryotes and bacteria. Most conspicuously, bacterial O-linked systems are uniformly dedicated to the modification of individual proteins or sets of highly related proteins. Prime examples include the subunits of S-layers, flagella, and Type IV pili, as well as non-pilus adhesins, such as autotransporters and 2-partner secretion pathway exoproteins (10). Evidence for glycosylation of multiple, structurally related protein substrates has been reported in Bacteroides fragilis, although the nature of the glycan linkage involved remains unknown (11). Two surface lipoproteins of Mycobacterium tuberculosis have been proven to be O-glycosylated, and an implicated transferase shares some structural similarity to eukaryotic protein mannosyltransferases (12, 13). However, whether this system is representative of bona fide general protein glycosylation has not been established.
The PilE pilin protein subunit of the Type IV pilus colonization factor expressed by the Gram-negative human pathogen Neisseria gonorrhoeae (Ngo) undergoes O-glycosylation at a single, defined serine residue (Ser-63) (14). Genetic analyses including interspecies complementation have revealed that this system (termed pgl for pilin glycosylation) is remarkably similar to the N-linked system of C. jejuni with regard to the use of a peptide-proximal 2,4 diacetamido-2,4,6 trideoxyhexose (DATDH) sugar and related biosynthetic pathways for generating lipid-linked glycan substrates (15). These systems then diverge at the step of oligosaccharide transfer with the Ser-targeting PglO OTase acting in Ngo and the sequon targeted PglB OTase in C. jejuni (supporting information Fig. S1). Thus, in contrast to eukaryotic O-linked glycosylation systems, the gonococcal O-linked system uses en bloc transfer of oligosaccharide from a lipid-linked donor rather than single UDP-monosaccharide donors and downstream glycan elaboration.
Results
Evidence for Multiple Glycoproteins in N. gonorrhoeae.
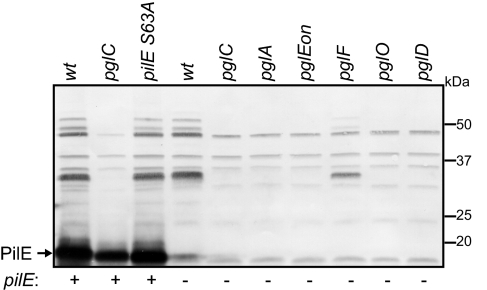
We made the observation that polyclonal rabbit antiserum raised against glycosylated PilE protein (in purified Type IV pili) reacted with multiple gonococcal proteins when examined by immunoblotting (Fig. 1). This pattern of immunoreactivity occurred independently of PilE expression and PilE glycosylation (as seen in a PilE S63A background) but was abolished in all pgl null mutants except that lacking PglF (encoding a putative flippase), in which the case the signal intensities were diminished. PglF has been modeled as a membrane translocase for the lipid-linked oligosaccharide, and we previously showed that PilE is hypoglycosylated in a pglF null mutant [residual glycosylation presumably being due to the presence of partially compensatory translocases in this background (14)]. In addition, specific immunoreactivity was absent in backgrounds expressing either the DATDH monosaccharide (pglA) or the O-acetylated (O-Ac) HexHexDATDH trisaccharide (pglEon) glycan precursor forms. These findings suggested that the immunoreactive proteins shared specific glycan-associated epitopes generated by the same glycosylation pathway that acts on PilE.
Fig. 1.
Evidence for multiple Ngo glycoproteins revealed via detection of glycan-associated epitopes. Immunoblotting of whole-cell lysates with rabbit serum (termed antiglycan) raised against purified PilE bearing the O-AcHexDATDH glycan reacts with multiple proteins in a pgl pathway–dependent fashion (see Fig. S1 for an overview of the pgl pathway). Note that the lack of immunoreactivity observed in the pglA and pglEon backgrounds is not associated with a lack of glycosylation but rather the presence of monosaccharide DATDH and trisaccharide O-AcHexHexDATDH glycans, respectively.
Identification of a Major Subset of N. gonorrhoeae Glycoproteins.
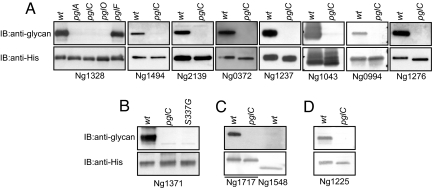
To examine these possibilities in more detail, a cell fraction enriched for the immunoreactive proteins was subjected to 2D gel electrophoresis and probed by immunoblotting with antiglycan antibodies (Fig. S2). Of the more than 15 immunoreactive proteins detected, 10 candidates were identified using MS analyses of corresponding spots excised from preparative gels. However, insufficient material was available by this method to unambiguously characterize glycosylation status. Therefore, gonococcal strains were individually engineered so that each protein carried a hexahistidine (6xHis) extension at its carboxy-terminus while remaining expressed from its endogenous locus and promoter. After affinity purification, it was confirmed that each of the 10 proteins retained pglC-dependent reactivity with glycan-specific antibodies, and in the case of Ng1328 this property was also dependent on pglA and pglO (but not pglF) (Fig. 2 A–C).
Fig. 2.
Affinity purification confirms the identities of candidate Ngo glycoproteins. (A–D) C-terminally 6xHis-tagged candidate proteins were affinity purified from wild-type (wt) and pgl backgrounds and tested for glycan-associated antigenicity by immunoblotting. (B) For Ng1371, a mutant form of the protein carrying a glycine substitution at Ser-337 was also purified and tested, whereas for Ng1717 (C), the Ng1548 paralogue (Fig. S3) was examined in parallel. (D) Ng1225 was identified as a potential glycoprotein on the basis of its lipoprotein processing site and proximal ASP-rich LCR (Table S1).
Mass Spectrometric Confirmation of Glycosylation Status via Glycopeptide Identification.
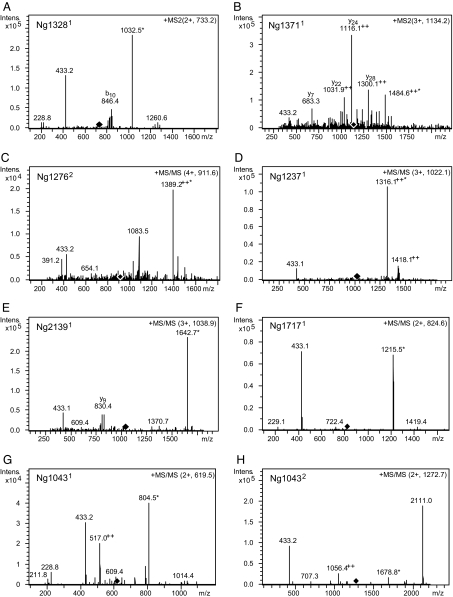
We next identified glycosylated peptides derived by proteolysis from these purified proteins using liquid chromatography-electrospray ionization–tandem mass spectrometry and collision-induced dissociation (CID). Data indicative of glycopeptides carrying the O-AcHexDATDH glycan moiety [as diagnostic oxonium ion fragments appearing at mass to charge ratio (m/z) 433.2 due to CID] were obtained from 7 of the 10 proteins (Fig. 3). In the case of Ng1043, 2 non-overlapping glycopeptides were detected (Fig. 3 G and H), and the signals seen for the larger indicated the presence of 2 glycans. Signals indicating the presence of 2 glycans were also detected on the glycopeptide found for Ng1276 (Fig. 3C).
Fig. 3.
Identification of glycopeptides derived from endoproteolytic cleavage of affinity-purified proteins using tandem MS. CID experiments of the glycopeptide-related molecular ions yield mainly cleavage at glycosidic bonds, revealing characteristic oxonium ions at m/z 433 and m/z 229 (corresponding to O-AcHexDATDH and DATDH glycans, respectively). Diamonds denote the m/z of the precursor ion fragmented in the spectrum. Superscript numbering appended to ORF designations indicates peptides bearing 1 (1) or 2 (2) glycans. The asterisk marks the unmodified peptide after loss of the glycan moiety resulting from CID. (A) MS/MS spectrum of [M + 2H] 2+ at m/z 733.2 derived from Ng1328 by AspN cleavage. (B) MS/MS spectrum of [M + 3H] 3+ at m/z 1134.2 derived from Ng1371 by tryptic cleavage. (C) MS/MS spectrum of [M + 4H] 4+ at m/z 911.6 derived from Ng1276 by tryptic cleavage. (D) MS/MS spectrum of [M + 4H] 4+ at m/z 1022.1 derived from Ng1237 by AspN cleavage. (E) MS/MS spectrum of [M + 3H] 3+ at m/z 1038.9 derived from Ng2139 by tryptic cleavage. (F) MS/MS spectrum of [M + 2H] 2+ at m/z 824.6 derived from Ng1717 by proteinase K cleavage. (G) MS/MS spectra of [M + 2H] 2+ at m/z 619.5 derived from Ng1043 by tryptic cleavage. (H) MS/MS spectra of [M + 2H] 2+ at m/z 1272.7 derived from Ng1043 by tryptic cleavage.
Evidence for Glycan Linkage at Serine Residues.
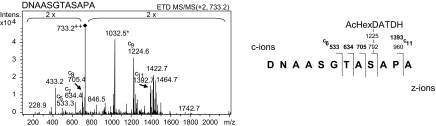
Consistent with the pgl-dependent glycosylation of Ser-63 of PilE, we noted that serine residues were present in all 8 glycopeptides, whereas threonine residues were represented in only 5 instances (see below). Because the Ng1371 glycopeptide only contained 2 serines, missense mutants bearing a substitution individually at each site were constructed and examined. Whereas substitution at Ser-327 with a non–hydroxyl-bearing amino acid had no effect on glycan antibody reactivity (data not shown), that at Ser-337 abolished reactivity (Fig. 2B). Directly locating the exact sites of glycosylation using conventional MS/MS CID techniques is difficult owing to the relative lability of glycosidic bonds. We therefore used ion trap MS with electron transfer dissociation (ETD). As shown for the 165-DNAASGTASAPA-176 glycopeptide from Ng1328, the ETD spectrum provided the necessary peptide backbone fragmentation to assign Ser-173 as bearing the glycan (Fig. 4). Its location to Ser-173 is derived by detection of fragment ions c9+ (DNAASGTAS) and c11+ (DNAASGTASAP) at m/z 1224.6 and 1392.7, respectively, representing the only c-ions of the c-ion series c6+ to c11+ with a mass increase of 432 Da, which corresponds to the glycan residue. Nonetheless, a missense form of Ng1328 with a non–hydroxyl-bearing amino acid replacement at this residue retained reactivity with the glycan antibodies (data not shown), showing that this protein carries an additional acceptor site. Taken together with the observations that serine is the only hydroxyl-bearing residue in Ng1043 and Ng2139 glycopeptides, these data strongly suggest that Ngo general protein glycosylation is primarily attributable to linkage at serine residues. However, we cannot formally rule out that threonine or other hydroxyl-bearing residues might also be targeted.
Fig. 4.
Identification of Ser-173 of Ng1328 as a glycan acceptor site using ETD. Fragmentation of the doubly charged peptide at m/z 733.2 shows the site of O-AcHexDATDH modification by detection of fragment ions c9+ and c11+ at m/z 1224.6 and 1392.7, respectively. These represent the only c-ions with a mass increase of 432 Da, which corresponds to the glycan moiety.
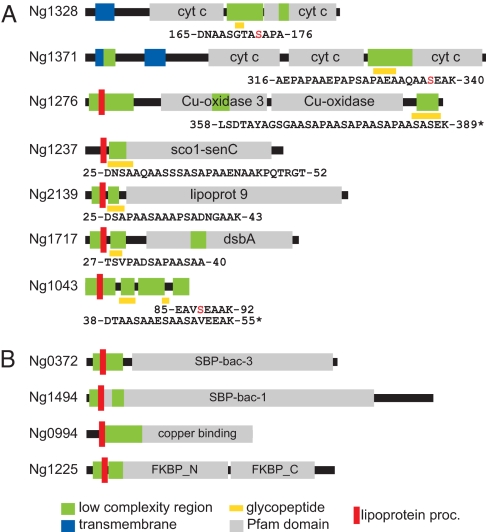
Glycan Occupancy Sites Are Associated with Low-Complexity Regions.
Examination of the glycopeptides revealed a remarkably conserved set of features, including an overabundance of alanine, serine, and proline residues (Fig. 5A). At the same time, the relative positions of the glycopeptides within the proteins were quite diverse, being found in the N-terminus in 4 cases, in the middle in 2 others, and at the C-terminus in another. To confirm the biased composition of the glycopeptides, glycoproteins were examined using the SEG algorithm to identify low-complexity regions (LCRs) (16). All 8 glycopeptides were predicted to be encompassed within LCRs (Fig. 5A), and although specific glycopeptides have yet to be identified from the 4 remaining substrates, each of these glycoproteins carries an alanine, serine, and proline (ASP)-rich LCR near their mature N-termini similar to those seen in the other glycoproteins (Fig. 5B and Table S1). To gain further evidence for the association between glycosylation and these ASP-rich LCR elements, the Ng1548 paralogue of Ng1717, which lacks the corresponding ASP-rich LCR domain (Fig. S3), was affinity tagged, purified, and found to lack reactivity with glycan antibodies (Fig. 2C). Finally, an in silico approach predicated on the presence of the ASP-rich LCR together with periplasmic targeting identified a number of candidate glycoproteins, and among these we were able to successfully affinity tag and purify Ng1225 (Fig. 5B and Table S1). Glycan antibodies reacted with affinity-purified Ng1225, and this recognition was abolished when the protein was derived from a pglC background (Fig. 2D).
Fig. 5.
Domain organization and structural architecture of Ngo glycoproteins. (A) Glycoproteins for which glycopeptides have been identified by MS/MS. Regions encompassed by glycopeptides are indicated by yellow rectangles, and specific residues are numbered according to those of unprocessed polypeptides. Serines identified as acceptor sites (via ETD, substitution mutation, or by virtue of being the sole hydroxyl-bearing residue) are labeled in red. Asterisks denote glycopeptides bearing multiple glycan moieties. (B) Glycoproteins for which glycopeptides have yet to be identified. Sequences of the LCRs shown for these proteins are found in Table S1. Further details and specific Pfam family identifiers are found in Table 1.
General Characteristics and Features of N. gonorrhoeae Glycoproteins.
Ngo glycoproteins share a number of intriguing features including their membrane tethering and translocation to or through the periplasm (Table 1). Trafficking to this compartment is likely a prerequisite for glycosylation because the PglO OTase is predicted to act in the periplasm (15, 17). Potential functional connections can also be discerned among them. For example, Ng1276 is a copper-containing nitrite reductase essential for growth under oxygen-limiting conditions that has been proposed to acquire electrons via Ng0994, a lipid-linked azurin (18). Ng1371 is a multiheme, c-type cytochrome of the cytochrome cbb3 oxidase complex (the sole predicted terminal respiratory oxidase in Ngo) and is believed to transfer electrons to the catalytic subunit (19), whereas Ng1328 is an integral-membrane, diheme c-type cytochrome that potentially channels electrons to both Ng1276 and Ng1371 (20). On the basis of their relative signal intensities in immunoblotting, Ng1276, Ng 1328, and Ng1371 represent the most abundant glycoproteins after PilE (Fig. S4). Ng1717 is an isoform of the disulfide oxidoreductase DsbA, whose reoxidation is linked via DsbB and quinones to the terminal oxidase of electron transport (21), which in this case is the cytochrome cbb3 oxidase. Finally, Ng1237 is a lipoprotein member of the Sco protein family whose constituents are implicated in copper binding and cytochrome oxidase biogenesis (22). The presence of a thioredoxin fold in these proteins suggests that they may have thiol/disulfide-based oxidoreductase activity and metal ion homeostasis functions, and an Ng1237 null mutant was shown to be hypersensitive to oxidative stress (23). Together, these findings establish a link between protein glycosylation, electron transport systems, and redox components. The remaining glycoproteins include a peptidyl prolyl-isomerase (Ng1225), 2 solute-binding proteins associated with ABC transport systems (Ng0372 and Ng1494), and 2 lipoproteins of unknown function (Ng1043 and Ng2139). We further note that with the exceptions of PilE as well as Ng1225 and Ng2139 [for which evidence for surface exposure exists (24, 25)], the glycoproteins are predicted to function in the periplasmic compartment, although their precise sites of localization await confirmation.
Table 1.
Features and properties of Ngo glycoproteins
| ORF | Protein name | Pfam domain(s) | Predicted mass (kDa) | Function | Target site associated with tandem repeat elements | Membrane association | Predicted localization |
|---|---|---|---|---|---|---|---|
| — | PilE | PF07963 PF00114 | Type IV pilus subunit | − | Transmembrane domain | Periplasm/cell surface | |
| Ng0372 | — | PF00497 | 29 | ABC transporter, periplasmic binding protein (amino acid) | ND | Lipoprotein | Periplasm |
| Ng0994 | Laz | PF00127 | 19 | Lipid-modified azurin Cu binding resistance to oxidative stress | ND | Lipoprotein | Periplasm |
| Ng1043 | — | — | 11 | — | + | Lipoprotein | Periplasm |
| Ng1225 | Mip | PF00254 PF01346 | 29 | Peptidyl-prolyl isomerase | ND | Lipoprotein | Periplasm/cell surface |
| Ng1237 | Sco | PF02630 | 23 | Cu homeostasis, resistance to oxidative stress | − | Lipoprotein | Periplasm |
| Ng1276 | AniA | PF00394 PF07732 | 41 | Cu containing nitrite reductase | + | Lipoprotein | Periplasm |
| Ng1328 | CycB | PF00034 | 29 | Cytochrome c5 | − | Transmembrane domain | Periplasm |
| Ng1371 | CcoP | PF00034 | 48 | Cytochrome cbb3 oxidase subunit | + | Transmembrane domain | Periplasm |
| Ng1494 | PotF | PF01547 | 41 | ABC transporter, periplasmic binding protein (polyamine) | ND | Lipoprotein | Periplasm |
| Ng1717 | DsbA | PF01323 | 25 | Thiol-disulfide isomerase | − | Lipoprotein | Periplasm |
| Ng2139 | Gna1946 | PF03180 | 31 | ABC transporter, periplasmic binding protein (metal) | − | Lipoprotein | Periplasm/cell surface |
ND, acceptor sites not yet defined.
Discussion
In this study, we discovered and characterized a general O-linked glycosylation system in bacteria. Additionally, our findings document an instance in which bacterial proteins carrying O-linked glycans are destined to carry out their roles within the cell rather than at the cell surface or beyond. Although we remain ignorant as to what properties might be imparted or modified by glycosylation, these findings imply that Ngo O-linked protein glycosylation acts in both intracellular and extracellular capacities and contributes at a system level to the biology of the organism. It seems unlikely, however, that Ngo will be the only bacterial species with a complex repertoire of O-linked glycoproteins. The closely related species Neisseria meningitidis expresses a very similar pilin glycosylation system, and immunoblotting with the glycan-recognizing antibodies used here reveals analogous patterns of reactive protein forms in some meningococcal backgrounds (B. Børud, unpublished data). In addition, LCRs similar in composition and localization to those seen in Ngo glycoproteins are discernible among orthologous counterparts in some proteobacterial species. It is also perhaps relevant to note that the ASP-rich LCRs defined here are reminiscent of those sites of glycan occupancy documented in M. tuberculosis lipoglycoproteins (12, 13).
Although further studies are required to delineate the specific targeting signals associated with the ASP-rich LCRs, it seems clear that there is no consensus amino acid sequence or obvious sequon. A tetrapeptide serine–alanine–proline–alanine motif is notably overrepresented within both the defined glycopeptides (in 6 of 8 instances and tandemly triplicated in Ng1276; Fig. 5A) and the LCRs of those proteins for which glycopeptides have yet to be identified (in 2 of 4 cases; Table S1). However, it is neither sufficient (as seen in the nonglycosylated Ng1371 mutant expressing a substitution at Ser-337) nor necessary (given that it is absent in PilE and Ng1043). Rather, the LCRs are themselves evocative of the structural features associated with acceptor sites in mucin-type glycoproteins. Commonalities include the biased overrepresentation of alanine, serine, and proline residues, the structural contexts of acceptor sites confined to interdomain regions, the clustering of multiple acceptor sites, and an association with tandem repeat elements (26). Relative surface exposure and accessibility of the acceptor sites are other likely shared determinants of these elements. It is important to note in this context that although PilE (the most abundant glycoprotein) lacks any discernible LCR, its single acceptor site maps on the protein surface as defined by x-ray diffraction (27). Taken together with findings suggesting that PglO exhibits relaxed specificity for lipid-linked oligosaccharide donors (17, 28), the promiscuity of PglO with regard to protein substrates demonstrated here significantly augments the potential of this system for exploitation in novel glycoprotein engineering strategies.
From a larger perspective, our findings reveal remarkable similarities in both the global contexts and structural signals dictating glycan acceptor sites of bacterial and eukaryotic O-linked glycosylation systems. They also extend the ways in which the Ngo O-linked system emulates the C. jejuni N-linked systems to include the targeting of multiple periplasmic substrates. Like in the C. jejuni system, these findings raise the obvious questions as to what biologic significance global protein glycosylation may have and what forces and processes have shaped glycoproteome content.
Materials and Methods
Bacterial Strains, Media, and Culture Conditions.
The bacterial strains used in this study are described in Table S2 and were grown on conventional GC medium as described previously (15). Carboxyterminal 6xHis-tagged alleles as well as missense alleles of Ng1328 and Ng1371 were generated using PCR-based splicing by overlap extension reactions (29). Purified PCR products were used to genetically transform Ngo strains (30), and correct transformants were screened for by PCR using diagnostic primer sets. Protein glycosylation null mutations were introduced into various strain backgrounds using transformation as previously described (15).
Purification of 6xHix-Tagged Proteins.
6xHis-tagged proteins were purified from Ngo using magnetic Ni-NTA beads (Qiagen). Frozen cell pellets were lysed by sonication in buffer containing 100 mM NaH2PO4, 10 mM Tris, 8 M urea, 0.05% Tween, 100 mM NaCl, 15 mM imidazole, 1× complete protease inhibitor mixture, EDTA-free (Roche), 1 mM PMSF, and 1% detergent (Triton X-100 for Ng1371, Ng0372, and Ng1494 and Sarkosyl for Ng1328, Ng1237, Ng1276, Ng2139, Ng1548, Ng1717, Ng1043, Ng0994, and Ng1225), pH 8.0, as described in the manufacturer's instructions. Proteins were eluted directly into SDS–sample loading buffer.
SDS-PAGE/2D Gel Electrophoresis and Immunoblotting.
For SDS-PAGE, whole-cell lysates were prepared from equivalent numbers of cells by heating cell suspensions to 65 °C in SDS–sample loading buffer. Procedures for SDS-PAGE and immunoblotting using alkaline phosphatase–coupled goat antirabbit and goat antimouse antibodies (Tago) have been described previously (31). For 2D gel analyses, cells were harvested from plates incubated overnight, lysed by sonication in 62.5 mM Tris-HCl (pH 6.8) with protease inhibitors (described above), and centrifuged at 200,000 × g for 2 h at 4 °C. The pellet from this step was resuspended in Tris buffer with 1% Triton X-100 and then centrifuged again. The supernatant from the second centrifugation was then fractionated by ammonium sulfate precipitation (a 20%–30% saturation cut) and concentrated using Trichloroacetic acid precipitation and acetone washes. Samples containing 80 μg protein dissolved in isoelectric focusing (IEF) buffer were subjected to IEF in a Protean IEF cell (Bio-Rad) using 11-cm ReadyStrip IPG strips (Bio-Rad) with nonlinear gradients of pH 3–10, as follows: 250 V for 15 min, 8,000 V for 2.5 h, and 8,000 V until 35,000 Vh were reached. After IEF, the strips were subjected to standard disulfide reduction with DTT and cysteine alkylation with iodoacetamide, followed by second-dimension electrophoresis using 12% ReadyGels (Bio-Rad). Gels were treated with MS-compatible silver stain (32) or immunoblotted as detailed above. For glycan detection, polyclonal antiserum no. 904 [ref. 33; termed antiglycan antibodies in this text] raised against purified Type IV pili bearing the O-AcHexDATDH glycan was used at a 1:1,000 dilution. For 6xHis-tagged protein detection, a penta-His–recognizing mouse monoclonal antibody (Qiagen) was used at a 1:1,000 dilution.
Bioinformatic Analysis.
Ngo gene and protein data were obtained through the DBGET web site (http://www.genome.jp/dbget/), an integrated database retrieval system operated by the Kyoto University Bioinformatics Center. Because the Ngo genome data archived there are derived from a strain different from that used here, all relevant gene sequences examined here were confirmed by DNA sequencing. Pfam protein domain designations were derived through the UniProt Knowledgebase (UniProtKB) web site (http://www.uniprot.org/help/uniprotkb). LCRs were identified using the SEG program (16).
MS/MS CID and ETD Analysis.
Each gel spot/slice was destained, and the encompassed proteins were subjected to reduction, alkylation, and digestion with either trypsin, AspN, or proteinase K (Roche/Sigma) at 37 °C overnight. The resulting peptides were extracted with 5% formic acid and acetonitrile, evaporated, and then redissolved in 0.1% formic acid. Samples were subjected immediately to mass spectrometric analyses or frozen at −80 °C. Conditions for these methods have been previously described (14). Reverse-phase (0.75 × 43-mm Zorbax 300SB-C18, 5-μm analytical column; Agilent Technologies) chip-based nanoflow liquid chromatographic MS and MS/MS (CID and ETD mode) analyses (nano-LC-chip MS/MS) of proteolytic peptides were performed using an Agilent 1100 LC/MSD Trap XCT Ultra equipped with a Chip/MS Cube interface. Alternatively, proteolytic peptides were directly applied onto a stainless steel target (Bruker Daltonics) as a mixture of 1:1 (vol/vol) with saturated a-cyano-4-hydroxy-cinnamic acid (Bruker Daltonics) solution (30% acetonitrile/70% 0.1% TFA). Analyses were performed using an Ultraflex TOF/TOF (time of flight) mass spectrometer equipped with a nitrogen laser (337 nm, 5 Hz) in the reflector mode for matrix-assisted laser desorption/ionization (MALDI)-TOF peptide mass fingerprint (PMF) or LIFT mode for MALDI-TOF/TOF analyses. MS/MS or PMF data were searched using the Mascot program (Matrix Sciences). All searches were performed against the Neisseria protein sequence database.
Supplementary Material
Acknowledgments.
We thank Morten Skaugen, University of Life Sciences, Aas, Norway, for access to MALDI-TOF MS equipment. This research was supported in part by Research Council of Norway grants 166931, 152020, and 183613 and funds from the Department of Molecular Biosciences and Center for Molecular Biology and Neurosciences at the University of Oslo.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809504106/DCSupplemental.
References
- 1.Spiro RG. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2003;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 2.Wacker M, et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc Natl Acad Sci USA. 2006;103:7088–7093. doi: 10.1073/pnas.0509207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grogan MJ, Pratt MR, Marcaurelle LA, Bertozzi CR. Homogeneous glycopeptides and glycoproteins for biological investigation. Annu Rev Biochem. 2002;71:593–634. doi: 10.1146/annurev.biochem.71.110601.135334. [DOI] [PubMed] [Google Scholar]
- 4.Hang HC, Bertozzi CR. The chemistry and biology of mucin-type O-linked glycosylation. Bioorg Med Chem. 2005;13:5021–5034. doi: 10.1016/j.bmc.2005.04.085. [DOI] [PubMed] [Google Scholar]
- 5.Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 6.Leavy TM, Bertozzi CR. A high-throughput assay for O-GlcNAc transferase detects primary sequence preferences in peptide substrates. Bioorg Med Chem Lett. 2007;17:3851–3854. doi: 10.1016/j.bmcl.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whelan SA, Hart GW. Identification of O-GlcNAc sites on proteins. Methods Enzymol. 2006;415:113–133. doi: 10.1016/S0076-6879(06)15008-9. [DOI] [PubMed] [Google Scholar]
- 8.Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 9.Igura M, et al. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 2008;27:234–243. doi: 10.1038/sj.emboj.7601940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szymanski CM, Wren BW. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 11.Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci USA. 2007;104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobos KM, Khoo KH, Swiderek KM, Brennan PJ, Belisle JT. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J Bacteriol. 1996;178:2498–2506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VanderVen BC, Harder JD, Crick DC, Belisle JT. Export-mediated assembly of mycobacterial glycoproteins parallels eukaryotic pathways. Science. 2005;309:941–943. doi: 10.1126/science.1114347. [DOI] [PubMed] [Google Scholar]
- 14.Aas FE, et al. Neisseria gonorrhoeae type IV pili undergo multisite, hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J Biol Chem. 2006;281:27712–27723. doi: 10.1074/jbc.M604324200. [DOI] [PubMed] [Google Scholar]
- 15.Aas FE, Vik A, Vedde J, Koomey M, Egge-Jacobsen W. Neisseria gonorrhoeae O-linked pilin glycosylation: Functional analyses define both the biosynthetic pathway and glycan structure. Mol Microbiol. 2007;65:607–624. doi: 10.1111/j.1365-2958.2007.05806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 17.Faridmoayer A, Fentabil MA, Mills DC, Klassen JS, Feldman MF. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol. 2007;189:8088–8098. doi: 10.1128/JB.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulanger MJ, Murphy ME. Crystal structure of the soluble domain of the major anaerobically induced outer membrane protein (AniA) from pathogenic Neisseria: A new class of copper-containing nitrite reductases. J Mol Biol. 2002;315:1111–1127. doi: 10.1006/jmbi.2001.5251. [DOI] [PubMed] [Google Scholar]
- 19.Pitcher RS, Watmough NJ. The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 20.Deeudom M, Koomey M, Moir JW. Roles of c-type cytochromes in respiration in Neisseria meningitidis. Microbiology. 2008;154:2857–2864. doi: 10.1099/mic.0.2008/020339-0. [DOI] [PubMed] [Google Scholar]
- 21.Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 22.Balatri E, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Solution structure of Sco1: A thioredoxin-like protein Involved in cytochrome c oxidase assembly. Structure. 2003;11:1431–1443. doi: 10.1016/j.str.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 23.Seib KL, Jennings MP, McEwan AG. A Sco homologue plays a role in defence against oxidative stress in pathogenic Neisseria. FEBS Lett. 2003;546:411–415. doi: 10.1016/s0014-5793(03)00632-x. [DOI] [PubMed] [Google Scholar]
- 24.Leuzzi R, et al. Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol Microbiol. 2005;58:669–681. doi: 10.1111/j.1365-2958.2005.04859.x. [DOI] [PubMed] [Google Scholar]
- 25.Pizza M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 26.Gerken TA, Owens CL, Pasumarthy M. Determination of the site-specific O-glycosylation pattern of the porcine submaxillary mucin tandem repeat glycopeptide. Model proposed for the polypeptide:galnac transferase peptide binding site. J Biol Chem. 1997;272:9709–9719. doi: 10.1074/jbc.272.15.9709. [DOI] [PubMed] [Google Scholar]
- 27.Parge HE, et al. Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 28.Faridmoayer A, et al. Extreme substrate promiscuity of the Neisseria oligosaccharyltransferase involved in protein O-glycosylation. J Biol Chem. 2008;283:34596–34604. doi: 10.1074/jbc.M807113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Gunn JS, Stein DC. Use of a non-selective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol Gen Genet. 1996;251:509–517. doi: 10.1007/BF02173639. [DOI] [PubMed] [Google Scholar]
- 31.Freitag NE, Seifert HS, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 32.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 33.Winther-Larsen HC, et al. Pseudomonas aeruginosa Type IV pilus expression in Neisseria gonorrhoeae: Effects of pilin subunit composition on function and organelle dynamics. J Bacteriol. 2007;189:6676–6685. doi: 10.1128/JB.00407-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.