Summary
The SOS response to DNA damage in E. coli involves at least 43 genes, all under the control of the LexA repressor. Activation of these genes occurs when the LexA repressor cleaves itself, a reaction catalyzed by an active, extended RecA filament formed on DNA. It has been shown that the LexA repressor binds within the deep groove of this nucleoprotein filament, and presumably cleavage occurs in this groove. Bacteriophages, such as, have repressors (cI) that are structural homologs of LexA and also undergo self-cleavage when SOS is induced. It has been puzzling that some mutations in RecA that affect the cleavage of repressors are in the C-terminal domain (CTD) far from the groove where cleavage is thought to occur. In addition, it has been shown that the rate of cleavage of cI by RecA is dependent upon both the substrate on which RecA is polymerized and the ATP analog used. Electron microscopy and three-dimensional reconstructions show that the conformation and dynamics of RecA’s CTD are also modulated by the polynucleotide substrate and ATP analog. Under conditions where the repressor cleavage rates are the highest, cI is coordinated within the groove by contacts with RecA’s CTD. These observations provide a framework for understanding previous genetic and biochemical observations.
Introduction
RecA, a key enzyme in homologous recombination, performs many cellular roles in bacteria1. RecA plays a fundamental role coordinating a comprehensive defense mechanism in Escherichia coli, referred to as the SOS response2. The SOS response is triggered by conditions which cause massive DNA damage and halt DNA replication in E. coli. This process has been known to promote cell survival by stimulating DNA repair via recombination, excision repair and mutagenesis, and by inhibition of cell growth3–5. RecA localizes to damaged chromosomal sites, where it facilitates DNA repair by means of homologous recombination6. It has recently been shown that RecA plays several roles in mutagenic translesion DNA synthesis, acting in concert with an error-prone DNA polymerase7.
The LexA protein functions as a repressor for a set of about 43 genes involved in the SOS response, including both recA and lexA8, 9. SOS is initiated when RecA is activated by an inducing signal in the form of a single-stranded DNA (ssDNA), and the ssDNA appears when DNA replication reaches a lesion10. The activated RecA-ssDNA-ATP complex acts as a co-protease to catalyze the self-cleavage of LexA at a specific site11, 12. While formation of the active RecA-DNA nucleoprotein complex requires ATP or an ATP analog, the self-cleavage reaction does not require ATP hydrolysis13. Cleaved LexA loses its repressor function, which results in the increased expression of SOS genes14.
LexA is not the only protein that undergoes self-cleavage upon binding to the active RecA filament. The other known LexA functional homologues are E. coli UmuD mutagenesis protein15–17, which is part of a heterotrimeric E. coli polymerase V (UmuD′2C) complex18, and several phage repressors, such as c2 repressor of P2219 and cI repressor of φ8020 or bacteriophage λ cI repressor21.
The λ cI repressor is a part of a phage molecular switch which transforms the phage from lysogenic to lytic growth. The λ cI mimics E. coli LexA, and senses the physiological condition of a host cell. Under SOS activation cI undergoes self-cleavage, catalyzed by the active RecA nucleoprotein filament. The cI protein contains two domains connected by a flexible “hinge” region which contains the cleavage site22, 23. The repressor’s self-cleavage allows the prophage to efficiently shift from lysogeny (when the majority of phage genes are repressed) to induction and lytic growth, at a time when the survival of a host cell is in question24. The RecA filament is believed to stabilize one particular conformation of λ cI. This is believed to facilitate the proper orientation of catalytic residues in the cI C-terminal domain with respect to the cleavage site in the hinge region23, 25, 26.
Three-dimensional reconstructions of electron micrographs of RecA complexed with either LexA27, 28 or UmuD29 show that both substrates bind deep within the RecA helical groove. Structural information about the intermolecular interactions in the RecA-cI complex, however, has been absent until now. The functional and structural similarities between the LexA, UmuD and cI λ proteins have led to a model in which all three proteins bind and undergo self-cleavage in the filament’s helical groove28, 29. However, biochemical and genetic studies have yielded a much more complicated and difficult to understand picture. RecA mutations producing a cleavage-deficient phenotype are located not only in the groove, where cleavage takes place, but also on the outer surfaces of the filament30. These results suggest that either there are large allosteric effects in RecA or that binding of the repressors to RecA is more complicated than indicated by previous EM studies that showed density only in the groove. We have therefore used electron microscopy and three-dimensional reconstruction to directly visualize the binding of the cI repressor to RecA filaments. We show that while the binding of the repressor is within the groove, the RecA CTD plays an important role in coordinating the cleavage of this repressor.
Results
The cI repressor contains an N-terminal DNA-binding domain (residues 1–92) that is not needed for either binding to RecA or for RecA-mediated cleavage of cI. In fact, removal of the N-terminal domain has been shown to accelerate RecA-mediated cleavage by the destabilization of cI dimer formation, since it is the monomeric form of cI that has been shown to undergo RecA-mediated cleavage31. The RecA-mediated cleavage of cI93–236 was determined in the presence of two different ATP analogs (non-hydrolyzable AMP-PNP and slowly hydrolyzed ATP-γ-S) and two different polynucleotide substrates (a 48-mer, (GTG)16, and a naturally occurring E. coli sequence 83-mer32) for RecA polymerization (Figure 1). With ATP-γ-S as a cofactor, the cleavage rate is significantly higher for the GTG 48-mer than it is for the naturally occurring 83-mer. These particular polynucleotide substrates were chosen as they have been used for the EM studies (below). It has previously been shown33 that cleavage of the cI fragment is faster on GTG-repeating than on all other triplet-repeating sequences, for both 15-mers and 48-mers. Cleavage was faster on 15-mers than on 48-mers, but the sequence-dependence remained the same. The use of the repeating GTG sequence33 comes from in vitro selection studies34 which sought to determine sequence preferences for RecA binding and pairing. The triplet-repeating nature of the sequence is due to the fact that in the nucleoprotein filament, one protomer of RecA is bound to three nucleotides of ssDNA. While the preference for GTG repeats is still not fully understood, the recent crystal structure of RecA bound to DNA shows that the DNA in this complex has a triplet structure with large extensions occurring between triplets35. Since every base is not equivalent in this complex, this would explain why when RecA is bound to oligonucleotides containing the Chi sequence (5′-GCTGGTGG-3′), the RecA protomers bind in a phased manner relative to the Chi sequence36. Since secondary structure formation of the ssDNA actually competes with RecA binding, the fact that GTG-repeating sequences give particularly tight binding to RecA and high levels of coprotease activity33 provides evidence that the GTG sequences are not forming unusual secondary structures.
Figure 1.
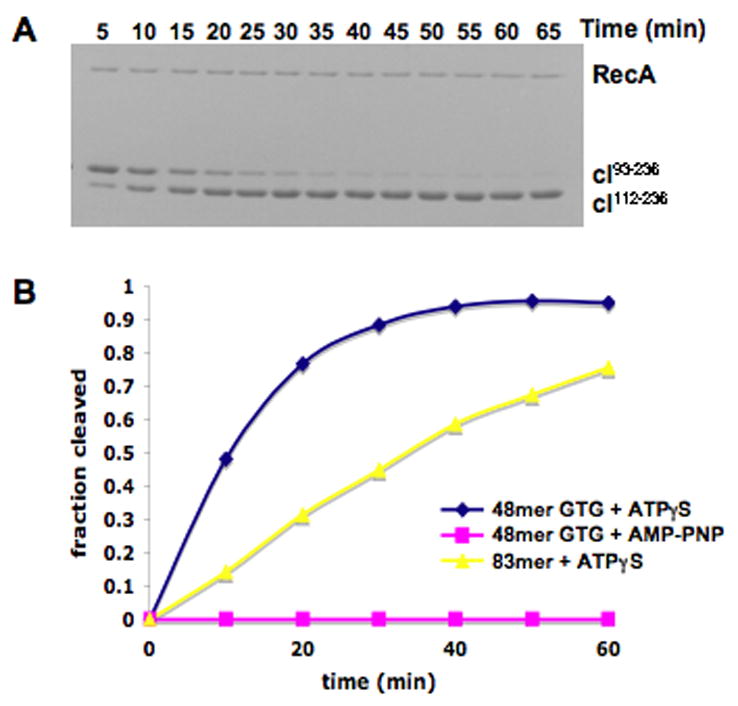
RecA mediated cleavage of lambda repressor. (A) SDS-PAGE gel showing the RecA-mediated cleavage of cI93–236 at the peptide bond between Ala111-Gly112 to form cI112–236. The reaction was performed in the presence of a 48-mer GTG repeating single stranded oligonucleotide and ATP-γ-S. (B) Comparison of cleavage rates in the presence of different ATP analogs (ATP-γ-S vs. AMP-PNP) and different oligonucleotide sequences (GTG-repeating 48-mer vs. 83-mer with a naturally occurring sequence). The fraction of repressor cleaved at each time point was determined as described in Methods.
The inability of RecA to cleave cI in the presence of AMP-PNP (Fig. 1) was not due to the absence of RecA filament formation, since short filaments are formed under these conditions as observed by EM (Fig. 3b). We can further exclude the possibility that AMP-PNP fails to induce the extended, active state of the RecA filament needed for LexA cleavage23, 37, since filaments formed on both ssDNA and dsDNA are extended in the presence of AMP-PNP27. When long ssDNA substrates are used, RecA filaments formed in the presence of AMP-PNP are also long (Fig. 2b), so we can eliminate the possibility that RecA does not polymerize in a cooperative manner in the presence of AMP-PNP.
Figure 3.
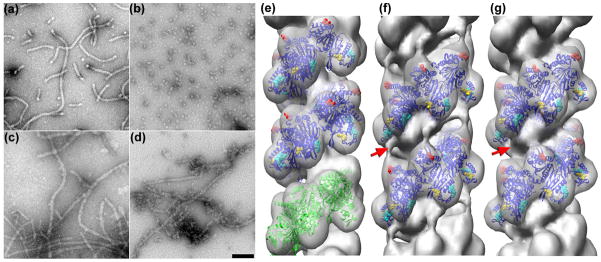
The binding of a non-cleavable cI fragment to RecA filaments. Electron micrographs of RecA polymerized on a repeating GTG sequence 48-mer in the presence of ATP-γ-S (a) show clear end-to-end stacking of the oligomers, as the filaments are much longer than the ~ 2.5 helical turns expected in the absence of stacking. When RecA is polymerized on this same 48-mer in the presence of AMP-PNP (b) very little end-to-end stacking is seen. The non-cleavable cI101–229TM fragment has been incubated with the RecA filaments formed under conditions in (a) and (b), and the resulting images are shown in (c) and (d), respectively. It can be seen that when the cI fragment is present (d) a substantial stacking of the oligomers takes place that is not found in the absence of cI (b). A three-dimensional reconsruction (e) of RecA polymerized on the 48-mer GTG sequence in the presence of ATP-γ-S (a), generated from 8,179 segments. An atomic model27 of the active RecA filament is shown in the blue ribbons, while three subunits from a crystal structure of the active RecA-DNA filament35 are shown in green ribbons. The RecA residues shown to be involved in cI cleavage in vivo30 are marked as follows: E4 and K8 in red, K280, E281 and K282 in cyan, and K310 and D311 in gold. Residues 280,281, 282, 310 and 311 are located in RecA’s CTD. A reconstruction (f) of the filaments in (c), RecA-(48-mer GTG)-ATP-γ-S + cI101–229TM, shows the bridge of density spanning the space between the CTD of one RecA protomer and the core of another RecA protomer one turn below (f, red arrow). A reconstruction (g) from the filaments shown in (d), RecA-(48-mer GTG)-AMP-PNP + cI101–229TM, shows the cI fragment density in the groove, but no bridge of density (g, red arrow).
Figure 2.
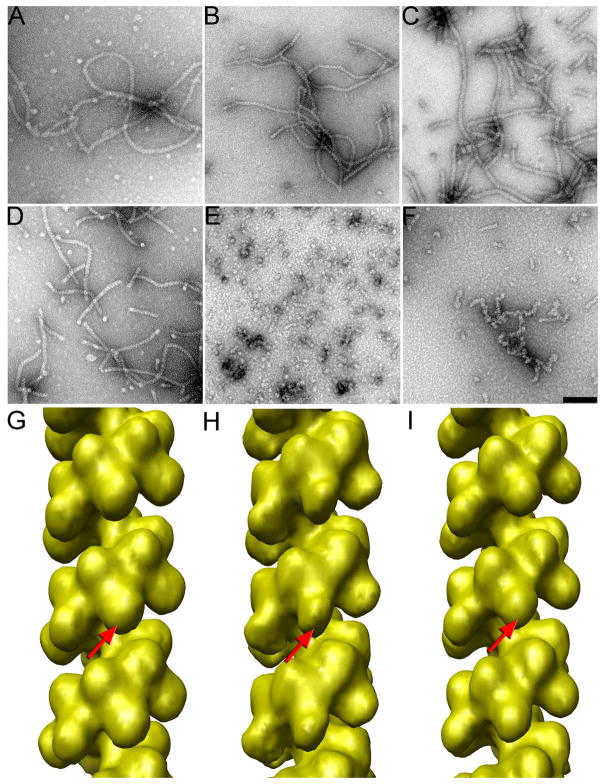
Properties of pure RecA filaments depend upon both nucleotide cofactor and polynucleotide substrate. Electron micrographs of negatively stained RecA-DNA preparations (a-f), and three-dimensional reconstructions (g-i). RecA has been polymerized on M13 ssDNA with ATP-γ-S (a) or with AMP-PNP (b). RecA has been polymerized with ATP-γ-S on long poly(dT) in (c). RecA polymerization on a random sequence 40-mer in the presence of ATP-γ-S (d) leads to a stacking of the short oligomers, as the filaments are much longer than the ~ two helical turns expected for filaments containing a 40-mer. However, when this same 40-mer is used for RecA polymerization with AMP-PNP (e), very little end-to-end stacking is observed. Similarly, when a 48-mer of poly(dT) is used with ATP-γ-S for RecA polymerization (f), poor end-to-end stacking is observed in contrast to the stacking observed for the random sequence 40-mer (d). The scale bar (f) is 1,000 Å, and applies also to (a-e). A three-dimensional reconstruction (g, from 6,228 segments) from the RecA-M13ssDNA-ATP-γ-S filaments (a) shows nearly the full density expected for the CTD (g, red arrow). In contrast, a three-dimensional reconstruction (h, from 3,482 segments) from the RecA-M13ssDNA-AMP-PNP filaments (b) shows significantly less density for the CTD (h, red arrow), consistent with disorder or structural heterogeneity of this domain under these conditions. A three-dimensional reconstruction (i, from 9,115 segments) from the RecA-poly(dT)-ATP-γ-S filaments (c) also shows less density due to the CTD (i, red arrow).
RecA filaments change with cofactors and substrates
To address why cI RecA-mediated cleavage rates might depend upon both the polynucleotide substrate and nucleotide cofactor, we have used EM to look at the filaments formed by RecA under these different conditions in the absence of cI (Fig. 2). When RecA is polymerized on long random sequence ssDNA in the presence of either ATP-γ-S (Fig. 2a) or AMP-PNP (Fig. 2b) very similar filaments are formed, as judged by casual inspection of micrographs. However, a three-dimensional reconstruction of the ATP-γ-S filaments (Fig. 2g) shows a more massive CTD when compared to the AMP-PNP reconstruction (Fig. 2h). The significance of this difference was determined by cross-correlation sorting of the image segments from both conditions (ATP-γ-S and AMP-PNP) against reference volumes having either a full CTD or one with half the mass of the CTD, and a third that was in between (data not shown). It was found that with ATP-γ-S most segments correlated with the reference having the full CTD, while with AMP-PNP most segments had the best correlation with the volume having half of the expected CTD. Reconstructions from these subsets, always starting with a featureless solid cylinder as an initial reference, confirmed that the sorting was valid. The less mass seen for the CTD in the presence of AMP-PNP must arise from disorder, and motions of the CTD are consistent with what has previously been observed by both EM27 and x-ray crystallography38.
The different effects of the two nucleotide cofactors can be clearly seen when a random sequence 40-mer is used for RecA polymerization with ATP-γ-S (Fig. 2d) or with AMP-PNP (Fig. 2e). Since the stoichiometry of binding is three bases per RecA35, a RecA filament covering the 40-mer should only contain 13 RecA subunits and be ~ two turns long (where each turn is ~ 90–95 Å). Electron micrographs of the RecA polymerized with AMP-PNP do show many such short filaments, and a few that may be several times this length (Fig. 2e). In contrast, RecA incubated with the same 40-mers in the presence of ATP-γ-S forms filaments that are many times this expected length (Fig. 2d), consistent with earlier observations of a “stacking” of short oligonucleotides into long filaments by RecA39. The ability of RecA to stack such short oligomers is a function of both the nucleotide cofactor and the sequence of the polynucleotide, since in the presence of ATP-γ-S and a 48-mer poly(dT) long filaments are rarely found (Fig. 2f). We cannot do three-dimensional reconstruction of such short filaments, so we have used RecA polymerized on long poly(dT) in the presence of ATP-γ-S (Fig. 2c) for such analysis. The poly(dT) that has been used contains a broad distribution of lengths, but a significant fraction is more than 1,000 nt in length. A three-dimensional reconstruction (Fig. 2i) from filaments formed on such poly(dT) shows less mass for the CTD than seen when RecA is polymerized on random sequence ssDNA with the same nucleotide (Fig. 2g). Interestingly, it has been observed33 that RecA-mediated cleavage of cI is also reduced when the RecA is polymerized on poly(dT) when compared to poly(GTG). The observations just described suggest that both the ability of short RecA polymers to stack with each other, and the rates of cleavage of cI, may be a function of the structure and dynamics of RecA’s CTD. To test this, we have looked at complexes of RecA filaments with the cI repressor.
Binding of cI to RecA filaments
Since the cI repressor is cleaved by RecA, we used the non-cleavable cI fragment26 cI101–229TM (where TM is Triple Mutant) to form stable complexes with RecA that could be examined by EM. The A152T and P158T mutations in this construct disrupt repressor dimer formation and thereby enhance RecA-binding and self-cleavage31, whereas the K192A mutation further enhances RecA-binding but completely prevents cleavage26. These mutations result in a cI repressor molecule with optimal RecA-binding properties for structural studies. Similar EM results were obtained with the cI93–229TM fragment (data not shown), consistent with the finding that residues 93–100 are not important for binding RecA26. The binding of cI101–229TM to RecA can be seen in a solution assay by its inhibition of cI93–236 cleavage26. Interestingly, when the non-cleavable fragment is present at a stoichiometry of ~ 1:2 with RecA, it provides nearly complete inhibition of RecA’s ability to cleave cI93–236. Previous studies of the binding of a non-cleavable LexA repressor27, 28 and the UmuD′2C complex29 to RecA filaments also found a saturating stoichiometry of 1:2, suggesting that these molecules may not be able to bind to the RecA filament in a 1:1 stoichiometry.
Two different conditions were used for forming RecA-DNA filaments that were complexed with cI101–229TM for analysis by EM. These were RecA polymerized on the 48-mer (GTG)16 in the presence of ATP-γ-S (Fig. 3c), and RecA polymerized on the same 48-mer but with AMP-PNP (Fig. 3d). As shown in Fig. 1, rapid cleavage of cI occurs with ATP-γ-S, but almost no cleavage of cI occurs with AMP-PNP under these conditions. As discussed above, RecA filaments would be expected to have only 16 RecA subunits in the absence of stacking, and would be expected to be ~ 240 Å long, with only about 2.5 helical turns. However, in the absence of cI substantially longer filaments are seen when ATP-γ-S is present (Fig. 3a), showing that short filaments must stack together. In contrast, when AMP-PNP is present very little stacking is observed in the absence of cI (Fig. 3b). When cI101–229TM is added to these AMP-PNP filaments (Fig. 3d), substantially longer filaments are seen, suggesting that cI101–229TM helps “glue” together the shorter filaments formed with AMP-PNP. The mechanism for this stacking of short filaments might involve two adjacent cI molecules interacting with each other, cI bridging two adjacent RecA protomers, or the binding of cI inducing a conformational change in RecA that favors annealing. The addition of cI101–229TM (Fig. 3c,d) also results in filaments having less modulation arising from the deep groove of the RecA helix, which appears similar to what has been observed for the binding of the LexA repressor to RecA27, 28.
A control three-dimensional reconstruction was made from the pure RecA on the 48-mer in the presence of ATP-γ-S using 8,179 segments, where each segment was ~ 420 Å long (Fig. 3e). Another reconstruction was made under the same conditions, but in the presence of cI101–229TM, using 9,699 segments (Fig. 3f). The decorated segments were observed to be more heterogeneous than the control pure RecA segments, so classification methods were used to generate more homogeneous subsets by using multiple reference volumes for sorting. Since some of the decorated filaments were expected to have poor occupancy, one of the reference volumes was naked RecA (Fig. 3e). Two other reference volumes had additional mass within the deep helical groove, or on the outside of the RecA filament. Only 3% of the decorated segments showed the highest correlation with the naked RecA reference, while 62% of the segments showed the highest correlation with RecA having additional mass in the groove. The 35% of the segments showing the highest correlation with RecA having additional mass bound to the outside of the filament were judged to be extremely heterogeneous and failed to converge to any reasonable three-dimensional reconstructions. We expect that this class was generated by variable binding of the cI fragment to the outside of the filaments. On the other hand, the class sorted as having additional mass within the helical groove (6,003 segments) was relatively homogeneous and yielded the reconstruction shown in Fig. 3f.
A similar approach to sorting and classification was used for the 5,874 segments extracted from filaments (Fig. 3d) formed on the GTG repeating 48-mer in the presence of AMP-PNP and the non-cleavable cI fragment. The occupancy by the cI fragment appeared lower than with ATP-γ-S, as 43% of the segments were classified as most similar to naked RecA, and 38% of the segments were sorted as having mass bound within the deep groove. As with the filaments formed with ATP-γ-S, the class sorted as having additional mass bound to the outside of the filament (19%) were judged to be very heterogeneous and did not yield any reasonable three-dimensional reconstruction. The reconstruction from the segments classified as having additional mass in the groove (2,232 segments) is shown in Fig. 3g. A comparison between the two volumes having the bound non-cleavable cI fragment shows that in the presence of ATP-γ-S (where the cleavable fragment is readily cleaved) there is a bridge of density (Fig. 3f, red arrow) extending between the lower portion of the CTD of one RecA protomer and the upper portion of the core of a RecA subunit one turn below. In the presence of AMP-PNP (where the cleavable fragment is not cleaved by RecA), this bridge of density is absent (Fig. 3g, red arrow). The absence of the bridge of density in the presence of AMP-PNP cannot be explained by a lower occupancy of cI in the RecA filament, since the mass in the grove that corresponds to cI is very similar to that in the presence of ATP-γ-S. We have tested the significance of this feature by doing statistical difference maps (data not shown). By comparing the reconstructions with AMP-PNP and with ATP-γ-S we have found that this feature has a value ~ six times the standard error of the difference map, and is highly significant and reproducible.
This suggests that the bridge of density, possibly formed by the non-cleavable cI fragment, represents a coordination of this fragment by RecA that occurs under conditions where the cleavable fragment is readily cleaved (in the presence of ATP-γ-S) and absent under conditions where very little cleavage occurs (in the presence of AMP-PNP). Since the cI crystal structure does not readily suggest how this density might arise from cI, there must be a significant conformational change in cI if this density arises from cI. An alternative explanation is that this density belongs to the ~ 25 C-terminal RecA residues that are not seen in most RecA crystal structures and have only recently been visualized40. In this interpretation, these residues would become ordered by cI under conditions where cleavage is favored (with ATP-γ-S). However, the C-terminal domain would probably need to rotate to bring these terminal residues into the correct orientation. Additionally, this bridge could arise from a conformational change in RecA itself, either the C-terminal domain of one subunit or the core of a subunit one turn below. The present resolution is simply too limited to be able to distinguish among these possibilities. In all cases (the density is due to cI or RecA), we suggest that the presence of this bridge is important to cleavage. To test this hypothesis, we have looked at a RecA double mutant that is defective in cI cleavage in vivo30.
The RecA E4A-K8A double mutant
A RecA mutagenesis study30 identified several patches of residues within RecA whose mutation led to complete deficiency in cI cleavage at the same time that RecA recombination and repair functions were not impaired (mutations that eliminate RecA function, such as by preventing polymerization, would obviously be deficient in cI cleavage). When these residues are mapped onto the pseudo-atomic model of RecA (Fig. 3e), two patches are on the CTD (K280, E281, K282 and K310, D311) while the third patch (E4, K8) is on the N-terminal portion of RecA, located on the opposite side of the helical groove. A fourth site (K232, E235) is in a cleft between RecA subunits and faces into the deep groove (not marked in Fig. 3e). The repressor bound within the groove would be contacting this site (Fig. 3f,g). RecA residues E4 and K8 directly face the bridge of density that is present with ATP-γ-S (Fig. 3f, red spheres) and absent with AMP-PNP, so we have looked at properties of the double mutant E4A-K8A.
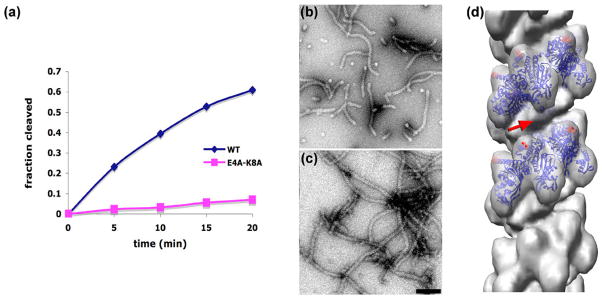
Cleavage rates of cI are dramatically reduced for the RecA-E4A-K8A mutant when compared to wt RecA (Fig. 4a). As expected from the in vivo results30, where this double mutation did not lead to any loss of recombination activity or impairment of DNA repair, the RecA-E4A-K8A formed filaments on the repeating GTG 48-mer in the presence of ATP-γ-S (Fig. 4b) that appeared indistinguishable from the wt RecA protein under these same conditions (Fig. 3a). The existence of these filaments also eliminates the trivial explanation that the loss of cI cleavage (Fig. 4a) might be due to the loss of RecA’s ability to form normal filaments. In addition, our preparation of E4A-K8A exhibits ssDNA-dependent ATPase activity comparable to wild type RecA (data not shown). The addition of cI101–229TM to these RecA filaments (Fig. 4c) does not lead to anything very different than observed when this same fragment is added to wt RecA filaments (Fig. 3c). A three-dimensional reconstruction (Fig. 4d) was generated after using a similar approach to sorting as described for the wt RecA. From the 7,770 segments cut from images of RecA-E4A-K8A filaments incubated with cI101–229TM, 51% were classified as having additional mass in the groove, while 26% were determined to be poorly occupied. The remaining 23%, classified as having extra density on the outside of the filaments, were determined to be very heterogeneous. The 3,980 segments classified as having additional density in the groove gave rise to a three-dimensional reconstruction (Fig. 4d) where the bridge of density present in the corresponding wt RecA reconstruction (Fig. 3f) is now absent. Since this bridge maps onto the region on the top of the RecA core where residues E4 and K8 are located, we interpret these results as showing that the E4A-K8A double mutation allows cI to still bind in the deep groove of the RecA-DNA filament, but fails to provide the proper coordination of cI that results in the observed bridge of density that spans the RecA groove. It is this full coordination of cI by RecA that is needed for efficient cleavage, as we also fail to see this bridge of density when filaments are formed with AMP-PNP (Fig. 3g), a nucleotide cofactor that does not support significant cI cleavage (Fig. 1).
Figure 4.
RecA residues E4 and K8 are directly involved in the coordination and cleavage of cI. The rate of cI93–236 cleavage is greatly reduced when the RecA-E4A-K8A mutant is used in comparison to wt RecA (a). The repeating GTG 48-mer has been used along with ATP-γ-S, as in Fig. 1. An electron micrograph of RecA-E4A-K8A polymerized on the repeating GTG 48-mer (b) shows a large degree of end-to-end stacking of these 48-mers, as the filaments are considerably longer than the ~ 2.5 helical turns that would be expected in the absence of stacking. When the non-cleavable cI101–229TM fragment is added to these RecA-E4A-K8A filaments (c) there are no dramatic differences. A three-dimensional reconstruction (d), generated from 3,980 segments of filaments as shown in (c), lacks the bridge of density that is seen with the wt RecA protein under the same conditions when the cI101–229TM fragment is bound (red arrow, Fig. 3f). Residues E4 and K8 are marked as red spheres in (d). The scale bar (c) is 1,000 Å.
Discussion
According to existing models of RecA-mediated cleavage of LexA27, 28, 37 and UmuD29, cleavage of these proteins occurs when they are stabilized in a particular conformation by binding within the helical groove of an active RecA-DNA filament. Surprisingly, alanine mutagenesis of RecA to identify regions of RecA involved in repressor cleavage mapped some of these sites outside the groove, with no evident relationship to structural studies showing binding within the groove30. Our structural and biochemical results provide an explanation for the puzzling mutagenesis data, at the same time as providing new insights into the role of RecA’s dynamic CTD.
While RecA and RadA/RAD51 proteins share highly conserved structural cores, the RecA protein CTD and the RadA/RAD51 N-terminal domain have no structural homology with each other. But they both display a large degree of dynamic behavior. It has been shown41 that the RadA/RAD51 N-terminal domain can move over distances greater than 20 Å, and a similar degree of mobility has been described for the RecA CTD27. Different crystal structures for the RecA protein have shown that the most variable part of the molecule is the CTD42, and rotations of the CTD by as much as 32° exist between different crystals38. The mobility of the CTD depends on both the ATP analog and the polynucleotide substrate on which the filament is formed (Fig 2). The three-dimensional reconstructions of the RecA filament show that the CTD lobe is bridging between two adjacent RecA protomers (Fig. 3e). Thus, the ordering of the CTD in the presence of ATP-γ-S provides a structural framework to explain the stacking of short filaments under these conditions, as the CTD is likely establishing an important contact between protomers that is part of the annealing mechanism in bringing short filaments together.
According to a pseudo-atomic model of the RecA filament built into our three-dimensional reconstructions (Fig. 3e–g), residues 280–282, and 310–311 do not interact directly with the cI repressor under conditions where it gets cleaved rapidly (Fig. 3f) or poorly (Fig. 3g). It is possible that these mutations alter the mobility of the CTD and thus allosterically influence the ability of the CTD to coordinate the cI repressor bound to the RecA filament. Alternatively, when the repressor is bound to the RecA filament, the CTD may be rotated so that these residues are directly involved in the direct interaction with the cI. Such a movement of the CTD is consistent with large displacements of the CTD that can be affected by other RecA-binding proteins (Wang et al., in preparation). The limited resolution in our maps does not allow us to choose between these two explanations.
Our results show a correlation between the degree of disorder of the CTD in the absence of cI, and the efficiency of repressor cleavage. In RecA-ssDNA filaments formed in the presence of AMP-PNP, the CTD is more disordered, and there is no cleavage of cI. Interestingly, a similar relationship has previously been reported43 for RecA filaments formed with ATP-γ-S, but in the presence of dsDNA instead of ssDNA. In early work, it was observed that the cleavage of cI is dramatically reduced when dsDNA is used instead of ssDNA as the polynucleotide cofactor13. This has been attributed to the notion that the second strand of the duplex, which is near the central axis of the filament, must overlap with the repressor binding site. Our results offer an alternative or additional explanation. In previous work43, we showed that RecA filaments formed in the presence of dsDNA have increased mobility of the CTD. Thus, the lack of robust cleavage activity in the presence of dsDNA could be due to the increased mobility of the CTD, which prevents it from forming the proper coordination with the repressor.
Materials and Methods
ADP, ATP-γ-S and AMP-PNPwere from Sigma-Aldrich, and poly(dT) was from USB. PhiX174 ssDNA was from New England Biolabs. IPTG and DTT were from Research Products International. The GTG-repeating 48-mer oligonucleotide, the 83-mer and the 40-mer were from Integrated DNA Technologies. All other reagents were Fisher Scientific ACS grade.
Protein expression and purification
For the RecA protein used in the coprotease assays of Figure 1, the gene for RecA was amplified by PCR from E. coli genomic DNA, cloned into the pET-9a vector between the NdeI and BamH1 restriction sites, and overexpressed in BL21 (DE3) E. coli cells. 1L cultures in L.B. were grown at 37° C to an OD600 of ~0.8, and expression was induced with 1 mM IPTG. RecA was purified as described previously44 with some modifications. After cell lysis and the polyethyleneimine and ammonium sulfate precipitations, the protein was re-suspended in 50 ml of R buffer (20 mM Tris-HCl pH 7.5, 0.1 mM EDTA and 10% glycerol) plus 100 mM KCl and dialyzed against the same buffer. The protein was loaded onto a 25 ml HiTrap Q HP column (GE Healthcare) and eluted with a linear gradient of 0.1–1.5 M KCl. Collected fractions were dialyzed into P buffer (20 mM potassium phosphate, pH 6.8, 150 mM NaCl, 0.1 mM EDTA and 10% glycerol), and loaded onto a HiTrap Heparin HP column (GE Healthcare). Bound protein was eluted with a linear gradient of 0.15–1.5 M NaCl and dialyzed into 20 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM DTT and 10% glycerol, concentrated to 30 mg/ml, and frozen in small aliquots at −80 °C. RecA protein concentration was determined by O.D. at 280 nm using the extinction coefficient calculated from its amino acid sequence. The E4A-K8A RecA mutant was expressed from the BLR (DE3) pLysS strain of E. coli and purified using the same procedures as WT RecA. The lambda repressor protein fragments cI93–236 and cI101–229TM, which contains the A152T, P158T, and K192A mutations, were expressed and purified as described previously26.
RecA-mediated cleavage
Coprotease reactions (100 μl) were performed at 37° C and included 2 μM RecA protein (or E4A-K8A RecA), 6 μM (nt) of the indicated single-stranded oligonucleotide, 1 mM ATP analog (ATP-γ-S or AMP-PNP), 50 mM Tris pH = 7.4, 50 mM NaCl, 2 mM MgCl2, and 10 μM of a λ repressor fragment (cI93–236). Reactions were initiated by addition of cI93–236 (at time = 0 min.) after pre-incubation of all other components for 15 minutes at 37° C. At the indicated time points, 7 μl of each reaction was removed and immediately mixed with SDS-PAGE loading buffer and heated to 95° C for 5 min. to stop the reaction. Reactions were analyzed on 15% SDS-PAGE gels stained with Coomassie blue. The fraction cleaved at each time point was calculated with the equation:
where Int.C and Int.UC are the net intensities of the bands corresponding to cleaved product (cI112–236) and uncleaved substrate (cI93–236), respectively, as measured by scanning densitometry. The value for Int.C is multiplied by 1.16 (145/125) to account for the smaller size of the product relative to the substrate.
Electron Microscopy and Image Analysis
RecA protein was prepared as described45. RecA-ssDNA complexes were formed in 25 mM triethanolamine-HCl (Fisher) buffer (pH 7.2) at 37°C for 10 minutes, with 3 μM RecA, a RecA to M13 ssDNA (Sigma) ratio of 80:1 (w/w), 1.3 mM AMP-PNP (Sigma) or ATP-γ-S (Boehringer), and 2mM magnesium acetate (Sigma). For experiments with cI, the repressor was added in a 3-fold molar excess to the RecA and incubated at 37° C for an additional 15 min. The samples were applied to glow-discharged carbon-coated grids and negatively stained with 2% (w/v) uranyl acetate. A Tecnai-12 electron microscope was used at an accelerating voltage of 80kV and a nominal magnification of 30,000x. Images were digitized at a raster of 4.16Å/px, and segments (100 pixels long) were processed using the SPIDER package46 and the IHRSR package47. The resolution of the reconstructions was judged to be ~ 23 Å using the FSC=0.5 criterion. Surfaces were thresholded to enclose ~ 115% of the expected molecular volumes.
Acknowledgments
This work was supported by NIH grants GM035269 (to E.H.E.) and GM067947 (to C.E.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Kowalczykowski SC. Biochemical and biological function of Escherichia coli RecA protein: behavior of mutant RecA proteins. Biochimie. 1991;73:289–304. doi: 10.1016/0300-9084(91)90216-n. [DOI] [PubMed] [Google Scholar]
- 2.Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- 3.Ennis DG, Ossanna N, Mount DW. Genetic separation of Escherichia coli recA functions for SOS mutagenesis and repressor cleavage. J Bacteriol. 1989;171:2533–2541. doi: 10.1128/jb.171.5.2533-2541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witkin EM. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 6.Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 7.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442:883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 8.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 10.Sassanfar M, Roberts JW. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212:79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- 11.Little JW. Autodigestion of lexA and phage repressors. Proc Nat Acad Sci, USA. 1984;81:1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y, Pfuetzner RA, Mosimann S, Paetzel M, Frey EA, Cherney M, Kim B, Little JW, Strynadka NC. Crystal structure of LexA: a conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/s0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- 13.Craig NL, Roberts JW. Function of nucleotide triphosphate and polynucleotide in E. coli RecA protein- directed cleavage of phage lambda repressor. J Biol Chem. 1981;256:8039–8044. [PubMed] [Google Scholar]
- 14.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 15.Peat TS, Frank EG, McDonald JP, Levine AS, Woodgate R, Hendrickson WA. Structure of the UmuD′ protein and its regulation in response to DNA damage. Nature. 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- 16.Little JW, Mount DW. The SOS regulatory system of Escherichia coli. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 17.Peat TS, Frank EG, McDonald JP, Levine AS, Woodgate R, Hendrickson WA. The UmuD′ protein filament and its potential role in damage induced mutagenesis. Structure. 1996;4:1401–1412. doi: 10.1016/s0969-2126(96)00148-7. [DOI] [PubMed] [Google Scholar]
- 18.Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. UmuD′(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci U S A. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Anda J, Poteete AR, Sauer RT. P22 c2 repressor. Domain structure and function. J Biol Chem. 1983;258:10536–10542. [PubMed] [Google Scholar]
- 20.Eguchi Y, Ogawa T, Ogawa H. Cleavage of bacteriophage phi 80 CI repressor by RecA protein. J Mol Biol. 1988;202:565–573. doi: 10.1016/0022-2836(88)90286-0. [DOI] [PubMed] [Google Scholar]
- 21.Bell CE, Frescura P, Hochschild A, Lewis M. Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding. Cell. 2000;101:801–811. doi: 10.1016/s0092-8674(00)80891-0. [DOI] [PubMed] [Google Scholar]
- 22.Pabo CO, Sauer RT, Sturtevant JM, Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci U S A. 1979;76:1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer RT, Ross MJ, Ptashne M. Cleavage of the lambda and P22 repressors by recA protein. J Biol Chem. 1982;257:4458–4462. [PubMed] [Google Scholar]
- 24.Johnson AD, Poteete AR, Lauer G, Sauer RT, Ackers GK, Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 25.Roberts JW, Roberts CW. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci U S A. 1975;72:147–151. doi: 10.1073/pnas.72.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ndjonka D, Bell CE. Structure of a hyper-cleavable monomeric fragment of phage lambda repressor containing the cleavage site region. J Mol Biol. 2006;362:479–489. doi: 10.1016/j.jmb.2006.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanLoock MS, Yu X, Yang S, Galkin VE, Huang H, Rajan SS, Anderson WF, Stohl EA, Seifert HS, Egelman EH. Complexes of RecA with LexA and RecX Differentiate Between Active and Inactive RecA Nucleoprotein Filaments. J Mol Biol. 2003;333:345–354. doi: 10.1016/j.jmb.2003.08.053. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, Egelman EH. The LexA repressor binds within the deep helical groove of the activated RecA filament. J Mol Biol. 1993;231:29–40. doi: 10.1006/jmbi.1993.1254. [DOI] [PubMed] [Google Scholar]
- 29.Frank EG, Cheng N, Do CC, Cerritelli ME, Bruck I, Goodman MF, Egelman EH, Woodgate R, Steven AC. Visualization of two binding sites for the Escherichia coli UmuD′(2)C complex (DNA pol V) on RecA-ssDNA filaments. J Mol Biol. 2000;297:585–597. doi: 10.1006/jmbi.2000.3591. [DOI] [PubMed] [Google Scholar]
- 30.Mustard JA, Little JW. Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of recA. J Bacteriol. 2000;182:1659–1670. doi: 10.1128/jb.182.6.1659-1670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gimble FS, Sauer RT. Lambda repressor mutants that are better substrates for RecA-mediated cleavage. J Mol Biol. 1989;206:29–39. doi: 10.1016/0022-2836(89)90521-4. [DOI] [PubMed] [Google Scholar]
- 32.De Zutter JK, Knight KL. The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J Mol Biol. 1999;293:769–780. doi: 10.1006/jmbi.1999.3200. [DOI] [PubMed] [Google Scholar]
- 33.Rajan R, Wisler JW, Bell CE. Probing the DNA sequence specificity of Escherichia coli RECA protein. Nucleic Acids Res. 2006;34:2463–2471. doi: 10.1093/nar/gkl302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tracy RB, Kowalczykowski SC. In vitro selection of preferred DNA pairing sequences by the Escherichia coli RecA protein. Genes Dev. 1996;10:1890–1903. doi: 10.1101/gad.10.15.1890. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–4. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 36.Volodin AA, Camerini-Otero RD. Influence of DNA sequence on the positioning of RecA monomers in RecA-DNA cofilaments. J Biol Chem. 2002;277:1614–1618. doi: 10.1074/jbc.M108871200. [DOI] [PubMed] [Google Scholar]
- 37.DiCapua E, Cuillel M, Hewat E, Schnarr M, Timmins PA, Ruigrok RW. Activation of recA protein. The open helix model for LexA cleavage. J Mol Biol. 1992;226:707–719. doi: 10.1016/0022-2836(92)90627-v. [DOI] [PubMed] [Google Scholar]
- 38.Krishna R, Prabu JR, Manjunath GP, Datta S, Chandra NR, Muniyappa K, Vijayan M. Snapshots of RecA protein involving movement of the C-domain and different conformations of the DNA-binding loops: crystallographic and comparative analysis of 11 structures of Mycobacterium smegmatis RecA. J Mol Biol. 2007;367:1130–1144. doi: 10.1016/j.jmb.2007.01.058. [DOI] [PubMed] [Google Scholar]
- 39.Register JC, Griffith J. RecA protein filaments can juxtapose DNA ends: an activity that may reflect a function in DNA repair. Proc Natl Acad Sci U S A. 1986;83:624–628. doi: 10.1073/pnas.83.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krishna R, Manjunath GP, Kumar P, Surolia A, Chandra NR, Muniyappa K, Vijayan M. Crystallographic identification of an ordered C-terminal domain and a second nucleotide-binding site in RecA: new insights into allostery. Nucleic Acids Res. 2006;34:2186–2195. doi: 10.1093/nar/gkl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galkin VE, Wu Y, Zhang XP, Qian X, He Y, Yu X, Heyer WD, Luo Y, Egelman EH. The Rad51/RadA N-Terminal Domain Activates Nucleoprotein Filament ATPase Activity. Structure. 2006;14:983–992. doi: 10.1016/j.str.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Xing X, Bell CE. Crystal structures of Escherichia coli RecA in a compressed helical filament. J Mol Biol. 2004;342:1471–1485. doi: 10.1016/j.jmb.2004.07.091. [DOI] [PubMed] [Google Scholar]
- 43.Haruta N, Yu X, Yang S, Egelman EH, Cox MM. A DNA pairing-enhanced conformation of bacterial RecA proteins. J Biol Chem. 2003;278:52710–52723. doi: 10.1074/jbc.M308563200. [DOI] [PubMed] [Google Scholar]
- 44.Lusetti SL, Wood EA, Fleming CD, Modica MJ, Korth J, Abbott L, Dwyer DW, Roca AI, Inman RB, Cox MM. C-terminal deletions of the Escherichia coli RecA protein. Characterization of in vivo and in vitro effects. J Biol Chem. 2003;278:16372–16380. doi: 10.1074/jbc.M212917200. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Egelman EH. Direct visualization of dynamics and cooperative conformational changes within RecA filaments that appear to be associated with the hydrolysis of ATP- -S. J Mol Biol. 1992;225:193–216. doi: 10.1016/0022-2836(92)91036-o. [DOI] [PubMed] [Google Scholar]
- 46.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 47.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]