Abstract
Rationale: Polymorphisms in the gene for transforming growth factor-β1 (TGFB1) have been associated with asthma, but not with airway responsiveness or disease exacerbations in subjects with asthma.
Objectives: To test for association between single nucleotide polymorphisms (SNPs) in TGFB1 and markers of asthma severity in childhood.
Methods: We tested for the association between nine SNPs in TGFB1 and indicators of asthma severity (lung function, airway responsiveness, and disease exacerbations) in two cohorts: 416 Costa Rican parent-child trios and 465 families of non-Hispanic white children in the Childhood Asthma Management Program (CAMP). We also tested for the interaction between these polymorphisms and exposure to dust mite allergen on asthma severity.
Measurements and Main Results: The A allele of promoter SNP rs2241712 was associated with increased airway responsiveness in Costa Rica (P = 0.0006) and CAMP (P = 0.005), and the C allele of an SNP in the promoter region (rs1800469) was associated with increased airway responsiveness in both cohorts (P ≤ 0.01). Dust mite exposure modified the effect of the C allele of exonic SNP rs1800471 on airway responsiveness (P = 0.03 for interactions in both cohorts). The T allele of a coding SNP (rs1982073) was associated with a reduced risk of asthma exacerbations in Costa Rica (P = 0.009) and CAMP (P = 0.005). Dust mite exposure also significantly modified the effect of the A allele of the promoter SNP rs2241712 on asthma exacerbations in both cohorts.
Conclusions: SNPs in TGFB1 are associated with airway responsiveness and disease exacerbations in children with asthma. Moreover, dust mite exposure may modify the effect of TGFB1 SNPs on airway responsiveness and asthma exacerbations.
Keywords: airway responsiveness, asthma, dust mite allergen, single nucleotide polymorphisms, transforming growth factor-β1
AT A GLANCE COMMENTARY
Scientific Knowledge on the Subject
DNA sequence variants in the gene for transforming growth factor-β1 (TGFB1) have been associated with airway inflammation and asthma.
What This Study Adds to the Field
Genetic polymorphisms in TGFB1 are associated with airway responsiveness and disease exacerbations in children with asthma. Dust mite exposure may modify the effect of TGF-β1 single nucleotide polymorphisms on airway responsiveness and disease exacerbations in children with asthma.
Asthma exacerbations are among the leading causes of morbidity in children and have resulted in increased healthcare expenditures in the pediatric population over the last decade (1). Children who experience asthma exacerbations are at high risk of recurrence (2–4), which may be partly explained by poorly characterized interactions between genetic variants and environmental exposures such as dust mite allergen (5).
Transforming growth factor-β1 (TGF-β1) is a multifunctional cytokine that plays a critical role in cell growth and differentiation, immune modulation, airway development, and inflammation. TGF-β1 is ubiquitously expressed in the lung and is involved in both normal cellular processes and numerous disease states. Several lines of evidence suggest that TGF-β1 contributes to the development of asthma, airway responsiveness, and airway remodeling (6).
Animal models have demonstrated the importance of TGF-β1 in allergen-induced T helper type 2 (Th2)-mediated airway inflammation (7), but the precise role of TGF-β1 in asthma has been difficult to define. In murine models, overexpression of the gene for TGF-β1 (TGFB1) leads to increased airway inflammation and subepithelial fibrosis (8, 9). Alternatively, recent data suggest that TGF-β1 antagonism results in a marked increase in airway responsiveness in mice with chronic airway inflammation induced by exposure to dust mite allergen, suggesting a potential role for gene-by-environment interactions (10). In humans, levels of TGF-β1 are higher in the bronchoalveolar lavage (BAL) fluid of subjects with asthma than in that of control subjects (11). Moreover, TGFB1 expression is increased in the asthmatic airway upon repeated exposure to low-dose allergen, suggesting that TGFB1 is involved in the late phase of airway inflammation (7). Given these observations, we hypothesized that variants in TGFB1 would influence disease severity in children with asthma. In addition, we hypothesized that significant interactions between these variants and dust mite allergen would affect asthma severity.
TGFB1 is located on chromosome 19q13.1 and harbors over 160 single nucleotide polymorphisms (SNPs). Previous twin studies have demonstrated a significant genetic contribution to circulating TGF-β1 levels, with heritability estimates ranging from 0.39 to 0.66 (12). In a variety of experimental approaches, polymorphisms in the promoter and exons of TGFB1 (Table 1) have been shown to influence interindividual differences in circulating TGF-β1 levels (12–19).
TABLE 1.
PREVIOUS FUNCTIONAL STUDIES OF POLYMORPHISMS IN TGFB1
| SNP | Alternative SNP Identification | Location | Allele | Function | Assay Performed | Study (Reference No.) |
|---|---|---|---|---|---|---|
| rs1800469 | −509C-T | Promoter | T | Increased serum level | Correlation of genotype with serum protein levels | Grainger, 1999 (12) |
| rs1800469 | −509C-T | Promoter | T | Increased transcription | EMSA, reporter construct and transient transfection in A549 human lung carcinoma cells and/or bronchial epithelial cells | Silverman, 2004 (13) |
| rs1800469 | −509C-T | Promoter | C | Increases serum concentration and hepatocyte transcription | Correlation of genotype with serum protein levels and reporter-gene constructs | Wang, 2008 (14) |
| rs1982073 | +869T-C | Exon | T | Increased serum concentration | Correlation of genotype with serum protein levels | Wang, 2008 (14) |
| rs1982073 | +869T-C | Exon | C | Increased secretion in HeLa cells | Cell culture and transfection of HeLa cells | Dunning, 2003 (15) |
| rs1982073 | +869T-C | Exon | T | Increased serum level | Correlation of genotype with serum protein levels | Hinke, 2001 (16) |
| rs1982073 | +869T-C | Exon | No correlation with level | Correlation of genotype with serum protein levels | Li, 2008 (17) | |
| rs1982973 | +869T-C | Exon | C | Increased serum level | Correlation of genotype with serum protein levels | Yokota, 2000 (18) |
| rs1800471 | +915G-C | Exon | G | Increased expression in lymphocytes | Cell culture and ELISA: correlation of genotype with levels in stimulated leukocytes | Awad, 1998 (19) |
To date, association studies of TGFB1 and disease severity in obstructive airways diseases (and their intermediate phenotypes) have yielded conflicting findings, which may be partially explained by differences in study design, sample ascertainment, environmental exposures, and sample size (leading to differences in power) (13, 20–22). To our knowledge, there has been no association study of TGFB1 and indicators of disease severity in children with asthma. We demonstrate that SNPs in TGFB1 (including putative functional variants) are associated with markers of disease severity (airway responsiveness and disease exacerbations) in two family-based studies of children with asthma. In addition, we show significant modification of the effect of SNPs in TGFB1 on asthma severity by indoor exposure to dust mite allergen. Some of these results have been previously reported in the form of an abstract (23).
METHODS
For Methods details, see the online supplement.
Study Population
As part of the Genetic Epidemiology of Asthma in Costa Rica cohort, 426 parent-child trios were recruited between February of 2001 and March of 2005. Details on subject recruitment and study protocols have been published elsewhere (24). In brief, children 6 to 14 years of age were included if they had asthma (a physician's diagnosis of asthma and two or more respiratory symptoms or asthma attacks in the previous year) and a high probability of having six or more great-grandparents born in the Central Valley of Costa Rica. All children completed a protocol including a questionnaire, pulmonary function testing, and methacholine challenge testing. Blood samples were obtained from each child and his/her parents for DNA extraction.
Replication Population
A total of 483 white (non-Hispanic) families participated in the Genetics Ancillary Study of the Childhood Asthma Management Program (CAMP). CAMP was a multicenter, randomized, double-blind, placebo-controlled trial established to investigate the long-term effects of inhaled corticosteroids and inhaled nedocromil (a nonsteroidal anti-inflammatory medication). Of 1,041 children randomized in the clinical trial, 968 children (and 1,518 of their parents) contributed DNA samples as part of the genetic ancillary study of CAMP. DNA was sufficient for all members of the 483 nuclear families of self-reported non-Hispanic white ancestry studied previously (25).
Children enrolled in the CAMP study had mild to moderate persistent asthma based on the demonstration of increased airway responsiveness (a PC20 [provocative concentration causing a 20% fall in FEV1] ≤12.5 mg/ml of methacholine) and at least two of the following: asthma symptoms at least twice weekly, use of inhaled bronchodilator at least twice weekly, or use of daily asthma medication for at least 6 months in the year preceding screening (26). Participating children were followed for a mean of 4.3 years. A complete description of the trial design, methodology, and primary outcomes analyses of the CAMP study have been previously published (27).
For the Genetics of Asthma in Costa Rica Study, approval was obtained from the Institutional Review Boards of the Hospital Nacional de Niños (San José, Costa Rica) and the Brigham and Women's Hospital (Boston, MA). Approval was obtained from the review boards of each CAMP institution before participant enrollment in the study. Informed consent was obtained from the parents of participating children, and assent was obtained from each child before enrollment.
Genotyping
Nine SNPs, including three variants for which there are functional data (Table 1), were genotyped in Costa Rican parent-child trios and in families of white children in CAMP. SNPs were selected from the HapMap (http://www.hapmap.org) and dbSNP (http://www.ncbi.nlm.nih.gov/SNP) databases to tag all common SNPs in TGFB1 and to include known functional variants. The nine SNPs genotyped in TGFB1 capture 80% or greater HapMap SNPs with minor allele frequency of 10% or greater in TGFB1 and its 10-kb flanks in European American trios at an r2 ≥ 0.97. SNP genotyping was performed using the Illumina Golden Gate Genotyping Assay (Illumina Inc., San Diego, CA). For purposes of quality control, duplicate genotyping was performed on approximately 5% of the samples. The pedigree data were assessed for evidence of parent-offspring genotype incompatibility using PedCheck (28). Hardy-Weinberg Equilibrium was tested in parents at each locus using an exact method (29). Genotype data quality was assessed by completion rates, discordance in duplicate genotyping, and evidence of Mendelian inconsistencies.
Association Analysis
Family-based tests of association were conducted using the Pedigree-Based Association Test (PBAT) program (version 5.3) implemented using GoldenHelix software (http://www.goldenhelix.com). We tested for the association between SNPs in TGFB1 and markers of asthma severity including lung function, airway responsiveness, and disease-related exacerbations. Given the results of previous studies, our analysis assumed a dominant genetic model (13). The analyses of lung function phenotypes (post-bronchodilator FEV1 and FEV1/FVC) and airway responsiveness were adjusted for age, sex, and height. Data on exacerbations for asthma in the previous year were collected from each participant in Costa Rica and CAMP at study entry and at randomization, respectively. The analyses of asthma exacerbations were adjusted for age, sex, and use of anti-inflammatory medications (inhaled steroids in CAMP and inhaled steroids or leukotriene inhibitors in Costa Rica). Standard phenotypic residuals were used as the offset choice for the quantitative trait analysis to maximize power. To account for study-wide multiple comparisons (testing nine markers and four nonindependent phenotypes), results were considered significant only when identical associations (i.e., same allele, same phenotype, same direction of genetic effect given the same genetic model) were observed in both populations with a P value < 0.05 and when the Bonferroni-adjusted Fisher's combined P value was ≤ 0.001 (0.05/36, to account for testing nine SNPs and four phenotypes).
Because of the availability of longitudinal data in CAMP, we used Poisson regression models to test for an association between variants in TGFB1 and the number of asthma-related exacerbations (including emergency department visits and hospitalizations) during the 4-year follow-up period. This analysis was adjusted for age, sex, and use of inhaled steroids.
Given the previous experimental evidence of modification of the effect of TGF-β1 on allergic airway inflammation and airway responsiveness by dust mite allergen (10), we tested for gene-by-environment interactions using the family-based association test of interaction (FBAT-I) implemented in PBAT (30). These analyses were adjusted for the same covariates used in the single-SNP association analyses. On the basis of previous findings and consistent with our previous work (31) exposure to dust mite allergen (Der p 1 level) was dichotomized at 10 μg/g or greater.
RESULTS
The baseline characteristics of the index children in Costa Rica and CAMP are presented in Table 2. Of the 426 Costa Rican parent-child trios, 10 were excluded from this analysis because of Mendelian inconsistencies (n = 9) or inadequate genotypic data (n = 1), leaving 416 trios. Of the 483 white families participating in CAMP, 18 were removed from this analysis because of Mendelian inconsistencies (n = 13) or inadequate genotypic data (n = 5), leaving 465 families (and 498 children) for replication studies. Genotypic quality was high for both populations, with an average completion rate of 97% and no noted discrepancies between the initial genotyping and the 5% of samples that underwent repeat genotyping. Parental genotypes in both populations were in Hardy-Weinberg equilibrium (P value adjusted for number of tests > 0.05) at all loci.
TABLE 2.
BASELINE PHENOTYPIC CHARACTERISTICS OF INDEX CHILDREN IN CAMP AND COSTA RICA
| Variable | Costa Rica (n = 416) | CAMP (n = 498) |
|---|---|---|
| Age, y | 8.84 (7.79–10.48)* | 8.76 (7.08–10.56) |
| Female sex | 158 (38) | 189 (38) |
| Baseline post-bronchodilator FEV1, L | 1.74 (1.49–2.03) | 1.77 (1.49–2.11) |
| Baseline post-bronchodilator, FEV1/FVC | 86.07 (81.94–89.95) | 86.0 (82.0–90.0) |
| Airway responsiveness† | 1.2 (0.7–1.4) | 1.1 (0.5–2.8) |
| Disease exacerbation, ≥1‡ | 21 (5) | 352 (33) |
| High dust mite allergen exposure, (≥10 μg/g) | 203 (49) | 45 (9) |
Parenthetical values are median range (interquartile range) or count (percentage) reported.
Airway responsiveness: PD20 (μmol of methacholine) used in the Costa Rican study. PC20 (mg/ml of methacholine) used in the CAMP study.
In Costa Rica, at least one hospitalization for asthma in the previous year. In CAMP, at least one emergency department visit or hospitalization for asthma in the year preceding randomization.
Despite differences in geographic origin and sample ascertainment, the baseline characteristics of the CAMP and Costa Rican probands were very similar. Consistent with the known sex distribution of asthma in childhood, there was a predominance of boys in both cohorts. Although baseline lung function and airway responsiveness were similar in the two populations, there were more asthma-related hospitalizations in the year preceding study enrollment in CAMP than in the Costa Rican cohort.
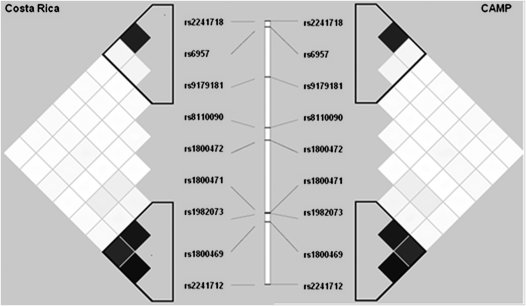
Figure 1 presents the patterns of linkage disequilibrium (LD) and haplotype block structure of TGFB1 in both cohorts. Regional LD surrounding TGFB1 was similar in both cohorts, with the nine genotyped SNP forming two haplotype blocks. However, there were differences in block boundaries and haplotype frequencies, which may be partially due to differences in minor allele frequencies s between the two populations. A high degree of LD between putative functional variants was noted in both populations, but the degree of LD between SNP rs1982073 and rs1800469 was higher in Costa Ricans (r2 = 0.87) than in CAMP participants (r2 = 0.71).
Figure 1.
Linkage disequilibrium structure based on R2 in the Costa Rica (left) and CAMP (right) participants. The nine single nucleotide polymorphisms genotyped are represented from 3′ (top) to 5′ (bottom).
The results of the family-based association analysis of SNPs in TGFB1 and airway responsiveness are shown in Table 3. The CC and CT genotypes of the promoter SNP rs1800469 and the AA and AG genotypes of the promoter SNP rs2241712 were associated with increased airway responsiveness in both populations (Fisher's combined P value ≤ 0.001 for each of the two SNPs). These associations remained significant after correction for multiple comparisons. Although alleles associated with airway responsiveness had relatively high frequencies, index children in both cohorts were ascertained on the basis of asthma, and thus these results may not be generalizable to nonasthmatic populations.
TABLE 3.
FAMILY-BASED ANALYSIS OF ASSOCIATION BETWEEN VARIANTS IN TGFB1 AND AIRWAY RESPONSIVENESS AND ASTHMA EXACERBATIONS IN CAMP AND COSTA RICA*
| Costa-Rica
|
CAMP
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenotype | dbSNP | Allele | Allele Frequency | Informative Families, n | Z Score | P Value | Allele | Allele Frequency | Informative Families, n | Z Score | P Value | Fisher's Combined P Value† |
| Airway responsiveness | rs1800469 | C | 0.57 | 138 | −2.87 | 0.004 | C | 0.70 | 110 | −2.24 | 0.01 | 0.0004 |
| rs2241712 | A | 0.56 | 128 | −3.43 | 0.0006 | A | 0.70 | 108 | −2.58 | 0.005 | 0.00004 | |
| Asthma hospitalizations‡ | rs1982073 | T | 0.55 | 165 | −2.58 | 0.009 | T | 0.59 | 219 | −2.41 | 0.005 | 0.0005 |
All analyses were conducted under the assumption of a dominant genetic model.
Combined P value ≤ 0.001, which is significant after correction for multiple comparisons.
In Costa Rica, at least one hospitalization for asthma in the previous year. In CAMP, at least one hospitalization or emergency department visit for asthma in the year preceding randomization.
The results of the analysis of association between polymorphisms in TGFB1 and asthma exacerbations in the previous year are shown in Table 3. In this analysis, the TT and CT genotypes of exonic SNP rs1982073 were significantly associated with a reduced risk of asthma hospitalizations in Costa Rica and CAMP (Fisher's combined P value = 0.0005).
The results of the association analysis of variants in TGFB1 and lung function phenotypes are shown in Table E1 (see online supplement). In this analysis, the TT and CT genotypes of SNP rs6957 (located in the 3′genomic region of the gene) were associated with higher post-bronchodilator FEV1 and FEV1/FVC in Costa Ricans (P = 0.01 and P = 0.03, respectively). However, these associations were not replicated in CAMP.
The family-based association analysis of haplotypes in TGFB1 and airway responsiveness and asthma exacerbations yielded nominally significant results in Costa Rica and in CAMP (see Table E2). However, there were differences in haplotypic frequencies, and haplotypic association findings were not consistently replicated across cohorts.
Because longitudinal data on the number of asthma-related hospitalizations and emergency department visits were available for the 4 year follow-up period of the original CAMP trial, we tested for association between TGFB1 variants and frequency of asthma exacerbations. Consistent with the results of our association analysis at randomization (baseline), white children in CAMP who had the TT or CT genotype for exonic SNP rs1982073 were 0.70 (95% CI, 0.60–0.92) times less likely to have an asthma-related emergency department visit or hospitalization during the 4-year trial than those with the CC genotype.
Exposure to high levels of dust mite allergen (≥10 μg/g) was more common in Costa Rican children than in children in CAMP (Table 2). The results of the family-based analysis of interactions between SNPs in TGFB1 and exposure to high levels of dust mite allergen on indicators of asthma severity are shown in Table 4. We found that dust mite allergen exposure significantly modified the effect of the C allele of exonic SNP rs1800471 on airway responsiveness (P for interaction = 0.03 in both cohorts). Furthermore, dust mite allergen exposure significantly modified the effect of the A allele of the promoter SNP rs2241712 on asthma exacerbations in both Costa Rica (P for interaction = 0.008) and CAMP (P for interaction = 0.03). An analysis stratified by dust mite allergen exposure confirmed SNP-specific affects on increased airway responsiveness and increased asthma exacerbations among children exposed to higher levels of dust-mite allergen (Table 4). Although SNP rs1800469 was also found to have a significant interaction with dust mite allergen on asthma exacerbations in Costa Rica, this was not replicated in CAMP.
TABLE 4.
FAMILY-BASED ANALYSIS OF POTENTIAL MODIFICATION OF THE EFFECT OF TGFB1 POLYMORPHISMS ON ASTHMA-RELATED OUTCOMES BY DUST MITE ALLERGEN EXPOSURE*
| Costa Rica Stratified Analysis
|
CAMP Stratified Analysis
|
||||||
|---|---|---|---|---|---|---|---|
| Phenotype | SNP (Allele) | Unexposed (51%) | Exposed (49%) | Costa Rica Interaction P Value | Unexposed (91%) | Exposed (9%) | CAMP Interaction P Value |
| Airway responsiveness | rs1800471 (C) | NS† | +0.02‡ | 0.03 | NS | +0.03 | 0.03 |
| Asthma exacerbations | rs2241712 (A) | NS | +0.001 | 0.008 | NS | +0.03 | 0.03 |
Definition of abbreviations: NS = nonsignificant; SNP = single nucleotide polymorphism.
Der p 1 levels in house dust (dust mite allergen exposure) were dichotomized at ≥10 μg/g. The proportion of unexposed/exposed children was similar for each single nucleotide polymorphism in a given cohort.
NS = P > 0.05.
Plus sign represents increased airways responsiveness or increased exacerbations after high exposure to dust mite.
DISCUSSION
To our knowledge, this is the first report of an association between variants in TGFB1 and airway responsiveness or disease exacerbations in subjects with asthma. In addition, this is the first study to show modification of the effect of TGFB1 polymorphisms on asthma-related outcomes by dust mite allergen exposure.
Previous studies of SNP rs1800469 (a putative functional variant in the promoter of TGFB1) and asthma per se have yielded conflicting results. For example, the TT genotype has been associated with an increased risk of asthma in some studies (13, 17, 20) but not in others (21, 32). We found that the CC and CT genotypes of SNP rs1800469 (which were correlated with relatively reduced circulating levels of TGF-β1 in two of three previous studies [Table 1]) were associated with increased airway responsiveness. Consistent with our findings in children with asthma, Ogawa and colleagues reported that the CC genotype of SNP rs1800469 was associated with increased airway responsiveness in 590 adults with COPD (33). If allele C of SNP rs1800469 is in fact correlated with reduced levels of TGF-β1 (see below), our findings and those of Ogawa and colleagues are consistent with those of murine models of allergic airway inflammation that suggest that antagonism of TGF-β1 results in increased airway responsiveness (34), and that intratracheal administration of TGF-β1 inhibits antigen-induced airway responsiveness (35).
The observed association between the CC and CT genotypes of SNP rs1800471 (which were correlated with decreased circulating TGF-β1 levels in a previous study [Table 1]) and increased airway responsiveness in subjects exposed to high levels of dust mite allergen is consistent with recent experimental findings (36) and suggests that dust mite allergen exposure modifies the effect of TGFB1 SNPs on airway responsiveness in children with asthma. We also demonstrate that dust mite allergen exposure modifies the effect of the promoter SNP rs2241712 on disease exacerbations in children with asthma. Other investigators have shown a significant interaction between passive exposure to smoking and the CC genotype of SNP rs1800471 in individuals with cystic fibrosis, in whom this interaction results in reduced lung function (37). Previous studies have also shown modification of the effect of variants in TGFB1 on asthma by environmental exposures other than dust mite allergen (intrauterine smoke exposure and oxidant stress by traffic-related emissions) (38).
In children, disease exacerbations may be better indicators of asthma severity than spirometric measures of lung function. In fact, previous disease exacerbations may be the single strongest predictor of future exacerbations in children with asthma (39). We found that the TT and CT genotypes of a coding polymorphism in TGFB1 (rs1982073) were inversely associated with asthma exacerbations.
A number of studies have attempted to elucidate the functional significance of genetic variants in TGFB1 in humans (summarized in Table 1). Whereas results of two studies (including one that assessed bronchial epithelial cells and human lung carcinoma cells) suggest that the T allele of SNP rs1800469 is correlated with enhanced transcription and increased serum levels of TGFB1, opposite results were recently demonstrated in experimental studies in hepatocytes. Conflicting results have also been shown for studies of the association between alleles in exonic SNP rs1982073 and serum levels of TGF-β1 (Table 1). Whereas the discrepant results of functional studies for SNP rs1800469 may be related to tissue-specific differences in gene expression, these findings (and those for SNP rs1982073) may be due to LD with other variants within or near TGFB1 and/or untested gene-by-gene or gene-by-environment interactions. Further work on these polymorphisms is required to fully understand their functional significance.
Elucidating the effects of TGF-β1 on asthma phenotypes is further complicated by the fact that it is a pleiotropic cytokine. For example, increased TGF-β1 levels are associated with increased airway remodeling but decreased airways responsiveness in transgenic mice over-expressing TGFB1 in the lungs (42). In humans, TGF-β1 has been shown to stimulate increased synthesis of fibronectin/collagen (26, 27), stimulate smooth muscle proliferation in the airways of people with asthma (by increasing both smooth muscle cell size and number) (43), and enhance immune responses against viral illnesses (44). Thus, genetic variants that lead to reduced levels of TGF-β1 could theoretically have paradoxical effects on airway remodeling and airway responsiveness in subjects with asthma. Consistent with a complex relationship between TGF-β1 and respiratory diseases, genetic variants that may result in increased levels of TGF-β1 have been associated with reduced risk of COPD (41, 45) but increased severity of lung disease in cystic fibrosis (40).
As previously noted, genetic association studies of TGFB1 and asthma and its intermediate phenotypes have yielded inconsistent findings. In addition to differences in LD patterns, gene-by-gene interactions, and/or gene-by-environment interactions among study populations, potential explanations for these discrepant results include differences in statistical power (most important), lack of control for multiple testing, and (for case-control studies) population stratification. Our study had adequate statistical power to detect associations of the magnitude previously reported, and we have reduced the potential for false positive results by replication of our findings in two family-based studies, which are not susceptible to population stratification. We recognize, however, that some of our nonreplicated results at the SNP level (e.g., for measures of lung function) may be due to underlying differences between our study populations (e.g., ancestral history, LD, and environmental exposures).
In summary, genetic variation in TGFB1 is associated with indicators of asthma severity (airway responsiveness and disease exacerbations) in two cohorts of children with asthma. Furthermore, in concordance with previous experimental data (36), we have demonstrated significant interactions between polymorphisms in TGFB1 and dust mite allergen exposure on airway responsiveness in children with asthma. Our findings provide further evidence that TGFB1 plays an important role in determining disease severity in children with asthma.
Supplementary Material
Acknowledgments
The authors thank all subjects for their ongoing participation in this study. The Genetics of Asthma in Costa Rica Study was supported by grants HL04370 and HL66289 from the U.S. National Institutes of Health (NIH). We acknowledge the CAMP investigators and research team, supported by NHLBI, for collection of CAMP Genetic Ancillary Study data. All work on data collected from the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of the Brigham and Women's Hospital under appropriate CAMP policies and human subject protections. The CAMP Genetics Ancillary Study is supported by U01 HL075419, U01 HL65899, P01 HL083069, R01 HL086601, and T32 HL07427 from the National Heart, Lung, and Blood Institute, National Institutes of Health. Additional support for this research came from grants P50 HL67664 from the National Institutes of Health and the National Heart, Lung, and Blood Institute.
The Genetics of Asthma in Costa Rica Study was supported by NIH grants HL04370 and HL66289. The CAMP Genetics Ancillary Study is supported by grants U01 HL075419, U01 HL65899, P01 HL083069, R01 HL086601, N01-HR16049, and T32 HL07427 from the National Institutes of Health.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.200808-1268OC on December 18, 2008
Conflict of Interest Statement: S.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.A.R. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. G.M.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. M.S.-Q. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.A. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.J.M. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.L.-S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.J.K. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.S.S. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.T.W. received a grant from AstraZeneca for the Asthma Policy Modeling Study in the amount of $900,065, from 1997 to 2003. S.T.W. has been a co-investigator on a grant from Boehringer Ingelheim to investigate COPD natural history which began in 2003. S.T.W. has been an advisor and chair of the advisory board to the TENOR Study for Genentech and has received $10,000 for 2005 to 2006. S.T.W. received a grant from Glaxo-Wellcome for $500,000 for genomic equipment from 2000–2003. S.T.W. was a consultant for Roche Pharmaceuticals in 2000 but received no financial renumeration for this consultancy. S.T.W. has also served as a consultant to Pfizer (2000–2003), Schering Plough (1999–2000), Variagenics (2002), Genome Therapeutics (2003), and Merck Frost (2002). J.C.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee Report. Allergy 2004;59:469–478. [DOI] [PubMed] [Google Scholar]
- 2.Monica J, Federico MJ, Wamboldt FS, Carter R, Mansell A, Wamboldt MZ. History of serious asthma exacerbations should be included in guidelines of asthma severity. J Allergy Clin Immunol 2007;119:50–56. [DOI] [PubMed] [Google Scholar]
- 3.Fuhlbrigge AL, Weiss ST, Kuntz KM, Paltiel AD. Forced expiratory volume in 1 second percentage improves the classification of severity among children with asthma. Pediatrics 2006;118:e347–e355. [DOI] [PubMed] [Google Scholar]
- 4.Kitch BT, Paltiel AD, Kuntz KM, Dockery DW, Schouten JP, Weiss ST, Fuhlbrigge AL. A single measure of FEV1 is associated with risk of asthma attacks in long-term follow-up. Chest 2004;126:1875–1882. [DOI] [PubMed] [Google Scholar]
- 5.Sporik R, Chapman MD, Platts-Mills TAE. House dust mite exposure as a cause of asthma. Clin Exp Allergy 1992;22:897–906. [DOI] [PubMed] [Google Scholar]
- 6.Duvernelle C, Freund V, Frossard N. Transforming growth factor-beta and its role in asthma. Pulm Pharmacol Ther 2003;16:181–196. [DOI] [PubMed] [Google Scholar]
- 7.de Blay F, Krieger P, Spirlet F, Moreau L, Duvernelle C, Kassel O, Kopferschmitt MC, Gasser B, Demangeat C, Pauli G, et al. Repeated inhalation of low doses of cat allergen that do not induce clinical symptoms increases bronchial hyperresponsiveness and eosinophil cationic protein levels. Int Arch Allergy Immunol 1999;120:158–165. [DOI] [PubMed] [Google Scholar]
- 8.Boxall C, Holgate ST, Davies DE. The contribution of transforming growth factor-beta and epidermal growth factor signalling to airway remodelling in chronic asthma. Eur Respir J 2006;27:208–229. [DOI] [PubMed] [Google Scholar]
- 9.Ihn H. Pathogenesis of fibrosis: role of TGF-beta and CTGF. Curr Opin Rheumatol 2002;14:681–685. [DOI] [PubMed] [Google Scholar]
- 10.Fattouh R, Midence NG, Arias K, Johnson JR, Walker TD, Goncharova S, Souza KP, Gregory RC Jr, Lonning S, Gauldie J, et al. Transforming growth factor-beta regulates house dust mite-induced allergic airway inflammation but not airway remodeling. Am J Respir Crit Care Med 2008;177:593–603. [DOI] [PubMed] [Google Scholar]
- 11.Simcock DE, Kanabar V, Clarke GW, O'Connor BJ, Lee TH, Hirst SJ. Proangiogenic activity in bronchoalveolar lavage fluid from patients with asthma. Am J Respir Crit Care Med 2007;176:146–153. [DOI] [PubMed] [Google Scholar]
- 12.Grainger DJ, Heathcote K, Chiano M, Snieder H, Kemp PR, Metcalfe JC, Carter ND, Spector TD. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet 1999;8:93–97. [DOI] [PubMed] [Google Scholar]
- 13.Silverman ES, Palmer LJ, Subramaniam V, Hallock A, Mathew S, Vallone J, Faffe DS, Shikanai T, Raby BA, Weiss ST, et al. Transforming growth factor-β1 promoter polymorphism C–509T is associated with asthma. Am J Respir Crit Care Med 2004;169:214–219. [DOI] [PubMed] [Google Scholar]
- 14.Wang H, Zhao YP, Gao CF, Ji Q, Gressner AM, Yang ZX, Weiskirchen R. Transforming growth factor β1 gene variants increase transcription and are associated with liver cirrhosis in Chinese. Cytokine 2008;43:20–25. [DOI] [PubMed] [Google Scholar]
- 15.Dunning AM, Ellis PD, McBride S, Kirschenlohr HL, Healey CS, Kemp PR, Luben RN, Chang-Claude J, Mannermaa A, Kataja V, et al. A transforming growth factorβ1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res 2003;63:2610–2615. [PubMed] [Google Scholar]
- 16.Hinke V, Seck T, Clanget C, Scheidt-Nave C, Ziegler R, Pfeilschifter J. Association of transforming growth factor-beta1 (TGF-beta1) T29 → C gene polymorphism with bone mineral density (BMD), changes in BMD, and serum concentrations of TGF-beta1 in a population-based sample of postmenopausal German women. Calcif Tissue Int 2001;69:315–320. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Romieu I, Wu H, Sienra-Monge J-J, Ramírez-Aguilar M, del Río-Navarro BE, del Lara-Sanchez IC, Kistner EO, Gjessing HK, London SJ. Genetic polymorphisms in transforming growth factor beta-1 (TGF-β1) and childhood asthma and atopy. Hum Genet 2007;121:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokota M, Ichihara S, Lin TL, Nakashima N, Yamada Y. Association of a T29→C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in japanese. Circulation 2000;101:2783–2787. [DOI] [PubMed] [Google Scholar]
- 19.Awad MR, El-Gamel A, Hasleton P, Turner DM, Sinnott PJ, Hutchinson IV. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factor-beta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation 1998;66:1014–1020. [DOI] [PubMed] [Google Scholar]
- 20.Mak JC, Leung HC, Ho SP, Law BK, Ho AS, Lam WK, Ip MS, Chan-Yeung MM. Analysis of TGF-beta(1) gene polymorphisms in Hong Kong Chinese patients with asthma. J Allergy Clin Immunol 2006;117:92–96. [DOI] [PubMed] [Google Scholar]
- 21.Buckova D, Izakovicova Holla L, Benes P, Znojil V, Vacha J. TGF-beta1 gene polymorphisms. Allergy 2001;56:1236–1237. [DOI] [PubMed] [Google Scholar]
- 22.Heinzmann A, Bauer E, Ganter K, Kurz T, Deichmann KA. Polymorphisms of the TGF-beta1 gene are not associated with bronchial asthma in caucasian children. Pediatr Allergy Immunol 2005;16:310–314. [DOI] [PubMed] [Google Scholar]
- 23.Sharma S, Raby BA, Soto-Quiros ME, Avila L, Murphy A, Klanderman BJ, Sylvia JS, Weiss ST, Celedon JC. Functional variants in TGF-beta1 are associated with increased airway responsiveness and asthma exacerbations in children with asthma. Am J Respir Crit Care Med 2008;177:A774. [Google Scholar]
- 24.Hunninghake GM, Soto-Quiros ME, Avila L, Ly NP, Liang C, Sylvia JS, Klanderman BJ, Silverman EK, Celedon JC. Sensitization to ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol 2007;119:654–661. [DOI] [PubMed] [Google Scholar]
- 25.Raby BA, Silverman EK, Kwiatkowski DJ, Lange C, Lazarus R, Weiss ST. Adam33 polymorphisms and phenotype associations in childhood asthma. J Allergy Clin Immunol 2004;113:1071–1078. [DOI] [PubMed] [Google Scholar]
- 26.The Childhood Asthma Management Program (CAMP) research group. Long-term effects of budesonide or nedocromil in children with asthma. N Engl J Med 2000;343:1054–1063. [DOI] [PubMed] [Google Scholar]
- 27.The Childhood Asthma Management Program (CAMP) research group. Design, rationale, and methods. Childhood asthma management program research group. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- 28.O'Connell JR, Weeks DE. Pedcheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haldane J. An exact test for randomness of mating. J Genet 1954;52:631–635. [Google Scholar]
- 30.Lake SL, Laird NM. Tests of gene-environment interaction for case-parent triads with general environmental exposures. Ann Hum Genet 2004;68:55–64. [DOI] [PubMed] [Google Scholar]
- 31.Hunninghake GM, Soto-Quiros ME, Lasky-Su J, Avila L, Ly NP, Liang C, Klanderman BJ, Raby BA, Gold DR, Weiss ST, et al. Dust mite exposure modifies the effect of functional IL10 polymorphisms on allergy and asthma exacerbations. J Allergy Clin Immunol 2008;122:99–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hakonarson H, Bjornsdottir US, Ostermann E, Arnason T, Adalsteinsdottir AE, Halapi E, Shkolny D, Kristjansson K, Gudnadottir SA, Frigge ML, et al. Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am J Respir Crit Care Med 2001;164:2036–2044. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa E, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Transforming growth factor-beta1 polymorphisms, airway responsiveness and lung function decline in smokers. Respir Med 2007;101:938–943. [DOI] [PubMed] [Google Scholar]
- 34.Alcorn JF, Rinaldi LM, Jaffe EF, van Loon M, Bates JH, Janssen-Heininger YM, Irvin CG. Transforming growth factor-β1 suppresses airway hyperresponsiveness in allergic airway disease. Am J Respir Crit Care Med 2007;176:974–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fu CL, Ye YL, Lee YL, Chiang BL. Effects of overexpression of IL-10, IL-12, TGF-beta and IL-4 on allergen induced change in bronchial responsiveness. Respir Res 2006;7:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fattouh R, Pouladi MA, Alvarez D, Johnson JR, Walker TD, Goncharova S, Inman MD, Jordana M. House dust mite facilitates ovalbumin-specific allergic sensitization and airway inflammation. Am J Respir Crit Care Med 2005;172:314–321. [DOI] [PubMed] [Google Scholar]
- 37.Collaco JM, Vanscoy L, Bremer L, McDougal K, Blackman SM, Bowers A, Naughton K, Jennings J, Ellen J, Cutting GR. Interactions between secondhand smoke and genes that affect cystic fibrosis lung disease. JAMA 2008;299:417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salam MT, Gauderman WJ, McConnell R, Lin PC, Gilliland FD. Transforming growth factor-β1 C-509T polymorphism, oxidant stress, and early-onset childhood asthma. Am J Respir Crit Care Med 2007;176:1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieu TA, Quesenberry CP, Sorel ME, Mendoza GR, Leong AB. Computer-based models to identify high-risk children with asthma. Am J Respir Crit Care Med 1998;157:1173–1180. [DOI] [PubMed] [Google Scholar]
- 40.Drumm ML, Konstan MW, Schluchter MD, Handler A, Pace R, Zou F, Zariwala M, Fargo D, Xu A, Dunn JM, et al. Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med 2005;353:1443–1453. [DOI] [PubMed] [Google Scholar]
- 41.Celedon JC, Lange C, Raby BA, Litonjua AA, Palmer LJ, DeMeo DL, Reilly JJ, Kwiatkowski DJ, Chapman HA, Laird N, et al. The transforming growth factor-beta1 (TGF-b1) gene is associated with chronic obstructive pulmonary disease (COPD). Hum Mol Genet 2004;13:1649–1656. [DOI] [PubMed] [Google Scholar]
- 42.Park HK, Park HW, Jeon SG, Shin ES, Gho YS, Cho SH, Kim YY, Kim YK. Distinct association of genetic variations of vascular endothelial growth factor, transforming growth factor-beta, and fibroblast growth factor receptors with atopy and airway hyperresponsiveness. Allergy 2008;63:447–453. [DOI] [PubMed] [Google Scholar]
- 43.Coutts A, Chen G, Stephens N, Hirst S, Douglas D, Eichholtz T, Khalil N. Release of biologically active TGF-beta from airway smooth muscle cells induces autocrine synthesis of collagen. Am J Physiol Lung Cell Mol Physiol 2001;280:L999–L1008. [DOI] [PubMed] [Google Scholar]
- 44.Williams AE, Humphreys IR, Cornere M, Edwards L, Rae A, Hussell T. TGF-beta prevents eosinophilic lung disease but impairs pathogen clearance. Microbes Infect 2005;7:365–374. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, Chau J, Young RP, Pokorny V, Mills GD, Hopkins R, McLean L, Black PN. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax 2004;59:126–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.