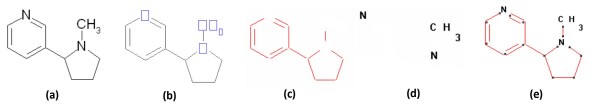
Abstract
Background
To search for chemical structures in research articles, diagrams or text representing molecules need to be translated to a standard chemical file format compatible with cheminformatic search engines. Nevertheless, chemical information contained in research articles is often referenced as analog diagrams of chemical structures embedded in digital raster images. To automate analog-to-digital conversion of chemical structure diagrams in scientific research articles, several software systems have been developed. But their algorithmic performance and utility in cheminformatic research have not been investigated.
Results
This paper aims to provide critical reviews for these systems and also report our recent development of ChemReader – a fully automated tool for extracting chemical structure diagrams in research articles and converting them into standard, searchable chemical file formats. Basic algorithms for recognizing lines and letters representing bonds and atoms in chemical structure diagrams can be independently run in sequence from a graphical user interface-and the algorithm parameters can be readily changed-to facilitate additional development specifically tailored to a chemical database annotation scheme. Compared with existing software programs such as OSRA, Kekule, and CLiDE, our results indicate that ChemReader outperforms other software systems on several sets of sample images from diverse sources in terms of the rate of correct outputs and the accuracy on extracting molecular substructure patterns.
Conclusion
The availability of ChemReader as a cheminformatic tool for extracting chemical structure information from digital raster images allows research and development groups to enrich their chemical structure databases by annotating the entries with published research articles. Based on its stable performance and high accuracy, ChemReader may be sufficiently accurate for annotating the chemical database with links to scientific research articles.
Background
In the scientific literature, there is a tremendous amount of information about the interaction of small molecules with specific targets, the influence of small molecules on biochemical pathways, the phenotypic effects of small molecules in different cell types, as well as the relationship of small molecules, targets, pathways and phenotypes to disease processes. However much of this information has yet to be compiled in the form that would allow using a molecule's chemical structure as an input to search for its potential relevance in a specific physiological, pathological or therapeutic area of interest. Two examples of information resources linking chemical structures with biomedical targets, pathways and phenotypes are PubMed [1] – the database of the scientific literature corpus – and PubChem [2] – a publicly available database of over 19 million chemical structures, each of which can have a cross-reference link to similar structures, bio-assay data, and bio-activity descriptions. If these resources can be used to construct a universal database encompassing all known chemical structures with links to specific targets, biochemical pathways, disease states and potential therapeutic applications, a powerful new tool for both biomedical research and drug discovery would emerge.
In general, one can envision two ways to parse scientific articles for chemical information: by searching for names or structure diagrams of chemical agents. The chemical structure diagrams in scientific articles are typically drawn manually using a program such as ChemDraw [3], ISIS/Draw [4], DrawIt [5], and ACD/ChemSketch [6]. Once a structure is drawn, the structural description can be translated into a computer readable format, such as ISIS, MOLfile, SMILES, or ROSDAL formats, which describes the atoms, bond orders, and connectivity patterns of atoms in molecules. However, the diagrams of chemical molecules in scientific journals and reference books are encoded as digitized images (e.g. BMP, TIFF, PNG or GIF), which in turn are embedded within lines of text in a form that is not readily translatable into a computer readable format. Therefore, most references to chemical agents in scientific research articles cannot be easily linked to other repositories of scientific knowledge, and are thus not amenable for analysis or searching using cheminformatic software.
An effective image searching capability would require converting the digital raster images of chemical diagrams into structured representations such as SMILE strings or atom connectivity tables in standard chemical file formats. Once a reliable structure recognition and conversion system is developed it can be used to scan pages of chemical literature and construct a database, as illustrated in Figure 1. The resulting database is one that can be queried with a cheminformatic search engine into which an investigator can input a chemical structure and pull any related information of interest. Novel drug candidates or newly synthesized molecules are usually referenced by chemical structure diagrams rather than molecule names. In addition, a single molecule may have a number of synonyms such that it could be referenced by different names in different articles. Thus, the capability of exploring research articles or patents where the chemical structure or similar compounds are drawn would complement existing text-based search engines for chemical information.
Figure 1.
Automated extraction of chemical structure information in scientific articles.
In the 1990s, several software programs were developed that could extract chemical structure diagrams in scientific articles and convert them to structured representations [7-9]. Recently, with the active development of cheminformatic tools for processing published chemical information [10], two more software programs were launched and continue to be updated [11,12]. This paper examines these existing systems and also reports our recent development of ChemReader – a comparable tool that can be specifically tailored for chemical database annotation (Table 1). Basic algorithms for recognizing lines and letters representing bonds and atoms in chemical structure diagrams are presented. In its present state, test results indicate that ChemReader outperforms Kekule, CLiDE and OSRA, side-by-side.
Table 1.
Comparison of existing machine vision system for chemical structure recognition. O and X denotes the availability of key features listed in the first column: O = Positive and X = Negative.
| Kekule | IBM OROCS | CLiDE | OSRA | chemOCR | ChemReader | |
| Written language | C++ | C | C++ | C++ | Java | C++ |
| Running Platform | MS windows | IBM OS/2 | MS windows | Linux/MS Windows/OS X | Independent | MS Windows |
| Batch mode | X | X | O | O | O | O |
| Bond streo | O | X | O | O | O | O |
| Abbreviation Interpretation | O | X | O | O (limited)1 | O | O |
| Chemical Knowledge | O | X | X | X | O | O |
| Document analysis | O | X | O | X | X | Under development |
| Automatic Extraction2 | X | O | O | X | X | Under development |
| Open source | X | X | X | O | X | X |
| Customizable extensibility | X | X | X | O | O | O |
1 OSRA has a hardcoded matching table to interpret only a few chemical abbreviations.
2 A functionality for extracting digitized images of chemical structure diagrams from scanned pages
Machine Vision Approaches for Digital Image Recognition
Machine-vision is concerned with the theory and method for processing the image data and identifying relevant image features effectively [13]. Machines see objects in different ways than human beings. Given digitized image data or multi dimensional data, machines extract features and classify patterns by examining each digital element (pixels) of each image. In general, a machine processes an image in the following steps:
• De-noising: Removing visual artifacts that decrease the ability to extract information from the images;
• Segmentation: Separating objects in the image;
• Feature extraction: Characterization of each segmented region by extracting topological features;
• Consistency analysis: Interpreting the entire image based on extracted local features; and
• Classification/Matching: Identifying the object in the image in relation to a reference set of objects.
Many applications of machine-vision have been developed and are used in various fields (e.g., automated diagnosis system in medicine, quality control in manufacturing industry, and security and intruder identification). There are also several applications performing tasks of extracting structural information from digital images of technical diagrams. Dori and Wenyin have developed the Machine Drawing Understanding System (MDUS), which can convert printed mechanical engineering drawings that are scanned and stored as raster digitized image files into standard file formats that can be read by Computer-Aided Design (CAD) software [14]. An automated conversion system of electronic circuit diagrams have been also developed [15].
Machine Vision Systems for Recognition of Chemical Structure Images
The essential components of chemical structure drawing can be categorized into bond lines and atom symbols. In all systems listed below, these two components in raw image data are first separated by a segmentation algorithm. Then bond lines in graphic segments can be processed by a line detection algorithm and atom symbols in text segments can be recognized by a character recognition algorithm. Finally, a graph representing the chemical structure is built from both results, and from this the structure information can be extracted and stored as a standard chemical file format.
To extract a chemical diagram from a document and convert it to a digital chemical file, any automated machine-vision based system would need to be able to execute all of the following tasks without manual intervention. The first step is to identify all the individual chemical diagrams in a document, and segment these diagrams into atoms and bonds connected to form an individual molecule. For this purpose, a document page containing chemical structure diagrams should be scanned to produce a digital raster image of the entire page. Before proceeding to process the scanned digital raster image, it is necessary to extract only a subarea of the page which contains a chemical structure diagram. Next, with the isolated chemical structure image, another algorithm is used to classify the graphic (bonds) and text components in those images. A conventional connected component algorithm is typically used to segment an image into sets of pixels connected with each other, and the relative size of each component gives information to distinguish a component between graphics (bond lines) and text (atom symbols).
Once lines and text have been separated from each other, the next step is to identify the length, position and direction of the lines, and the characters of the text. There are several types of bonds used in the chemical structure diagram: single, double, triple, wedged, dotted, dashed, and dashed-wedged. Since the basic graphical elements composing such bonds are lines, Hough transform [16] and vectorization algorithms, which are widely used in machine-vision systems, are employed for the line detection schemes. Different bond types can be distinguished by considering detected line length, width and arrangement patterns. For character recognition, text components are conveyed into a character recognition engine where they are analyzed using artificial neural networks or feature based approaches.
The last step of chemical structure extraction involves establishing the connectivity of the atoms, in terms of which atoms are linked to each other, and the number of bonds between them. Based on the result of previous steps, a graph representing the chemical structure is constructed. From the result of character recognition, the detected chemical symbols for atom types or molecular groups are assigned to nodes. The detected lines enable the construction of the entire structure of the grap. In some cases, a character string at a node could be an abbreviation (e.g., OMe for a methyl-ester). In such cases, it is necessary to interpret the chemical meaning of the abbreviation in order to build a complete chemical standard file. A database of chemical abbreviations which frequently appear in the chemical structure diagram can be used for this purpose. By looking up the abbreviation in the database, the abbreviation can be translated directly to a digital chemical representation. If there is no matching entry, the system can flag the structure as potentially misrecognized. At the final step, the compiled chemical structure graph is translated into a chemical standard file such as Molfile, SMILE strings.
Kekule
The first commercial program to read and interpret digital raster images of chemical structures was Kekule [7], developed by Joe R. McDaniel and Jason R. Balmuth of Fein-Marquart Associates Inc. in Baltimore, MD. The program requires at least a 150 dpi image resolution. In Kekule, the area of a page that contains a chemical structure diagram needs to be manually identified. In terms of interesting features, Kekule has a built-in algorithm to fix character recognition errors. For this purpose, a neural network is used for generating potential characters with scoring information estimating the likelihood that a specific character corresponds to a certain atom. Even when an incorrectly recognized character has a higher score than the correct candidate, Kekule can fix character-to-atom conversion errors by considering the valence and chemical neighbors of the atom. Still, manual correction at the post-processing step is often required, due to an average accuracy of 0.74 per structure diagram.
Optical Recognition Of Chemical graphics (OROCS)
For converting chemical structure images to computer-readable format, another program called OROCS [8], was developed at the IBM Almaden Research Center, San Jose, CA. The most interesting feature of the OROCS system is that is has an algorithm for automated extraction of chemical structure diagrams from scanned document images. In order to isolate chemical structure diagrams from other elements – such as text, figures and pictures on a page-the document is segmented by a conventional connected components algorithm. If the size of a segment is larger than a threshold, it is potentially regarded as a chemical structure, and the polygonal shapes of chemical structure diagrams are used to make a final decision. The methodology implemented in OROCS was granted a U.S. patent in 1992 [17].
Chemical Literature Data Extraction (CLiDE)
Amongst the chemical structure extraction efforts to date, the Chemical-Literature Data-Extraction Project (CLiDE) [9] is available commercially. CLiDE not only aims at extracting chemical structures but also abstracting chemical information from text. By employing the Documental Format Description Language (DFDL) which can describe logical relationships of objects and elements in a document, CLiDE builds logical associations between chemical structures and the text segments of document [18]. Unlike OROCS and like Kekule, CLiDE does not have an automated process to discriminate chemical structure diagram from graphical objects, so manual separation of chemical diagrams is necessary. As well as Kekule and OROCS, CLiDE requires at least a 300 dpi resolution in scanned images at the scanning step and manual correction at the post processing step to achieve reliable output. However, the drawn chemical structure diagrams are typically embedded in Word documents as GIF or JPG formats, whose the resolution is usually 72–96 dpi. Therefore, these software systems might be impractical tools for fully automated extraction of chemical structure information.
chemOCR
Recently, a new program, called chemOCR [11], has been developed and made available. Focusing on overcoming the most common errors generated by prior systems, chemOCR adopted a chemical rule-based expert system for the extraction of chemical structure diagrams. The most interesting features, at the post-processing stage, is that chemOCR uses a graph-matching algorithm to select the best-matching chemical structure fragment against sub-graphs of chemical structures stored in a database. With this approach, even if several errors occur during detecting lines or recognizing characters, the errors can be corrected by simply replacing unrealistic chemical fragments of a molecule with known sub-structural motifs present in the database of chemical substructures. In their own testing, chemOCR showed high correct recognition rates ranging from 67 to 97%, and thus outperformed CLiDE which could process only 25 images out of 100 successfully.
Optical Structure Recognition (OSRA)
OSRA [12], another recently released program is free and open source software written by the CADD group at the National Cancer Institute. OSRA attempts to generate three output structures by varying parameters for the de-noising stage, and then picks one as an output based on its own empirical confidence function. Since most machine vision algorithms could yield quite different interpretations of the same input with a slightly different parameter setting, this iterative processing of the same input could improve the overall ratio of correct outputs, so long as the confidence function is reliable enough.
Results
ChemReader – Overview
ChemReader is a software developer toolkit for translating digital raster images of chemical structures into standard, chemical file formats that can be searched and analyzed with other open source or commercial cheminformatic software. Its intention is to allow tailoring of each step of the extraction of chemical diagrams, to optimize annotating a database of chemical structures from references in the scientific literature, as illustrated in Figure 1. Recognizing the shortcomings of the other systems discussed in the previous section, ChemReader aims to achieve very high recognition accuracy and robust performance sufficient for fully automated processing of research articles. In addition, ChemReader possesses a graphical user interface (GUI) that allows each step of the algorithm to be tested independently.
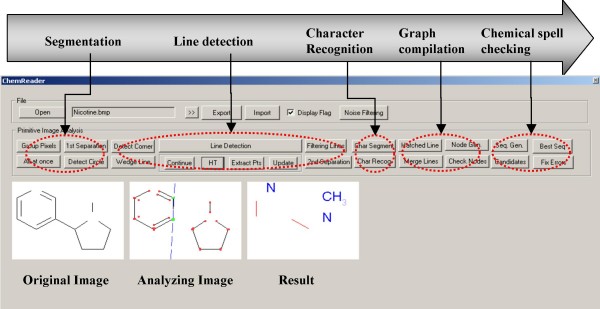
Figure 2 shows the basic recognition steps of chemical structure diagram extraction with ChemReader. The chemical structure drawing is a binary image which consists of a long sequence of bits that give pixel-by-pixel values. In the first step, the pixels are grouped into components based on pixel connectivity. Next, these connected components are classified as text or graphics. Text components are transferred to a character recognition algorithm and converted to chemical (atom) symbols. Graphical components representing bond connectivity are analyzed using the (Generalized) Hough Transformation, Corner Detection algorithm, and a few other geometric operations detailed below. Finally, from recognized chemical atom symbols and bonds, the whole of the structural information is assembled and displayed graphically for verification by the user. Figure 3 shows the GUI of ChemReader. The current version of ChemReader can read most of common image formats including GIF, JPG, BMP and PNG.
Figure 2.
Recognition of chemical structure diagram images in ChemReader. (a) input image, (b) character-line separation, (c) bond recognition, (d) character recognition, and (e) topology construction and data output.
Figure 3.
Graphical User Interface (GUI) of ChemReader.
Pre-processing
The first step in ChemReader involves an image processing for re-sizing and de-noising. The chemical structure diagrams are drawn with different settings in the drawing software, such as default bond lengths or character font sizes. Moreover, the image size and format are subjected to variations while transferred to the final destination, for example, a journal article or a web page. Thus it is necessary to resize and de-noise the input image so that the chemical structure diagram within the input image has bond lengths and character sizes optimally adjusted to ChemReader's recognition algorithms. With the first run of line detection as explained below, the length of the single bond is estimated. If the estimated bond length is shorter or larger than a certain threshold (currently 25 pixels), the image is resized such that bonds extracted in the next stages can have ChemReader's preferred length. For this purpose, GREYCstoration [19], a free implementation of image regulation algorithm [20] is used.
Separation of lines and characters
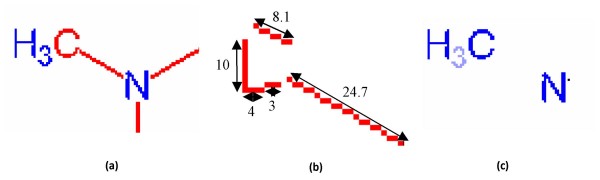
The second step is disassembling connected components based on pixel connectivity. In ChemReader, the 8-connectivity algorithm was used. Subsequently, the connected components are classified into characters and graphics. To detect characters, a character detection algorithm searches for objects with similar heights and areas. The most populated area/height combination will, in general, represent text components [21]. Most text components can be separated from the rest of the chemical structure using this method.
If a text component is not separated from a graphic component (e.g., because of a printer error) but is aligned with a successfully-separated text component (referred to as a "seed string"), the glued character component is separated from the graphics by extending the seed string [22] in the direction in which the seed string characters are aligned. In order to distinguish the small isolated lines or circles representing bonds from the text components, the relative location and horizontal/vertical run profile of each component are also checked. For example, the letter 'l' is often wrongly identified as a graphic component. However, since it always appears next to other letters, the letter 'l' can be correctly identified as a letter and not a bond by considering the relative location of each letter. If text components cannot be identified in this manner, they can often be corrected in subsequent steps.
Line Detection Algorithm
Most bonds in a chemical structure drawing are simple straight lines. Therefore, a robust line detection algorithm is the key software component for extracting bond features from a chemical structure diagram. In digital image processing, the Hough Transform (HT) is a standard technique used for this purpose. It detects lines by mapping the image in the Cartesian space to the polar Hough space using the normal representation of a line in x-y space (Figure 4(a) and 4(b)):
Figure 4.
Hough Transformation for bond detection. (a) Cartesian Image Space, (b) Polar Hough Space, (c) Example of HT applied to a chemical structure image, and (d) Hough Space corresponding to (c).
xi cos θi + yi sin θi = ri
Since a pixel corresponds to a sinusoidal curve in the Hough space, collinear pixels in the x-y space have intersecting sinusoidal lines. Therefore, all possible lines passing through every arbitrary pair of pixels in a chemical diagram image are identified by checking the intersection points of curves in the Hough space. Figure 4(c) and 4(d) shows the detected line and the corresponding Hough space. The density of a point (r*, θ *)in Hough Space (Figure 4(d)) would represent the likelihood of finding a corresponding line in the actual chemical diagram image (Figure 4(c)).
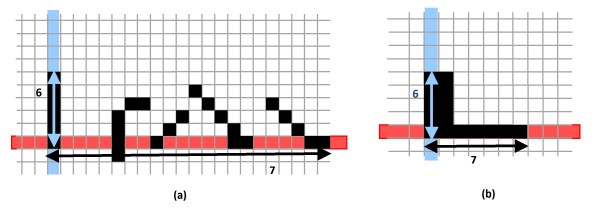
However, the conventional HT does not use pixel-connectivity and line-width information as important features for line-extraction. So, while the human eye only recognizes a linear grouping of pixels as a line (vertical (blue) line in Figure 5(a)), the Hough transform would assign greater weight to a broken, aligned sequence of pixels (illustrated by the horizontal (red) line in Figure 5(a)). In addition, while the human eye may only be able to see dark (thick) lines that are two or more pixels in width, the Hough transform will assign a greater weight to a long, narrow sequence of single pixels that may be less visible to the human eye (red line, Figure 5(b)). To correct these problems, a modified Hough-Transform [23] is used for line detection. In the modified HT, each pair of pixels is assigned a weight based on the probability that the two pixels originate from a single line-segment. The weight could be defined as
Figure 5.
Considering line thickness and connectivity in the HT. Vertical lines could have priority over horizontal lines in a modified HT due to their connectivity (a) and thickness (b).
where nij is the number of pixels that have distance less than a half of the thickness of the line connecting Pi and Pj, xij is the number of black pixels in nij pixels, and p0 is the total number of black pixels in the image space divided by the image size. The pixel pairs assigned by this method can be selected randomly to reduce computational time and memory usage. Since the ends of line segments can be recognized as corner pixels, those pixels which are identified by the wedge-bond detection-algorithm (described below) can also be used for general line detection. The line detection algorithm terminates when the assigned pixel pair results in a short line segment compared to the previously detected line segments.
While running the Hough Transform, it is possible that a text component can be recognized as multiple short line segments if it is not successfully separated and identified as a text component before line detection. This type of error often occurs if a text (character) component is glued to a graphic component (Figure 6(a)). Due to the complex shape of characters, the length of line segments from a character is much shorter than the length of chemical bonds (Figure 6(b)). Thus, these short line segments are examined to determine if they are in fact a graphic or a text component (Figure 6(c)).
Figure 6.
Recovering process for characters glued to graphics. A sub image which has (a) a character component connected to a graphic component, (b) line detection result of (a), and (c) Correctly separated characters.
Bond Type Identification
In low resolution (fuzzy) images, Hough Transformation often fails to distinguish a double or triple bond from a single bond. With thickness-based bond correction, a single line detected can be interpreted as a double or triple bond by considering the thickness of the bond, as well as the pattern of dark-white transitions perpendicular to the line.
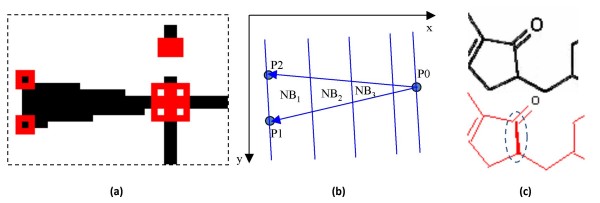
Stereochemical 'wedge-bonds' are detected after separating text from graphic components. Using a corner-detection algorithm [24], ChemReader examines every possible combination of 3 corner points which could be a set of 3 vertices constituting a wedge bond. To verify whether it is a wedge, the following three conditions are checked (Figure 7):
Figure 7.
Detection of streochemical wedge bond. (a) Detected corner points (Red Block) around a wedge bond, (b) a combination of 3 corner points, NBi = Number of Black Pixels in each region, and (c) wrongly detected wedge.
• Area of the triangle = Number of black pixels in the triangle
• Almost isosceles triangle shape
• NB1 > NB2 > NB3, where NB is the number of black pixels (see Figure 7)
In the case where a normal (non-stereochemical) bond is unusually thick or a double bond cannot be resolved as two separate lines, the wedge-bond detection can lead to a bond misrecognition error (Figure 7(c)). To correct this error, the width of wedge bonds (captured by the length between P1 and P2; see Figure 7) is compared to the average width of normal bonds after extracting the normal bonds.
Yet another challenging detection problem involves identifying individual dashed/dotted bonds that are drawn to indicate conjugated bonds or stereochemistry in chemical diagrams. These dashed or dotted bonds can be detected from residual pixels that are neither a part of the character component nor classified as a normal or wedged bonds (Figure 8). To detect dotted bonds, an algorithm is run to find short line segments having uniform length and interval, as well as being collinear in the direction perpendicular to the direction of the short line-segments.
Figure 8.
Sequential steps for bond detection. (a) Original Image, (b) Detected corner points after removing character components, (c) Detected normal and wedge bonds, and (d) left pixels before dashed bond detection.
Finally, dashed bond detection is performed on any left-over (unextracted) pixels in the original image (Figure 9). Applying each center of connected components of left pixels to a conventional Hough Transform, ChemReader can successfully detect a line which is orthogonal to a dashed line segment, and recognize that the line as a dashed bond.
Figure 9.
Left over pixels before hatched bond detection. (a) Original Image, (b) left pixels before hatched bond detection, and (c) Most voted line from HT and line segments orthogonal to it.
Ring Structure Identification
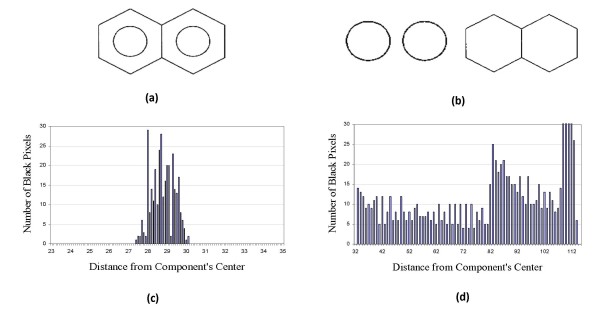
Another interesting bond recognition problem occurs in aromatic systems, where a circle is often used to represent the conjugated electron system of the benzene ring. To identify these circles, an algorithm looks for the pixels of a connected component that are distributed with almost the same distance from the center of the component (Figure 10). With this algorithm, the presence of circular features can be detected by checking whether the standard deviation of distances from the center of an object is smaller than a certain threshold.
Figure 10.
Detection of aromatic ring bond. (a) Chemical structure of Naphthalene, (b) Connected components and distribution of distances of pixels from component's center for (c) Circle bonding and for (d) Non circle bonding.
In low resolution images, it is often observed that a detected line have a different position, length or direction from the actual bond. This is especially the case for the bonds in a hexagonal or pentagonal ring structure because the pixels of the neighbor bonds can act as noise in the Hough Transform (HT). Accumulated errors of line detection around a ring structure would cause significant errors in constructing the topology of the chemical structure. This problem could be solved by detecting Pentagonal or Hexagonal ring structures directly using the Generalized Hough Transformation (GHT) [25]. With GHT, ChemReader detects ring structure as a skeleton for processing chemical structure diagrams, so the topology of molecules can be constructed more accurately and efficiently.
Text (Character) Recognition
All separated character components are sent to an open-source library for optical character recognition [26]. Employed character recognition algorithm is based on template matching of features such as holes at middle, upper, lower positions, pixel densities of sub-regions, and white-black transitions through a line. Currently the library is used without any customization, which leads to relatively high recognition error as will be discussed in the following section. Figure 11 shows common character recognition errors in the chemical structure diagram.
Figure 11.
Common character recognition errors. (a) low resolution, (b) broken character, (c) glued to a graphic component, and (d) glued characters.
Chemical Spell Checker
The GOCR library outputs the recognition results of each character without any chemical interpretation. The results can contain non-existing chemical symbols or valences. To correct these errors, a chemical "spell checker," a recovery process similar to the conventional OCR error correction, is implemented in ChemReader. The characters recognized by the GOCR library are grouped by their relative adjacency and each character group is regarded as a chemical word representing either an atomic symbol or chemical abbreviation, which is subject to "spell checking" based on the chemical dictionary.
The implementation of the chemical spell checker is based on a dictionary lookup approach and chemical rules regarding valences. In addition to the GOCR library, a character recognition algorithm based on the pixel-by-pixel distance between input character segments and all potential segments in the character library is employed to produce candidate characters and the associated confidence scores. A total of ten candidate characters and their confidence scores are assigned to each input character segment. In general, chemical words in chemical structure diagrams fall into one of three types: simple molecular formulas representing a combination of nonmetal and hydrogen atoms (e.g. NH2), user defined symbols like X, Y and R, and chemical abbreviations (e.g., three letters for amino acids or "Me" for methyl group). Given candidate characters and their confidence scores, the chemical spell checker tries to find a most-likely chemical word based on a predefined, frequently-used chemical symbol table containing 770 frequently used chemical abbreviations and fundamental chemical rules on molecular formulas containing nonmetal and hydrogen atoms. The calculation of the likelihood is based on following equation:
where S and T denote the extracted chemical word consisting of m character segments S1S2...Sm and the true chemical word within the chemical dictionary or possible molecular formula string, respectively, where Sim(Si, Ti) is the confidence score of character recognition, given as the similarity between input character segment Si and comparing candidate character Ti. Sim(Si, Ti) is also defined as
where M denotes the number of pixels in a character segment, and IX(j) is a normalized grayscale intensity at the jth pixel of the character segment X. Before the calculation of similarity, the comparing candidate character is always resized so that both input and comparing segments have the same size. Since the exact frequency of each chemical symbols in chemical structure diagram is not known priori we assume that P(T) is equi-probable for all T ∈ Chem_Dictionary and ∑T ∈ Chem_Dictionary P(T) = 1. With this chemical spell checker, the accuracy of chemical symbol recognition increased to 87%, up from 66% without spell checking.
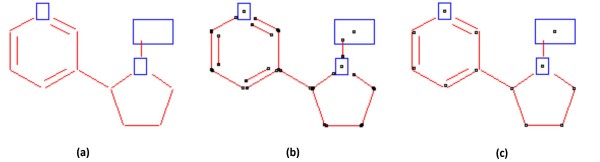
Topology Construction and Data Output
For data output, a graph representing the chemical structure is compiled based on the detected bonds and the recognized atomic or chemical symbols. Figure 12 shows a procedure to construct a chemical-structure graph. First, every end point of the identified bonds and center points of the identified chemical symbols are labeled as a node (Figure 12(a) and 12(b)). Next, among these nodes, the ones located within a certain distance are merged into a single node (Figure 12(c)). Based on this graph data structure, a node-edge connectivity-table is generated, which finally can be converted into a standard chemical file format [27] or SMILE string [28].
Figure 12.
Topology construction procedure. (a) detected bonds (lines) and symbols (rectangle), (b) created nodes (bold dots), and (c) final nodes and edges.
Testing
Three sets of the images of chemical structure diagrams collected from different sources were used to test the current ChemReader and compare it to OSRA V1.01, CLiDE V2.1 Lite, and Kekule V2.0 demo (Table 2). Since a new version, CLiDE Pro had been introduced [29], as it was not available at the time of testing, CLiDE V2.1 was used in this test (Full version has additional functionalities relating document analysis but they were not required in this test). Also, the results for chemOCR could not be obtained since we could not receive responses from SCAI to our requests for a demo version. Fifty images in Set I are obtained by querying pharmaceutically significant molecules to Google Image Search http://images.google.com/ so that the images have different drawing styles, sizes, font and resolutions. Set II consists of 100 ligand images randomly selected from the GLIDA database [30]. Since it requires the original structure information for result analysis, only ligand molecules with links to the PubChem database are considered while collecting ligand structure images. The images in Set III are collected from 121 journal articles. They often have non-chemical structure components such as descriptive text or symbols which represent neither atom nor chemical abbreviations, and thus those images are discarded. For the analysis, we obtain the original structure information of molecules in Sets I and II from the PubChem Database. For Set III, the original connection tables for test images are obtained by drawing structures manually using the ChemDraw software. The recognition results by ChemReader, OSRA, CLiDE, and Kekule are saved as either Molfiles or SMILE strings with graphical output images for analysis.
Table 2.
Image sets for performance test.
| Number of Images | Image Source | |
| Set I | 50 | Google image search |
| Set II | 100 | Ligand images at GLIDA database |
| Set III | 212 | Journals at PubMed database |
The performances of chemical structure recognition are analyzed in two aspects: the fraction of correct outputs and the capability to recognize chemically important substructure patterns. The first measure, the fraction of correct outputs shows straightforwardly the accuracy of the software. Although an error exists in the output molecule, it wouldn't be regarded as a totally useless one if chemically significant features-of-interests are well-recognized. For example, the misassignment of atom charge or bond-stereo would not be so critical for finding molecules similar to the recognized structure in a database. Thus, we compute the statistical measures, precision and recall rates in order to evaluate the software's capability for extracting chemically significant substructure patterns. Precision is the fraction of the extracted patterns that are correct whereas recall is the fraction of the correct patterns that are extracted. Structural patterns defined in the PubChem Substructure Fingerprint [31] are used in this test. The identity between the original molecule and output chemical structure is determined using an exact matching function in ChemAxon's JChem toolkits [32]. Also, the PubChem fingerprint is computed using an open-source code provided by the NIH Chemical Genomic Center (NCGC) [33].
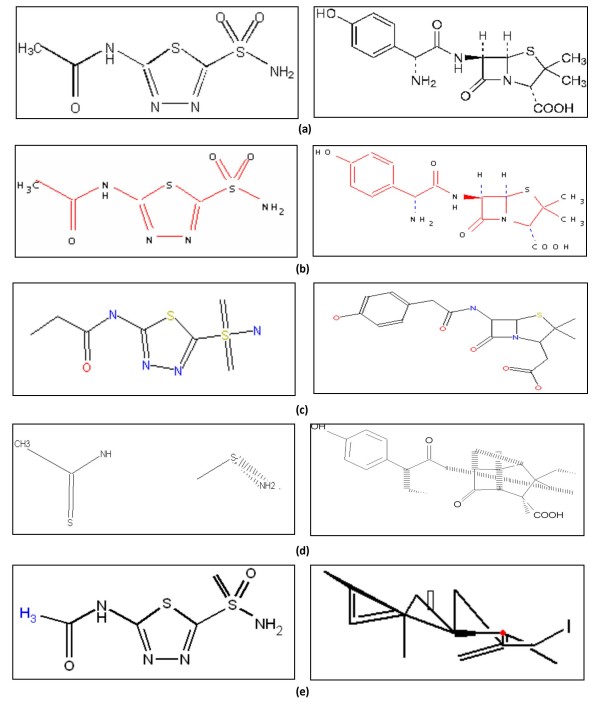
Table 3 shows a summary of testing results in terms of the fraction of correct outputs and the average Tanimoto similarity between the software outputs and the correct answers based on the PubChem Substructure Fingerprint. In all Sets (I, II, and III,) ChemReader performs the recognition process outstandingly compared to the other software programs (Figure 13). As the Kekule demo version does not have a batch mode, it's not tested for Set II and III. Figure 14 illustrates a few input and output examples. OSRA shows the most comparable performance to ChemReader in Sets I and II, but drops its correct outputs to under 20% in Set III while ChemReader keeps those over 30%. Set III contains chemical structure images within journal articles which have usually many abbreviations representing chemical group symbols. Also, as those figures are embedded in the middle of text, the sizes and resolutions of images are degraded in general. To process such images successfully, it would be necessary to have a chemical spell checker that can interpret many chemical symbols and fix errors that occur in the machine-vision algorithms. This explains the reason why the accuracy gap between ChemReader and OSRA is larger in Set III than in Sets I and II.
Table 3.
Summary of performance testing results for three sets of images.
| Set I (Total: 50) | Set II (Total: 100) | Set III (Total: 212) | ||||
| Correct | Similarity | Correct | Similarity | Correct | Similarity | |
| ChemReader | 25 (50%) | 0.860 | 33 (33%) | 0.877 | 64 (30.2%) | 0.740 |
| OSRA V1.0.1 | 20 (40%) | 0.798 | 25 (25%) | 0.785 | 36 (17%) | 0.526 |
| CLiDE Lite V2.1 | 2 (4%) | 0.489 | 2 (2%) | 0.490 | 14 (6.6%) | 0.294 |
| Kekule demo V2.01 | 6 (12%) | 0.428 | - | - | - | - |
1 Kekule was not tested for Set II and III due to lack of batch mode functionality in the demo version
Figure 13.
Percent of correct outputs and Average Tanimoto similarity scores over total outputs.
Figure 14.
Output examples. (a) input images, and results by (b) ChemReader, (c) OSRA, (d) CLiDE, and (e) Kekule.
The average Tanimoto similarity scores can be seen as the extent of correctly including chemically important features in the output structure. The more missed (false negative) or misinterpreted (false positive) features the output structure has, the smaller similarity score will become. It is noted that ChemReader's outputs show a high average-similarity score ranging from 0.74 to 0.88 even though only about 30% of outputs are perfectly correct. This indicates that ChemReader can be effective in annotating chemical structure database by linking published research articles to relevant entries in the database. Since those links would likely be created based on a molecular similarity rather than perfect matching, high similarity scores would imply the high accuracy in automated chemical database annotation.
From the generated binary PubChem Substructure Fingerprint, precision and recall rates for each substructure pattern are computed for detailed analysis. Table 4 shows the average precision and recall rates over seven types of patterns which are already categorized in the PubChem Fingerprint specification. The first and second groups of PubChem substructures are involved in testing for the presence or count of atoms or ring systems. Items in the third and fourth groups examine the presence of several specific bonded atom pairs and atom nearest neighbor patterns, respectively, regardless of bond type. In the 5th, 6th and 7th groups of substructure patterns, bond types and aromaticity are specific such that the exact presence of described SMILE or SMART pattern is examined. The main difference between ChemReader and other software programs presented here is that ChemReader's recall rate is significantly higher than others over all types of patterns while precision rates are similar. It indicates that ChemReader has the advantage on extracting substructure features with high accuracy (precision and recall rates) over the other programs, which would be essential for an automated annotation system for a chemical database.
Table 4.
Estimated Precision (P) and Recall (R) rates for classification of substructure patterns.
| ChemReader | OSRA | CLiDE | Kekule | |||||
| P | R | P | R | P | R | P | R | |
| 1. Hierarchic Element Counts | 0.95 | 0.92 | 0.95 | 0.83 | 0.95 | 0.59 | 0.88 | 0.57 |
| 2. Rings | 0.87 | 0.83 | 0.84 | 0.73 | 0.76 | 0.37 | 0.61 | 0.51 |
| 3. Simple atom pairs | 0.93 | 0.92 | 0.93 | 0.83 | 0.94 | 0.60 | 0.95 | 0.56 |
| 4. Simple atom nearest neighbors | 0.94 | 0.86 | 0.89 | 0.73 | 0.83 | 0.41 | 0.89 | 0.43 |
| 5. Detailed atom neighborhoods | 0.89 | 0.89 | 0.93 | 0.71 | 0.92 | 0.40 | 0.92 | 0.052 |
| 6. Simple SMARTS patterns | 0.93 | 0.84 | 0.92 | 0.68 | 0.90 | 0.36 | 0.93 | 0.42 |
| 7. Complex SMARTS patterns | 0.84 | 0.75 | 0.78 | 0.62 | 0.60 | 0.27 | 0.67 | 0.33 |
Discussion
We have examined several examples of the existing software programs that can be utilized for linking the databases of small molecules with the relevant scientific research articles, by matching the chemical structure diagrams in the articles with the structures in the database. In their current states, these programs have limitations to the extent that they generally need manual feeding of images and they have significant error rates. As an alternative, we have developed ChemReader – a machine-vision-based software program designed for the development of customized chemical diagram extraction tools in industry or academic laboratories. Compared to commercially or publicly available software, ChemReader function is transparent, in the sense that algorithm performance can be followed step-by-step. In side-by-side comparison with Kekule, CLiDE and OSRA, ChemReader makes more correct outputs and extracts chemically important substructure patterns with higher recall and precision rates.
To develop ChemReader into a fully-automated system, there are still several challenges that remain to be addressed. For automated extraction of chemical structures and relevant information from scientific articles, it would be important to rapidly distinguish between a diagram of a chemical structure and a non-chemical structure diagram, or a photograph, among the extracted images. Gkoutos et al. have reported a method to classify chemical images based on the use of the Kohonen network [18] with promising results. Such functionality still has to be incorporated into ChemReader. Finally, since the translation of chemical structure from a raster image to a standard chemical file format is highly error prone as seen in the test, output structures should be thoroughly inspected before utilization. Besides manual curation resulting in high cost of system operation, filtering method which can detect "unreadable" images or wrong outputs and filtered them out at the pre-processing or post-processing stages might be effective to improve the performance of machine vision systems for recognizing chemical structures. In this manner, accuracy can be increased at the expense of throughput. However, since ChemReader is already able to correctly recognize far more images than OSRA, CLiDE or Kekule, this may be a viable course of action for the future of ChemReader's development.
We postulate that, in its current state, ChemReader may be sufficiently accurate for annotating chemical-structure databases with links to scientific research articles. An error at the level of chemical-structure recognition does not necessarily lead to an error in the annotation, since incorrectly extracted molecules may not find a match in the chemical-structure databases. Furthermore, a useful database annotation scheme does not necessarily require perfect matches between database entries and scientific articles. In fact, the ability to link to similar but not identical structures may be important when the intent is to synthesize drug leads that are not identical to the molecule in question, and to identify related compounds in the scientific literature. Since not every chemical database entry may be represented as chemical-structure diagrams in published research articles, the ability to link to similar but not identical molecules may also be useful to increase the number of links between database entries and research articles.
Conclusion
The availability of ChemReader as a cheminformatic tool would allow research and development groups to enrich their knowledge bank of molecules and chemical structures. We are planning that ChemReader becomes commercially available in the near future, with removal of open source parts such as GOCR and Greycstoration. Like ChemReader, other image-based search engines are being developed in other academic disciplines. In mechanical engineering, for example, search engines are being developed for searching catalogues of three dimensional components for mechanical products. Compared to other image-based search engines, image-based cheminformatic search engines are simpler because chemical structures are two dimensional objects with well-defined connectivity patterns (grammars) determined by the atoms and their valences. Indeed, chemical-structure recognition algorithms may be most akin to character- and text-recognition algorithms. Like words in a dictionary, a chemical-structure database can serve as a training set of molecules that can be used to identify the most common chemical substructures present in all known chemical compounds. Based on the frequency of different substructures and using neighboring substructure information, computational techniques borrowed from statistical linguistics may be incorporated to generalize the chemical spellchecker to check structural "spelling", which will further optimize ChemReader's performance.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JP developed most recent versions of ChemReader software, conducted comparison tests, and drafted the manuscript. GRR conceived of the study, has provided and evaluated the key requirements of ChemReader, and revised the draft manuscript. KAS conceived of the study, and has provided and evaluated the key requirements of ChemReader. MN collected and labeled the sample images for the comparison tests and participated in the key development of ChemReader. NL wrote the initial version of ChemReader and conducted the preliminary testing. KS conceived of the study, has participated in the algorithmic development of ChemReader, and revised the draft manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by NIH grants P20-HG003890 to Kerby Shedden and Gus R. Rosania.
Contributor Information
Jungkap Park, Email: jungkap@umich.edu.
Gus R Rosania, Email: grosania@umich.edu.
Kerby A Shedden, Email: kshedden@umich.edu.
Mandee Nguyen, Email: nguyenmm@umich.edu.
Naesung Lyu, Email: naesunglyu@gmail.com.
Kazuhiro Saitou, Email: kazu@umich.edu.
References
- PubMed http://www.ncbi.nlm.nih.gov/entrez/query/static/overview.html#Introduction
- PubChem http://pubchem.ncbi.nlm.nih.gov/help.html#PubChem_Overview
- ChemDraw http://www.cambridgesoft.com/software/ChemDraw/
- ISIS/Draw http://www.symyx.com/products/software/decision-support/isis-draw/index.jsp
- DrawIt http://www.chemwindow.com
- ACD/ChemSketch http://www.acdlabs.com/products/chem_dsn_lab/chemsketch/
- McDaniel JR, Balmuth JR. Kekule: OCR – Optical Chemical (Structure) Recognition. J Chem Inf Comput Sci. 1992;32:373–378. [Google Scholar]
- Casey R, Boyer S, Healey P, Miller A, Oudot B, Zilles K. Proceedings of the Second International Conference on Document Analysis and Recognition: 20–22 October 1993. Tsukuba, Japan; 1993. Optical Recognition of Chemical Graphics; pp. 627–632. [Google Scholar]
- Ibison P, Jacquot M, Kam F, Neville AG, Simpson RW, Tonnelier C, Venczel T, Johnson AP. Chemical Literature Data Extraction: The CLiDE Project. J Chem Inf Comput Sci. 1993;33:338–334. [Google Scholar]
- Rosania GR, Crippen G, Woolf P, States D, Shedden K. A Cheminformatic Toolkit for Mining Biomedical Knowledge. Pharmaceutical Research. 2007;24:1791–1802. doi: 10.1007/s11095-007-9285-5. [DOI] [PubMed] [Google Scholar]
- Algorri ME, Zimmermann M, Friedrich CM, Akle S, Hofmann-Apititus M. Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBS): 23–26 August 2007. Lyon, France; 2007. Reconstruction of Chemical Molecules from Images; pp. 4609–4612. [DOI] [PubMed] [Google Scholar]
- OSRA: Optical Structure Recognition http://cactus.nci.nih.gov/osra/
- Snyder WE, Qi H. Machine Vision. New York: Cambridge University Press; 2004. [Google Scholar]
- Dori D, Wenyin L. Automated CAD Conversion with the Machine Drawing Understanding System: Concepts, Algorithms, and Performance. IEEE Transactions on Systems, Man and Cybernetics. 1999;29:411–416. [Google Scholar]
- Fahn CS, Wang JF, Lee JY. A Topology-Based Component Extractor for Understanding Electronic Circuit Diagrmas. Computer Vision, Graphics, Image Process. 1988;44:119–138. [Google Scholar]
- Richard OD, Peter EH. Use of the Hough transformation to detect lines and curves in pictures. Communications of the ACM. 1972;15:11–15. [Google Scholar]
- Boyer SK, Casey RG, Miller AM, Oudot B, Zilles KS. Apparatus and method for optical recognition of chemical graphics. US Patent No 5,157,736. 1992.
- Gkoutos GV, Rzepa H, Clark RM, Adjei O, Johal H. Chemical Machine Vision: Automated Extraction of Chemical Metadata from Raster Image. J Chem Inf Comput Sci. 2003;43:1342–1355. doi: 10.1021/ci034017n. [DOI] [PubMed] [Google Scholar]
- GREYCstoration: open source algorithms for image denoising and interpolation http://cimg.sourceforge.net/greycstoration/
- Tschumperle D. Fast Anisotropic Smoothing of Multi-Valued Images using Curvature-Preserving PDE's, International Journal of Computer Vision. International Journal of Computer Vision. 2006;68:65–82. [Google Scholar]
- Fletcher LA, Kasturi R. A Robust Algorithm for Text String Separation from Mixed Text/Graphics Images. IEEE Trans on Pattern Analysis and Machine Intelligence. 1998;10:910–918. [Google Scholar]
- Tombre K, Tabbone S, Pelissier L, Lamiroy B, Dosch P. Text/Graphics Separation Revisited. Proceedings of 5th International Workshop on Document Analysis Systems: 19–21 August 2002; Princeton. 2002. pp. 200–211.
- MCK Yang, Lee JS, Lien CC, Huang CL. Hough Transform Modified by Line Connectivity and Line Thickness. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1997;19:905–910. [Google Scholar]
- Sojka E. Proceedings of the 18th Spring Conference on Computer Graphics: 24–27 April 2002; Budmerice, Slovakia. Alan Chalmers: ACM; 2002. A New Algorithm for Detecting Corners in Digital Images; pp. 55–62. [Google Scholar]
- Ballard DH. Generalizing the Hough transform to detect arbitrary shapes. Pattern Recognition. 1981;13:111–122. [Google Scholar]
- GOCR Open source character recognition http://jocr.sourceforge.net/
- Dalby A, Nourse JG, Hounshell D, Gushurst AKI, Grier DL, Leland BA, Laufer J. Description of Several Chemical Structure File Formats Used by Computer Programs Developed at Molecuar Design Limited. J Chem Inf Comput Sci. 1992;32:244–255. [Google Scholar]
- Weininger D. SMILES, a Chemical Language and Information System. 1. Introduction to Methodology and Encoding Rules. J Chem Inf Comput Sci. 1988;28:31–36. [Google Scholar]
- Introducing CliDE Pro, Fall 2008 ACS National Meeting & Exposition, August 17th–21th, Philadelphia, USA http://www.simbiosys.ca/science/presentations/2008-acs-08/ACS_CLiDEPro.ppt
- GLIDA: GPCR-Ligand Database http://pharminfo.pharm.kyoto-u.ac.jp/services/glida/
- PubChem Substructure fingerprint ftp://ftp.ncbi.nlm.nih.gov/pubchem/specifications/pubchem_fingerprints.txt
- JChem, ChemAxon Ltd http://www.chemaxon.com/
- PubChem Fingerprint for JChem, NIH Chemical Genomics Center http://www.ncgc.nih.gov/pub/openhts/