Abstract
The Pleistocene was a dynamic period for Holarctic mammal species, complicated by episodes of glaciation, local extinctions, and intercontinental migration. The genetic consequences of these events are difficult to resolve from the study of present-day populations. To provide a direct view of population genetics in the late Pleistocene, we measured mitochondrial DNA sequence variation in seven permafrost-preserved brown bear (Ursus arctos) specimens, dated from 14,000 to 42,000 years ago. Approximately 36,000 years ago, the Beringian brown bear population had a higher genetic diversity than any extant North American population, but by 15,000 years ago genetic diversity appears similar to the modern day. The older, genetically diverse, Beringian population contained sequences from three clades now restricted to local regions within North America, indicating that current phylogeographic patterns may provide misleading data for evolutionary studies and conservation management. The late Pleistocene phylogeographic data also indicate possible colonization routes to areas south of the Cordilleran ice sheet.
The major climatic changes that occurred toward the end of the late Pleistocene had an important influence on the evolution and distribution of extant taxa. However, the genetic consequences of these events have been difficult to determine (1–6). The brown bear is a large, Holarctic carnivore whose distribution was dramatically altered by late Pleistocene events. In North America, the brown bear has had a limited history, appearing in eastern Beringia only 50–70,000 years ago and spreading into the contiguous United States about 13,000 years ago (7, 8). Previous research on mitochondrial control region sequences from 317 extant brown bears found that they defined one European (I) and three distinct North American (II, IIIa/IIIb, IV) clades (9) (Figs. 1 and 2a). Bears with sequences from the basal and divergent clade II were restricted to the Admiralty, Baranof, and Chichagof (ABC) islands. Sequences from populations in northern Canada and Alaska fell in a clade comprising two closely related groups (IIIa and IIIb), whereas populations in southern Canada and the contiguous United States belonged to a southern clade (IV). Of 22 localities surveyed, none had sequences from more than one clade, with one exception, where IIIa and IIIb cooccurred (ref. 9, Fig. 2a). Sequences from the western Alaskan clade (IIIa) also were found in Asia and northern and eastern Europe, suggesting a recent connection across the Bering Land Bridge (9, 10).
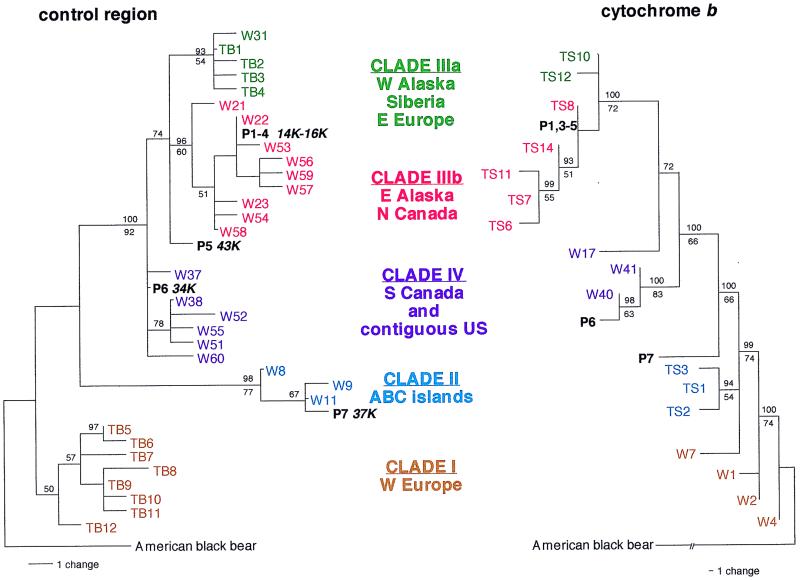
Figure 1.
Maximum likelihood trees of brown bear control region and cytochrome b sequences. For simplicity, we use the clade I–IV nomenclature that was established previously (9), although in our smaller dataset several clades appear paraphyletic, particularly in the less well resolved cytochrome b tree. Extant sequences are in color [W (9), TS (10), TB (11)], and permafrost sequences (P1–P7) are in black. Reliability values (above nodes) and bootstrap percentages (below nodes) with values greater than 50% are indicated. Corrected radiometric dates (12) and reference numbers are as follows: P1, UCR3742/CAMS-51806 (15,370 ± 60 bp); P2, UCR3741/CAMS-51805 (14,980 ± 60 bp); P3, UCR3743/CAMS-54128 (13,760 ± 50 bp); P4, UCR3745/CAMS-54129 (15,680 ± 50 bp); P5, UCR3746/CAMS-54130 (42,850 ± 850); P6, UCR3744/CAMS-51808 (35,970 ± 660); and P7, Beta16162 (36,500 ± 1,150).
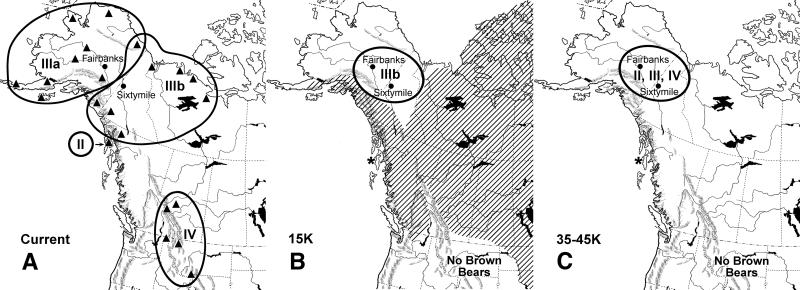
Figure 2.
Current (A) and past geographic distribution of brown bear control region sequence clades 15,000 (B) and 35,000–45,000 (C) years ago. Locations of modern samples (9) are indicated by triangles. The asterisk indicates a brown bear fossil dated to greater than 40,000 years ago on Prince of Wales Island (16), and no fossils are known in the contiguous United States before 13,000 years. The approximate extent of glacial ice sheets 15,000 ago is superimposed on current continental boundaries in hatching (17).
The recent appearance of brown bears in North America and their division into four genetically distinct populations with highly divergent mitochondrial control region sequences implied separate invasions of North America from long-isolated Old World populations (9, 10). Clade II was postulated to represent the earliest invasion, followed by clades IV and IIIa/IIIb. Furthermore, the precise correspondence between geographic and phylogenetic divisions suggests three evolutionary significant units for conservation (9, 13) (clades II, III, and IV). In contrast, nuclear microsatellite data in Alaskan brown bears do not support long-term genetic isolation of the three Alaskan clades, suggesting the possibility of sex-biased dispersal (14).
Mammalian remains have been recovered from vast permafrost deposits in central Alaska and northwestern Canada as a consequence of placer gold-mine operations (8). More than 100,000 skeletal elements are preserved in collections at the American Museum of Natural History (AMNH), New York, and the Canadian Museum of Nature (CMN), Ottawa. Mitochondrial and nuclear DNA sequences have been amplified from several individual permafrost specimens, and cold conditions are thought to favor DNA survival (15). Therefore, museum collections of permafrost-preserved bones potentially represent an extensive genetic record of mammal populations over the past 50,000 years. To directly record changes in genetic diversity through the late Pleistocene, we examined nine unassociated, permafrost-preserved brown bear bones from localities about 400 km apart, near Fairbanks, Alaska, and Sixtymile, Yukon Territory (Fig. 2a).
Methods
DNA Isolation and Sequencing.
Nine permafrost-preserved bones obtained from the AMNH (New York) and the CMN (Ottawa) were analyzed. From the following specimens, DNA was amplified successfully: (i) Fairbanks, Alaska: P1, AMNH 95679, right ulna from Lower Goldstream; P2, AMNH 95630, right humerus from Lower Goldstream; P3, AMNH 95665, right radius from Gold Hill; P4, AMNH 95629, left humerus from Fairbanks Creek; P5, AMNH 95664, radius from Cripple Creek; (ii) Sixtymile, Yukon Territory: P6, CMN 42381, right ulna; and P7, CMN 38279, right humerus. Using a Dremel tool, 1-cm2 fragments approximately 0.5 cm in depth were removed near the midshaft of long bones. Samples were powdered in a sterilized coffee grinder or steel mortar and pestle and decalcified in 10–30 vol of 0.5 M EDTA (pH 8) overnight at room temperature. The sediment was collected by centrifugation and digested with proteinase K/DTT overnight at 50–55°C. The samples were extracted twice with phenol and once with chloroform, and the DNA was recovered with Centricon-30 (Amicon) devices. We amplified a 502-bp fragment of the 5′ end of cytochrome b by using three overlapping primer sets: CarnCB51, GACCACATCCGAAAATCYC-3′, and CarnCB52, CCGTAGTTAACGTCTCGGC-3′ (225 bp); CarnCB53, CCTATTCCTAGCCATACACTACA-3′, and CarnCB58, CCAATGTTTCATGTTTCTGGGA-3′ (181 bp); and CarnCB55, ATCTGCCTATTCATGCACGTA-3′, and CarnCB58, GAAGCCYCCTCAGATTCAYTC-3′ (231 bp); and one control region segment of 173 bp (18). Amplification from 1–3 μl of the extract was carried out in a 25-μl volume using AmpliTaq or AmpliTaq Gold (Perkin–Elmer) with the following cycling conditions: AmpliTaq, 40 cycles of 92°C for 40 sec, 50–52°C for 1 min, and 72°C for 1 min using wax-mediated hot start, and AmpliTaq Gold, 60 cycles of 95°C for 1 min, 48–55°C for 1 min, and 72°C for 1 min with mineral oil instead of wax. PCR products were sequenced by using an Applied Biosystems PRISM Dye Terminator kit and Applied Biosystems 377 sequencer.
Authentication.
DNA extraction and PCR setup occurred in spatially isolated facilities dedicated to low-copy DNA research. Contamination was monitored throughout with extraction and PCR blanks (19). No DNA from modern brown bears had been amplified previously; all modern sequences were from GenBank or provided by Lisette Waits (University of Idaho, Moscow). Sequences of three fragments were replicated independently from three separate extractions at Oxford University and University of California, Los Angeles (control region of P3 and P7 and CarnCB53–58 of P3). Control region and cytochrome b sequences from P1 and P6 also were replicated at University of California, Los Angeles, by using separate extractions. Replication also was provided by the overlapping positions in the three sequenced segments of cytochrome b.
Carbon Dating.
We radiometrically dated the permafrost specimens to determine whether a succession of populations belonging to the different clades could explain the distribution of sequences (9, 10). Total amino acids from samples P1–P6 were extracted (20) and then converted to graphitic carbon at the University of California, Riverside Radiocarbon Laboratory (21). The date was determined by accelerator mass spectrometry (AMS) at the Center for Accelerator Mass Spectrometry at Lawrence Livermore National Laboratory, University of California. P7 previously had been AMS dated by the CMN.
Phylogenetic Analysis.
Maximum-likelihood trees and support values were calculated with paup* 4.01b (22) by using the Puzzle quartet algorithm and gamma parameters of 0.24 (control region) and 0.73 (cytochrome b) as determined empirically from the data. The confidence in estimated relationships was determined by using reliability values (23) and bootstrap percentages from 1,000 replicate trees, and only values greater than 50% are indicated. The likelihood scores are 497.5 and 614.6 for the control region and cytochrome b trees, respectively. Phylogenetic analyses used maximum parsimony, maximum likelihood, and neighbor-joining, the latter two with models of substitution including Jukes-Cantor, Tamura-Nei, and HKY85. Only one transversion was observed in pairwise comparisons of late Pleistocene and recent brown bear sequences; hence, no correction was applied for transition/transversion bias. We also constructed constraint trees that had the same topology of the four clades given in ref. 9 to determine the effect on the phylogenetic placement of the permafrost sequences.
Results
From six of the specimens (P1, P3–P7), we succeeded in amplifying one control region fragment of 173 bp and three cytochrome b fragments totaling 502 bp. A control region fragment also was obtained from a seventh specimen (P2). Two other permafrost bear bones failed to yield any DNA that could be amplified. Four control region and three cytochrome b haplotypes were found. All replicated sequences were identical. We could compare 132 bp of the control region and 258 bp of the cytochrome b sequences with published data (9–11) and found 32 and 23 polymorphic sites, respectively. The permafrost and modern North American bear control region sequences differed by 0–16 transitions, and the cytochrome b sequences differed by 0–9 transitions (Table 1). With the exception of one synonymous cytochrome b substitution, all polymorphic sites in the permafrost sequences were also polymorphic in extant brown bear sequences and no indels were observed.
Table 1.
Average Tamura-Nei percent sequence divergence (above diagonal) and average number of substitutions (below diagonal) between control region (above slash) and cytochrome b (below slash) sequences from late-Pleistocene brown bears and the four sequence clades (Fig. 1)
| P1–P4 | P5 | P6 | P7 | Clade I | Clade II | Clade III | Clade IV | |
|---|---|---|---|---|---|---|---|---|
| P1–P4 | — | 3.7 /0 | 3.7 /2.1 | 24.6 /2.5 | 19.5 /3.2 | 23.1 /3.2 | 2.5 /0.8 | 6.1 /2.2 |
| P5 | 4 /0 | — | 1.8 /2.1 | 23.6 /2.5 | 14.1 /3.2 | 24.5 /3.2 | 4.0 /0.8 | 3.7 /2.2 |
| P6 | 4 /5 | 2 /5 | — | 17.5 /3.8 | 11.0 /3.7 | 17.3 /3.7 | 4.0 /2.9 | 1.6 /1.2 |
| P7 | 14 /6 | 14 /6 | 12 /9 | — | 9.5 /2.4 | 1.5 /2.1 | 25.2 /3.2 | 23.2 /3.1 |
| Clade I | 11.1 /7.8 | 10.1 /7.8 | 10.1 /8.8 | 7.9 /5.8 | — | 11.8 /1.1 | 17.9 /4.0 | 14.1 /3.2 |
| Clade II | 13.8 /7.7 | 13.8 /7.7 | 11.8 /8.7 | 1.8 /5 | 9.5 /2.7 | — | 22.5 /3.3 | 20.6 /3.1 |
| Clade III | 2.4 /2 | 4.1 /2 | 4.1 /7 | 14.1 /7.7 | 11.5 /9.4 | 14.3 /8 | — | 5.8 /3.0 |
| Clade IV | 5.8 /5.3 | 3.8 /5.3 | 1.8 /3 | 13.8 /7.3 | 10.0 /7.8 | 13.9 /7.4 | 5.8 /7.1 | — |
Phylogenetic analysis of the control region and cytochrome b sequences indicated that the permafrost sequences could be assigned to three clades (II, III, IV) that now occur only in geographically widely separated populations (Table 1 and Figs. 1 and 2a). Sequences from permafrost specimens P1–P4 were identical for both regions (the cytochrome b sequence of P2 could not be determined). However, these specimens differ with respect to geographic distribution and radiometric dates (below) and, therefore, cannot all be first-order relatives. Sequences P1–P4 grouped with clade IIIb sequences, found in extant bears from eastern Alaska and northern Canada (Fig. 2a), and differ from them by 0–3 substitutions in 132 bp of the control region and 0–3 substitutions in 258 bp of cytochrome b sequence (Table 1). Permafrost bear sample P5 also was genetically most similar to clade IIIa/IIIb (Table 1), although it joined at the base of this clade in the control region tree (Fig. 1). In contrast, permafrost specimens P6 and P7 were genetically most similar to clades IV and clade II, respectively (Table 1), and grouped with these clades in both trees (Fig. 1). The concordant phylogenetic assignment of specimens in the control region and cytochrome b trees (Fig. 1) further supports their authenticity. When the trees were constrained to the previously published topology (9), the position of the late Pleistocene bear sequences were unchanged, although the position of extant bear sequences within clades differed. We found no rate heterogeneity when using Tajima's test (24).
Radiometric dating showed that the four permafrost specimens with sequences similar to those of extant bears from Alaska (P1–P4, clade IIIb) were the youngest and fell within a narrow range of radiometric dates from 13,760 ± 50 to 15,680 ± 50 years ago (Fig. 1). The permafrost sequence basal within this clade (P5) was 42,600 ± 850 years old. The sequences associated with clades now found only in southern Canada and the contiguous United States (clade IV, P6) and the ABC islands (clade II, P7) were 35,970 ± 660 and 36,500 ± 1150 years old, respectively (Fig. 1). Therefore, all the permafrost specimens with sequences from clades that are no longer found in Alaska predate the last glacial maximum (LGM), which occurred between 14,000 and 18,000 years ago (8, 17, 25, 26). In contrast, bears with the clade IIIb sequences that are found in the area today were already in place by about 15,000 years ago (Fig. 2).
Discussion
The molecular and radiometric data do not support suggestions that clades II, IIIa/IIIb, and IV represent separate invasions from Old World populations (9, 10). Instead, about 36,000 years ago, before the last glaciation, all three clades were represented in east Beringian brown bears, and, therefore, the geographical partitioning of mtDNA haplotypes in extant North American populations (9) is a relatively recent event. Although mtDNA data reflects only female dispersal events, our data agree with previous nuclear microsatellite data (14) and raise doubts that clades II, III, and IV are evolutionarily significant units that should be managed separately for conservation (9).
The most likely explanation for the current geographic segregation of mitochondrial haplotypes is that the founding populations of each area contained representatives of only one clade. This process of lineage sorting has imparted a geographic structure to sequence phylogenies that does not reflect a long history of isolation, as might be assumed (27–29). Recent molecular analyses of brown bear populations in Japan, Mongolia, and Tibet (30, 31) support the idea that the ancestral Beringian population possessed a diverse array of haplotypes and that subsequent lineage sorting has taken place in isolated populations. Mainland Asian populations were found to contain a diverse range of control region sequences, grouping with clades II, IIIa, and IV, whereas on Hokkaido Island a subset of three divergent clade IIIa and IV sequences were found in a geographic pattern as marked as that in North American bears (9).
Although the permafrost sample size is limited, our analysis suggests that extant and postglacial brown bear populations are less diverse than those existing before the LGM. Before the last glaciation, all three clades are found in our sample of brown bears. However, by 15,000 years ago, the east Beringian population appears to be considerably less diverse, with four samples from a 2,000-year range yielding identical clade IIIb sequences that are still found in the area today. This suggests that significant changes in brown bear demography occurred in that region between 36,000 and 16,000 years ago, but not afterward. Interestingly, these events precede the end-Pleistocene megafaunal extinctions in North America by some 5,000 years (7, 8, 25), suggesting that the genetic effects of climate change during the LGM were more significant in bears than those associated with the megafaunal extinction event.
The mitochondrial sequences found in pre- and post-LGM brown bear populations constrain biogeographical explanations for the colonization of North America by this species. Bears from lineage IIIb, which were present in eastern Beringia after the LGM, do not appear to have spread to the contiguous United States via the inland, ice-free corridor that opened subsequently (8, 17, 25, 26), suggesting bears and other taxa may have used a coastal route. During the LGM, the expanding Cordilleran ice sheet and deteriorating climatic conditions are likely to have fragmented eastern Beringian populations and reduced genetic diversity. Small, ice-free refugia are known to have existed on the adjacent Pacific Northwest coast, and paleontological records of bears are known from islands in the area (16, 32–34). Bear populations that survived the last glacial maximum on island refugia could reinvade the mainland across straits revealed by lower sea levels, such as Haida Gwaii (32–34). By a process of lineage sorting and founder effect (27–29), small populations in coastal refugia could have become dominated by bears with mitochondrial lineage II to the north (on the ABC islands) and lineage IV to the south. The latter then could have served as the source of bears that spread across southern Canada and the contiguous United States sometime before 13,000 years ago (Fig. 2). This coastal refugia hypothesis can be tested by examining DNA from cave-preserved late Pleistocene bear bones in putative glacial refugia (16) and, if confirmed, would add considerable support for a similar route of human colonization into the Americas.
This direct view of population genetics during the late Pleistocene suggests that major changes in the genetic diversity and geographic distribution of North American large mammals occurred before 16,000 years ago, well before the end-Pleistocene North American megafaunal mass extinction (7, 8, 25). The permafrost DNA sequences also reveal that an association between phylogenetic structure and geography does not necessarily imply long-term genetic isolation or incipient speciation, as has been suggested for North American Pleistocene vertebrates (1–6, 27, 28). Finally, this study shows that museum collections of permafrost remains are a unique source of genetic information about paleoecological events during the last major climate change.
Acknowledgments
We thank the CMN and AMNH for permission to sample their collections. We are grateful to Charles Marshall and the Oxford University Museum of Natural History for laboratory facilities. C. R. Harington, R. Tedford, B. Van Valkenburgh, L. Waits, and R. Ward provided helpful comments and assistance. This research was supported by the National Science Foundation, Natural Environment Research Council, and Royal Society.
Abbreviations
- CMN
Canadian Museum of Nature
- AMNH
American Museum of Natural History
- LGM
last glacial maximum
Note Added in Proof
Recent studies of permafrost remains have demonstrated that it is possible to amplify single copy nuclear sequences, including nuclear copies of mitochondrial genes (35). The latter is unlikely to be a factor in the current study as several different primer pairs and sequence regions produced consistent results.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF225566–AF225572).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.040453097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.040453097
References
- 1.Bermingham E, Rohwer S, Freeman S, Wood C. Proc Natl Acad Sci USA. 1992;89:6624–6628. doi: 10.1073/pnas.89.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hewitt G M. Biol J Linn Soc. 1996;58:247–276. [Google Scholar]
- 3.Klicka J, Zink R M. Science. 1997;277:1666–1669. [Google Scholar]
- 4.Avise J C, Walker D. Proc R Soc London Ser B. 1998;265:457–463. doi: 10.1098/rspb.1998.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klicka J, Zink R M. Proc R Soc London Ser B. 1999;266:695–700. [Google Scholar]
- 6.Wooding S, Ward R. Mol Biol Evol. 1997;14:1096–1105. doi: 10.1093/oxfordjournals.molbev.a025719. [DOI] [PubMed] [Google Scholar]
- 7.Kurtén B, Anderson E. Pleistocene Mammals of North America. New York: Columbia Univ. Press; 1980. [Google Scholar]
- 8.Guthrie R D. Frozen Fauna of the Mammoth Steppe. Chicago: Univ. of Chicago Press; 1990. [Google Scholar]
- 9.Waits L P, Talbot S L, Ward R H, Shields G F. Cons Biol. 1998;12:408–417. [Google Scholar]
- 10.Talbot S L, Shields G F. Mol Phylogenet Evol. 1996;5:477–494. doi: 10.1006/mpev.1996.0044. [DOI] [PubMed] [Google Scholar]
- 11.Taberlet P, Bouvet J. Proc Roy Soc London Ser B. 1994;255:195–200. doi: 10.1098/rspb.1994.0028. [DOI] [PubMed] [Google Scholar]
- 12.Stuiver M, Polach H A. Radiocarbon. 1977;19:355–363. [Google Scholar]
- 13.Moritz C. Trends Ecol Evol. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 14.Paetkau D, Shields G F, Strobeck C. Mol Ecol. 1998;7:1283–1292. doi: 10.1046/j.1365-294x.1998.00440.x. [DOI] [PubMed] [Google Scholar]
- 15.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 16.Heaton T H, Talbot S L, Shields G F. Quat Res. 1996;46:186–192. [Google Scholar]
- 17.Dawson A G. Ice Age Earth. London: Routledge; 1992. [Google Scholar]
- 18.Hanni C, Laudet V, Stehelin D, Taberlet P. Proc Natl Acad Sci USA. 1994;91:12336–12340. doi: 10.1073/pnas.91.25.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krings M, Stone A, Schmitz R W, Krainitski H, Stoneking M, Pääbo S. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 20.Taylor R E. Radiocarbon Dating: An Archaeological Perspective. New York: Academic; 1987. [Google Scholar]
- 21.Kirner D L, Taylor R E, Southon J R. Radiocarbon. 1995;37:697–704. [Google Scholar]
- 22.Swofford, D. L. (1998) paup4d65: Phylogenetic Analysis Using Parsimony and Other Methods (Sinauer, Sunderland, MA), Test Version.
- 23.Strimmer K, Von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 24.Tajima F. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pielou E C. After the Ice Age: The Return of Life to Glaciated North America. Chicago: Univ. of Chicago Press; 1991. [Google Scholar]
- 26.Anderson B G, Borns H W., Jr . The Ice Age World. Oslo: Scandinavian Univ. Press; 1997. [Google Scholar]
- 27.Avise J C, Neigel J E, Arnold J. J Mol Evol. 1984;20:99–105. doi: 10.1007/BF02257369. [DOI] [PubMed] [Google Scholar]
- 28.Hudson R R. Oxford Surv Evol Biol. 1990;7:1–44. [Google Scholar]
- 29.Templeton A R, Crandall K A, Sing C F. Genetics. 1995;132:619–633. doi: 10.1093/genetics/132.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda R, Murata K, Aiurzaniin A, Yoshida M C. Hereditas. 1998;128:277–280. doi: 10.1111/j.1601-5223.1998.00277.x. [DOI] [PubMed] [Google Scholar]
- 31.Matsuhashi T, Masuda R, Mano T, Yoshida M C. Mol Biol Evol. 1999;16:676–684. doi: 10.1093/oxfordjournals.molbev.a026150. [DOI] [PubMed] [Google Scholar]
- 32.Byun S A, Koop B F, Reimchen T E. Evolution. 1997;51:1647–1653. doi: 10.1111/j.1558-5646.1997.tb01487.x. [DOI] [PubMed] [Google Scholar]
- 33.Mann D H, Hamilton T D. Quat Sci Rev. 1995;14:449–471. [Google Scholar]
- 34.Harington C R. Can J Earth Sci. 1975;12:903–919. [Google Scholar]
- 35.Greenwood A D, Capelli C, Possnert G, Pääbo S. Mol Biol Evol. 1999;16:1466–1473. doi: 10.1093/oxfordjournals.molbev.a026058. [DOI] [PubMed] [Google Scholar]