Abstract
Recently, meta-analysis has been widely utilized to combine information across comparative clinical studies for evaluating drug efficacy or safety profile. When dealing with rather rare events, a substantial proportion of studies may not have any events of interest. Conventional methods either exclude such studies or add an arbitrary positive value to each cell of the corresponding 2×2 tables in the analysis. In this article, we present a simple, effective procedure to make valid inferences about the parameter of interest with all available data without artificial continuity corrections. We then use the procedure to analyze the data from 48 comparative trials involving rosiglitazone with respect to its possible cardiovascular toxicity.
Keywords: Continuity correction for zero events, Exact inference procedure, Odds ratio, Risk difference
1. INTRODUCTION
Meta-analysis provides a framework for combining information across multiple independent, but “similar” clinical studies to make inferences about a common parameter (Egger and Smith, 1997a; Egger and others, 1997b). Almost all existing methods rely on large-sample approximations to the distributions of the combined point estimators. Such approximations may be inaccurate and lead to invalid conclusions when the individual study sample sizes are small, the number of studies is not large, or the event rates are low (Brown and others, 2001). Moreover, when the events of interest are very rare, the standard procedures either apply continuity corrections to the studies with zero events or simply exclude them from the analysis (Sankey and others, 1996; Bradburn and others, 2007). For example, recently Nissen and Wolski (2007) (N&W) performed a meta-analysis to examine whether rosiglitazone, a drug for treating type 2 diabetes mellitus, significantly increases the risk of myocardial infarction (MI) or cardiovascular disease (CVD)–related death. Of 48 trials satisfied the inclusion criteria for their analysis, 10 studies have no MI events and 25 studies have no CVD-related deaths. N&W simply excluded those studies from their analysis. On the other hand, different continuity corrections may result in different conclusions (Sweeting and others, 2004). An interesting yet challenging methodology question is how to use all available data without assigning an arbitrary number to the empty cells in meta-analysis.
In this article, we present a simple procedure to construct “valid” confidence intervals (CIs) for a common parameter using all available data. The new proposal does not rely on the large-sample approximation or arbitrary continuity corrections to obtain interval estimates. We reanalyze the CVD-related event data of N&W with the proposed method. A numerical study is also conducted to compare the performance of the new proposal to existing procedures.
2. COMBINING INDIVIDUAL CIS FOR A COMMON PARAMETER OF INTEREST
Suppose that we are interested in constructing a 100(1 − α)% 1-sided CI (a,∞) for Δ, a common parameter, based on all data from n independent studies. For a given η, there are n study-specific 1-sided η-level CIs for Δ. Now, for any fixed value of Δ, say 0, we examine whether 0 is the true value of Δ. If yes, then on average, 0 should belong to at least 100η% of the above n intervals. The decision on whether the interval (a,∞) should include 0 can be made easily via a simple hypothesis testing procedure. To this end, let yi = 1, if 0 belongs to the observed η interval from the ith study, and yi = 0, otherwise. Then, we include 0 in (a,∞) if
| (2.1) |
where wi is a study-specific positive weight, c is chosen such that pr{T(η) < c} ≤ α,
| (2.2) |
is the null counterpart of t(η), and {Bi,i = 1,…,n} are n independent Bernoulli random variables with a “success” probability of η. We repeat this process with all other possible values for Δ and obtain the final CI (a,∞). Here, the weight wi may be the sample size for the ith study.
One may further improve the aforementioned interval estimate for Δ by utilizing multiple intervals with various levels of confidence from each study. Specifically, let Jij = (aij,∞) denote the ηj-level 1-sided CI based on the ith study, for j = 1,…,K. Without loss of generality, we assume that 0 < η1 < ⋯ < ηK < 1 and ai1 ≥ ⋯ ≥ aiK. For any given Δ, we would include Δ in the final combined interval (a,∞) if
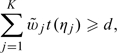 |
(2.3) |
where 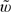 j is a positive weight for the ηj-level intervals and t(ηj) is obtained by replacing yi and η in (2.1) with yij and ηj, respectively. Here, the critical value d is chosen such that
j is a positive weight for the ηj-level intervals and t(ηj) is obtained by replacing yi and η in (2.1) with yij and ηj, respectively. Here, the critical value d is chosen such that
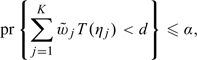 |
(2.4) |
where T(ηi) is obtained by replacing Bi and η in (2.2) by Bij and ηj, respectively, and {(Bi1,…,BiK)′,i = 1,…,n} are n independent random vectors whose components are correlated Bernoulli variables such that Bi1 ≤ Bi2 ≤ ⋯ ≤ BiK and pr(Bij = 1) = ηj. The final interval (a,∞) may be obtained by repeating the process for all possible values of Δ. We justify the validity of the proposed procedure in the Appendix. For the present case, one may let 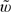 j be {ηj(1 − ηj)} − 1 (Wei and Johnson, 1995; Xu and others, 2003).
j be {ηj(1 − ηj)} − 1 (Wei and Johnson, 1995; Xu and others, 2003).
Similarly, we can obtain combined 100(1 − α)% 1-sided CI, ( − ∞,b). It follows that (a,b) would be a 100(1 − 2α)% 2-sided CI for Δ. A point estimator for Δ may be obtained as 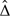 such that
such that 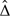 belongs to the intersection of all nonempty 2-sided CIs for Δ.
belongs to the intersection of all nonempty 2-sided CIs for Δ.
3. EXAMPLE AND NUMERICAL STUDY
First, we illustrate the new procedure with the rosiglitazone data set described in Section 1. For convenience, we provide the data from 48 studies in Table I of the supplementary material available at Biostatistics online (http://www.biostatistics.oxfordjournals.org). It is important to note that N&W did not utilize the information from studies which reported zero events of interest. Thus, their analysis only included 38 studies for the MI end point and 23 studies for the CVD-related mortality.
Table 1.
The empirical coverage levels of 95% CIs for the OR obtained based on (i) the MH method with 0.5 imputation (MH-0.5) and (ii) Peto's procedure with event rates ranging from extremely low to moderately low. The results are based on 1000 simulated data sets consisting of 48 2 × 2 tables with the observed sample sizes listed
| OR | (a) pCi∼U(0,0.01%) |
(b) pCi∼U(0,0.1%) |
(c) pCi∼U(0, 10%) |
|||
| MH-0.5 | Peto | MH-0.5 | Peto | MH-0.5 | Peto | |
| 1 | 1.000 | 0.987 | 0.995 | 0.958 | 0.948 | 0.947 |
| 2 | 0.974 | 0.968 | 0.878 | 0.961 | 0.944 | 0.862 |
| 5 | 0.741 | 0.972 | 0.233 | 0.975 | 0.957 | 0.000 |
With respect to MI, N&W obtained a 95% CI of (1.03,1.98) with a p-value of 0.03 for the odds ratio (OR) between rosiglitazone and the control arm (in favor of the control). With respect to mortality, the 95% CI is (0.98,2.74) with a p-value of 0.06, an almost statistically significant result in favor of the control arm. The findings by N&W are summarized in Figure I of the supplementary material available at Biostatistics online.
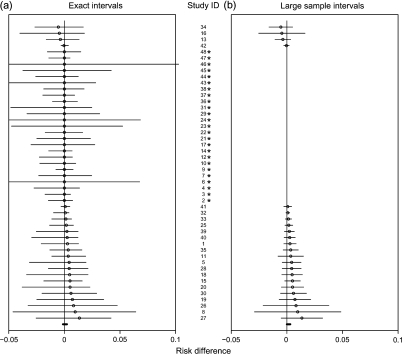
Fig. 1.
The 95% CIs of the RD for CVD death (rosiglitazone minus control) with 48 studies (small circles are the observed RDs).
While it is unclear how to utilize studies with zero events without arbitrary continuity corrections to obtain an overall assessment for OR in meta-analysis, we are able to use all data to make inference about the risk difference (RD), as a measure of the group difference. To this end, we let Δ be the RD (rosiglitazone minus control) in our analysis. We construct 95% CIs for Δ based on the data from 48 studies via the proposed procedure. Here, we let {ηj} be 20 equally spaced levels from 0.1 to 0.95. Moreover, for each study, the ηj-level CI for Δ is obtained via the mid-p-value method (Chan and Zhang, 1999; Hwang and Yang, 2001). The cutoff point d in (2.4) is determined by randomly generating 50 000 independent samples {(Bi1,…,BiK)}. For comparison, we also obtain the corresponding Mantel–Haenszel (MH) weighted CI estimates for RD.
For the mortality end point, we present the forest tree diagram in Figure 1(a). The thick line segment right above the x-axis is the combined 95% CI ( − 0.13,0.23)%. The point estimator of RD is 0.063%, and the p-value for testing RD = 0 is 0.83. On the other hand, in Figure 1(b), the 95% MH final interval for RD is (0.00,0.31)% with a p-value of 0.05 after excluding 25 studies with no events.
For the MI endpoint, the final combined 95% interval is ( − 0.08,0.38)%. The point estimator for RD is 0.183% with a p-value of 0.27. Again, if we exclude 10 studies with no events and use the asymptotic inference as in N&W, the final 95% MH interval is (0.02,0.42)% with a p-value of 0.03 (see Figure II in the supplementary material available at Biostatistics online).
Numerical studies were conducted to examine the performance of the new proposal and the standard large-sample interval estimation procedures. The current literature suggests that the large-sample MH-type combining techniques for the OR or RD are relatively more reliable than other methods (Bradburn and others, 2007). To save space, we only report the results with the MH-type procedures as the main comparators in the study.
The first part of our simulation study was designed to examine the performance of various existing interval estimation procedures for Δ = OR. We mimicked the data structure presented in Table I of the supplementary material available at Biostatistics online and generated 48 studies with the observed sample sizes. Let pci and pri be the incidence rates for the control and rosiglitazone groups in the ith study, respectively. For each given OR, {pci,i = 1,…,48} were generated randomly from a uniform distribution U(0,ξ). Then, we let logit(pri) = log(OR) + logit(pci) to generate data for the treated group. In Table 1, we present the empirical coverage levels of 95% CIs obtained based on (i) the MH method with 0.5 imputation (MH-0.5) for studies with zero event in one arm and (ii) the Peto's method utilized in N&W. All results are based on 1000 replications. It is clear that either method may not be valid when the true OR is away from one. Through our simulation studies, we also find that the existing exact method for the OR conditional on all marginal totals of 2×2 tables works well.
In the second part of our simulation, we use the RD as the contrast measure Δ. We generate pci from the uniform distribution and pri = RD + pci. The results are reported in Table 2. When the underlying event rates for both groups are low, the MH method without imputation has serious coverage problems. For this case, both our exact and MH-0.5 methods are conservative. However, the average interval lengths from exact method are shorter than that of the MH-0.5 method. On the other hand, when the underlying event rates are not very low, all procedure have proper coverage levels and comparable interval lengths.
Table 2.
The empirical coverage levels and average lengths of 95% CIs for RD obtained based on (i) the MH-0 method, (ii) the MH-0.5 method, and (iii) the new exact method for cases with extremely to moderately low event rates. The results are based on 1000 simulated data sets consisting of 48 2 × 2 tables with the observed sample sizes listed in Table 1
| Empirical coverage level (average length × 1000) | |||||||
| (a) pc ∼ U(0, 0.01%) |
(b) pc ∼ U(0, 10%) |
||||||
| RD | MH-0 | MH-0.5 | Exact | RD | MH-0 | MH-0.5 | Exact |
| 0 | 0.761 (0.29) | 1.000 (2.10) | 1.000 (1.45) | 0. | 0.959 (10.3) | 0.959 (10.3) | 0.995 (12.3) |
| 0.01% | 0.896 (0.44) | 1.000 (2.13) | 1.000 (1.51) | 5% | 0.947 (12.2) | 0.947 (12.2) | 0.959 (14.8) |
| 0.05% | 0.930 (0.78) | 0.999 (2.22) | 0.999 (1.91) | 10% | 0.957 (13.7) | 0.957 (13.7) | 0.958 (16.8) |
| 0.50% | 0.955 (2.28) | 0.998 (3.69) | 0.987 (3.67) | ||||
4. REMARKS
Unlike the standard large-sample meta-analysis procedures, the proposed simple method provides valid exact inferences about the parameter of interest under any fixed-effects framework. It effectively utilizes all data without artificial imputation. Our method may be regarded as a generalization of the combination method of confidence distributions studied by Singh and others (2005). As other exact procedures, the proposed method can be over conservative in some cases. It would be interesting to examine if one can improve the efficiency of the proposed exact method by considering a priori (or random-effect) distribution for the nuisance parameter, such as the control success rate in the 2 × 2 tables, as suggested by the associate editor. The computer code for implementing the procedure is available at “http://biosun1.harvard.edu/display=’block'>∼tcai/MetaCode.r”.
FUNDING
National Institutes of Health (U54LM008748; R01AI052817; R01HL89778).
Supplementary Material
Acknowledgments
Conflict of Interest: None declared.
Appendix
A.1. Justification of validity for the final combined interval
Assume that the n studies in our meta-analysis are realizations of a random sample from a population whose distribution is generated by a random quantity Π. For example, for the 2 × 2 tables, Π consists of Δ0, the underlying event rate for the control arm, and possibly the sample sizes for 2 arms of the study. Let πi be the realization of Π for the ith study, i = 1,2,…,n. Note that one may further assume that the number n of studies is a random component N of Π. Given πi, the data Xi were generated for i = 1,…,n. The 1-sided CI Jij = (aij,∞) for Δ0 satisfies the condition
| (A.1) |
where the probability is generated by Xi. Now, for any given Δ, we test the null hypothesis that Δ = Δ0. Let Yij = 1, if Δ∈Jij; Yij = 0, otherwise. Consider the test statistic 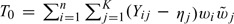 . We then derive the critical value d in (2.3) and (2.4) from the null counterpart of T0,
. We then derive the critical value d in (2.3) and (2.4) from the null counterpart of T0,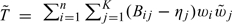 . Note that if Δ = Δ0, (A.1) implies that
. Note that if Δ = Δ0, (A.1) implies that
| (A.2) |
Under this condition, we will show that
| (A.3) |
Equation (A.3) directly implies that conditional on any set of realizations {π1,…,πn}, pr(T0 < d|Δ = Δ0,π1,…,πn) ≤ α, where the probability is generated by {Xi,i = 1,…,n}. It follows that pr(T0 < d|Δ = Δ0) ≤ α, where the probability is generated under the random pairs {(Xi,Πi),i = 1,…,n}. Here, Xi is a random quantity associated with Πi and {Πi} is a random sample from the population Π.
To show (A.3), first let 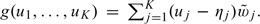 Since for each individual study, the K CIs are nested, g(yi1,…,yiK) or g(Bi1,…,BiK) can only assume K + 1 possible distinct values:
Since for each individual study, the K CIs are nested, g(yi1,…,yiK) or g(Bi1,…,BiK) can only assume K + 1 possible distinct values: 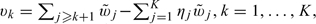 where y is the observed value of Y and v0 > v1⋯ > vK. Furthermore, g(yi1,…,yiK) = vs if and only if yij = I(j > s). Similarly, g(Bi1,…,BiK) = vs if and only if Bij = I(j > s). It follows that for v∈(vs,vs − 1],
where y is the observed value of Y and v0 > v1⋯ > vK. Furthermore, g(yi1,…,yiK) = vs if and only if yij = I(j > s). Similarly, g(Bi1,…,BiK) = vs if and only if Bij = I(j > s). It follows that for v∈(vs,vs − 1],
Under (A.2), pr(Yis = 1) ≥ ηs = pr(Bis = 1) = pr{g(Bi1,…,BiK) ≥ v}. Consequently, pr{g(Yi1,…,YiK) ≥ v} ≥ pr{g(Bi1,…,BiK) ≥ v} and (A.3) follows.
References
- Bradburn MJ, Deeks JJ, Berlin J, Localio AR. Much ado about nothing: a comparison of the performance of meta-analysis methods with rare events. Statistics in Medicine. 2007;26:53–77. doi: 10.1002/sim.2528. [DOI] [PubMed] [Google Scholar]
- Brown L, Cai T, Dasgupta A. Interval estimation for a binomial proportion. Statistical Science. 2001;16:101–103. [Google Scholar]
- Chan I, Zhang Z. Test-based exact confidence interval for the difference of two binomial proportions. Biometrics. 1999;55:1202–1209. doi: 10.1111/j.0006-341x.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- Egger M, Smith GD. Meta-analysis: potentials and promise. BMJ. 1997a;315:1371–1374. doi: 10.1136/bmj.315.7119.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Smith GD, Phillips AN. Meta-analysis: principles and procedures. BMJ. 1997b;315:1533–1537. doi: 10.1136/bmj.315.7121.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Yang M. An optimality theory for mid p-values in 2×2 contingency tables. Statistical Sinica. 2001;11:807–826. [Google Scholar]
- Nissen SE, Wolski K. Effect of rosiglitazone on the risk of myocardial infarction and death from cardiovascular causes. The New England Journal of Medicine. 2007;357:1–100. doi: 10.1056/NEJMoa072761. [DOI] [PubMed] [Google Scholar]
- Sankey S, Weissfeld L, Fine M, Kapoor W. An assessment of the use of the continuity correction for sparse data in meta analysis. Communications in Statistics: Simulation and Computation. 1996;25:1031–1056. [Google Scholar]
- Singh K, Xie M, Strawderman W. Combining information from independent sources through confidence distributions. The Annals of Statistics. 2005;33:159–183. [Google Scholar]
- Sweeting MJ, Sutton AJ, Lambert PC. What to add to nothing? Use and avoidance of continuity corrections in meta-analysis of sparse data. Statistics in Medicine. 2004;23:1351–1375. doi: 10.1002/sim.1761. [DOI] [PubMed] [Google Scholar]
- Wei LJ, Johnson W. Combining dependent tests with incomplete repeated measurements. Biometrika. 1995;72:359–364. [Google Scholar]
- Xu X, Tian L, Wei LJ. Combining dependent tests for linkage or association across multiple phenotypic traits. Biostatistics. 2003;4:223–229. doi: 10.1093/biostatistics/4.2.223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.