Abstract
Background
Aicardi‐Goutières syndrome (AGS) is an autosomal recessive, early onset encephalopathy characterised by calcification of the basal ganglia, chronic cerebrospinal fluid lymphocytosis, and negative serological investigations for common prenatal infections. AGS may result from a perturbation of interferon α metabolism. The disorder is genetically heterogeneous with approximately 50% of families mapping to the first known locus at 3p21 (AGS1).
Methods
A genome‐wide scan was performed in 10 families with a clinical diagnosis of AGS in whom linkage to AGS1 had been excluded. Higher density genotyping in regions of interest was also undertaken using the 10 mapping pedigrees and seven additional AGS families.
Results
Our results demonstrate significant linkage to a second AGS locus (AGS2) at chromosome 13q14–21 with a maximum multipoint heterogeneity logarithm of the odds (LOD) score of 5.75 at D13S768. The AGS2 locus lies within a 4.7 cM region as defined by a 1 LOD‐unit support interval.
Conclusions
We have identified a second AGS disease locus and at least one further locus. As in a number of other conditions, genetic heterogeneity represents a significant obstacle to gene identification in AGS. The localisation of AGS2 represents an important step in this process.
Keywords: AGS2, Aicardi‐Goutières syndrome, interferon α, intracranial calcification, 13q14–21
Aicardi‐Goutierès syndrome (AGS; MIM 225750) is an autosomal recessive encephalopathy characterised by cerebral atrophy, leukodystrophic changes, intracranial calcification, chronic cerebrospinal fluid (CSF) lymphocytosis, raised levels of interferon α (IFN‐α) in the CSF, and negative serological investigations for common prenatal infections (MIM 225750).1,2 Clinically, AGS can usefully be considered as a Mendelian mimic of congenital viral infection. Recognition of the condition is therefore important because of the possibility of misdiagnosis as a non‐genetic disorder and counselling of a falsely low risk of recurrence.
The features of AGS may have a prenatal onset or develop over the first few months of life.2,3 Typically, neurodegeneration is associated with microcephaly, spasticity, dystonic posturing, and psychomotor retardation. Death frequently occurs within the first decade. However, we are aware of several children in their teenage years with apparently non‐progressive disease. Systemic abnormalities include fever, raised levels of immunoglobulins and autoantibodies, thrombocytopenia, abnormal liver function with hepatosplenomegaly, and chilblain‐like cutaneous lesions.2,4,5,6,7
Levels of CSF IFN‐α are consistently elevated in the early stages of AGS and significantly higher than those recorded systemically.2,8 Such raised levels of CSF IFN‐α are not always accompanied by CSF lymphocytosis.9 IFN‐α does not cross the blood‐brain barrier, so when 100 IU/ml of IFN‐α is experimentally released into the blood, <1 IU/ml is detected in the CSF.10 It is likely then that in AGS, IFN‐α is produced intrathecally, possibly by astrocytes or microglia.11,12
The pathological finding of wedge shaped infarctions together with patchy myelin loss and calcified deposits in the media, adventitia, and perivascular space of small blood vessels suggests that AGS may represent a genetic cerebral angiopathy.13 IFN‐α is known to have an inhibitory effect on angiogenesis and astrocyte specific chronic overproduction of IFN‐α in transgenic mice recapitulates the neuropathological findings seen in AGS.14,15,16,17 These observations raise the possibility that the AGS phenotype results from exposure of the developing brain to high levels of IFN‐α. Identification of the causative genetic defect in AGS may, therefore, provide novel insights into IFN‐α metabolism.
Previously, we identified linkage to an interval on 3p21 in 48% of AGS families tested.18 Locus heterogeneity was considered the explanation for a failure to identify genetic linkage in a previous study.19 Since then, we have refined the AGS1 critical interval to a 3.47 cM region by demonstrating that AGS and Cree encephalitis are allelic disorders.9
Given that half of the families in our cohort were unlinked to the AGS1 locus, we performed a genome‐wide scan using 10 families incompatible with linkage to AGS1. Herein, we report the identification of a second AGS locus at chromosome 13q14–21 resulting from an analysis of this data set.
Methods
Subjects
For inclusion in the study, affected individuals had to demonstrate a compatible neurological phenotype with intracranial calcification and CSF lymphocytosis (>5 cells/mm3) and/or raised levels of IFN‐α in the CSF (>2 IU/ml; measured with a biological assay) as well as negative investigations for common prenatal infections.2
Seventeen families (15 consanguineous and two non‐consanguineous comprising three affected siblings) satisfied the inclusion criteria (tables 1 and 2). Blood samples were obtained with consent from affected children, their parents, and unaffected siblings where possible, for DNA extraction, genotyping, and subsequent linkage analysis. The study was approved by the Leeds Health Authority/United Teaching Hospitals NHS Trust Research Ethics Committee.
Table 1 Clinical characteristics of affected individuals from families consistent with linkage to AGS2.
| Family/patient | Ethnicity | Age at presentation | Birth OFC in centiles (gestation in weeks) | Postnatal OFC in centiles (age in months) | Brain calcification | CSF WCC/mm3* (age in months) | IFN‐α IU/ml in CSF/ serum† (age in months) |
|---|---|---|---|---|---|---|---|
| 1/IV:3 | Algerian | Birth | 2nd–9th (40) | <<0.4th (18) | PV; BG | 28 (1) | 100/NA (1) |
| 1/IV:4 | 2 months | 50th–75th (39) | <0.4th (44) | PV; BG | NA | NA | |
| 1/IV:5 | Prenatal | 75th (37) | 0.4th–2nd (2) | BG | NR | 25/NA (1) | |
| 2/V:1 | Algerian | 9 months | 75th–91st (40) | NR | BG | 43 (10) | 16/4 (10); <4/<2 (60) |
| 2/V:2 | 3 months | Normal | Normal | BG | 105 (5) | 60/8 (5) | |
| 3/IV:1 | Irish | 12 months | 50th (40) | 75th (108) | BG; WM | 16 (12); 8 (108) | <2/NA (108) |
| 3/IV:3 | <12 months | 50th (40) | 3rd (21) | BG; WM | 13 (1); 13 (21) | 25/<2 (21) | |
| 4/IV:1 | Moroccan | 7 months | NR | NR | BG | 8 (7) | 6/<2 (7) |
| 5/IV:1 | Italian | 2 months | 9th (40) | NR | BG | 25 (2) | >100/NA (2) |
| 6/II:1 | Dutch | 3 weeks | NR | <3rd (132) | BG; WM | 22 (8) | NA |
| 6/II:2 | 3 weeks | 9th (39) | 30th (84) | BG, WM | 13 (4) | 18/NA (3) | |
| 6/II:3 | 2–3 weeks | 25th–50th (38) | 30th (60) | BG | 13 (2) | 37/NA (48) | |
| 7/II:1 | French | 4 months | 75th–91st (40) | NR | BG | 0 (78) | <2/<2 (78) |
| 7/II:2 | 5 months | NR | NR | BG | 22 (6) | 32/12 (6) | |
| 7/II:3 | 8 months | NR | NR | BG | 39 (9) | 50/6 (9) | |
| 8/IV:1 | Spanish | 1 month | Normal | Microcephaly (18) | BG | 97 (24) | NA |
| 8/IV:3 | 10 days | Normal | Microcephaly (18) | BG | 38 (18) | NA |
BG, basal ganglia; CSF, cerebrospinal fluid; NA, not analysed; NR, not recorded; OFC, occipito‐frontal circumference; PV, periventricular; WCC, white cell count; WM, white matter.
*Abnormal ⩾5 cells/mm3; †normal levels <2 IU/l.
Table 2 Clinical characteristics of affected individuals from families inconsistent with linkage to AGS2.
| Family/ patient | Ethnicity | Age at presentation | Birth OFC in centiles (gestation in weeks) | Postnatal OFC in centiles (age in months) | Brain calcification | CSF WCC/mm3* (age in months) | IFN‐α IU/ml in CSF/ serum† (age in months) |
|---|---|---|---|---|---|---|---|
| 9/IV:1 | Pakistani | Antenatal | 9th–25th (38) | NR | BG; WM; PV | 63 (2) | NA |
| 10/V:1 | Algerian | Birth | 25th–50th (40) | <<0.4th (132) | BG | 26 (<1) | 60/16 (<1); 90/<10 (2) |
| 10/V:3 | Birth | 9th (40) | <<0.4th (72) | BG | 10 (<1) | 60/30 (<1); 30/24 (3) | |
| 11/IV:1 | Moroccan | 4 months | NR | <<0.4th (72) | BG | 23 (6); 11 (12) | 6/<2 (12) |
| 11/IV:2 | Birth | NR | <<0.4th (36) | BG; WM | 36 (3) | NA | |
| 12/VI:1 | Hungarian | 2 months | NR | Microcephaly (25) | PV | 22 (25) | NA |
| 12/VI:2 | <12 months | NR | Microcephaly (48) | PV | 80 (48) | NA | |
| 13/IV:1 | Pakistani | 6 months | NR | <<3rd | BG | 21 (6) | 32/4 (6) |
| 13/IV:2 | Birth | 50th (40) | NR | BG | NR | 200/9 (birth); NA/18 (3) | |
| 14/IV:1 | Dutch | 8 months | Normal | 3rd (60) | BG | 14 (13) | NA |
| 14/IV:3 | 12 months | Normal | 75th–91st (36) | BG | 550 (30) | NA | |
| 15/IV:2 | Pakistani | 8 months | Normal | 75th–91st (36) | BG | 0 (84) | <2/<2 (84) |
| 15/IV:3 | 6 months | 50th (34) | <0.4th (9) | BG | 4 (11) | 3/<2 (11) | |
| 16/IV:1 | Belgian | 4 months | 25th (38) | NR | BG; PV | 38 (4) | NA |
| 17/IV:1 | Spanish | Birth | <0.4th (37) | <<0.4th (84) | BG | 25 (<1) | NA |
| 17/IV:2 | 1 week | <0.4th (40) | <<0.4th (13) | BG; WM | 52 (1) | 70/NA (1) |
BG, basal ganglia; CSF, cerebrospinal fluid; NA, not analysed; NR, not recorded; OFC, occipito‐frontal circumference; PV, periventricular; WCC, white cell count; WM, white matter.
*Abnormal ⩾5 cells/mm3; †normal levels <2 IU/l.
Genotyping
All included families were initially genotyped across the AGS1 critical region. These families either gave logarithm of the odds (LOD) scores of <−2 across this interval or, where the family was too small to allow for statistical exclusion, consanguineous affected individuals showed a number of heterozygous markers within the critical region (data not shown).
Figure 4 Genotype data for families 16 and 17 unlikely to be linked to the AGS2 locus.
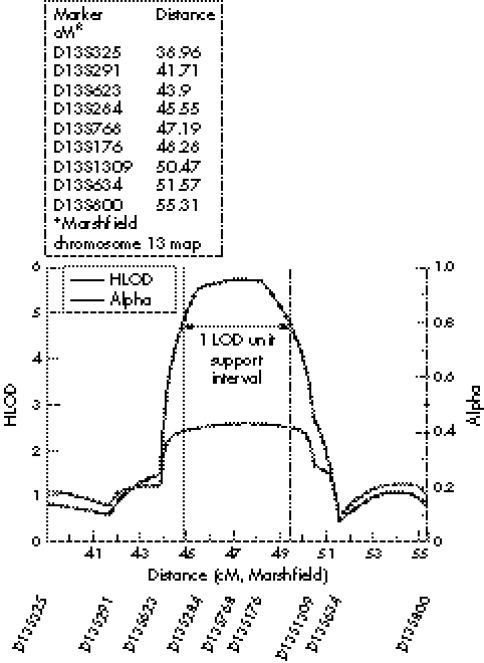
Figure 5 Graph of multipoint HLOD scores across AGS2.
A genome‐wide scan was performed in 10 pedigrees (families 1, 2, 3, 6, 7, 8, 11, 15, 16, and 17) by the National Heart, Lung and Blood Institute genotyping facility (NHLBI; http://research.marshfieldclinic.org/genetics/) using 404 polymorphic markers spaced at approximately 10 cM intervals.20 For higher density genotyping across the AGS2 interval, information regarding marker order and genetic distance was obtained from the Marshfield linkage maps and the Human Genome Browser (http://genome.ucsc.edu/) May 2004 freeze. After individually optimised PCR amplification, markers were analysed using previously described methods.18
Linkage analysis
A model of autosomal recessive inheritance with full penetrance was used with a disease allele frequency estimated at 1 in 500. Marker allele frequencies were calculated from transmitted and non‐transmitted parental alleles with a minimum marker allele frequency set at 0.05. Genetic map distances were taken from the Marshfield map. Pedigree allele inconsistencies were identified using PedCheck.21 Two point analysis was performed with the LINKAGE program.22 Multipoint LOD scores and heterogeneity testing were computed by means of the GENEHUNTER program, version 2.0 beta.23
Results
An initial genome screen was performed using eight consanguineous families and two non‐consanguineous pedigrees. Under the hypothesis of locus heterogeneity, the highest heterogeneity LOD (HLOD) score obtained after two point linkage analysis of this data set was at D13S768 (HLOD score of 2.41 with recombination fraction [θ] = 0.05) (data not shown).
Further genotyping using polymorphic microsatellite markers around D13S768 was undertaken in the original 10 families and an additional seven consanguineous pedigrees satisfying the inclusion criteria (figs 1 and 2). Two point LOD scores for the individual pedigrees are given in table 3 with the highest individual score obtained in family 1. Under the hypothesis of locus heterogeneity, a maximum multipoint HLOD score of 5.75 was obtained at marker D13S768 with α = 0.43 (where α is the population of linked families) (fig 3). Construction of a 1 LOD‐unit support measure suggests the AGS2 critical region encompasses an approximately 4.7 cM interval (fig 3).
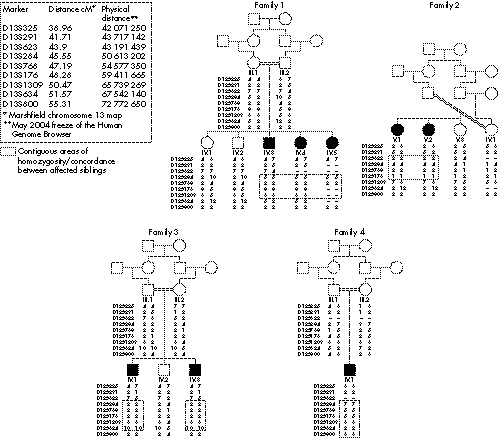
Figure 1 Genotype data for families 1–5 consistent with linkage to AGS2.
Figure 2 Genotype data for families 6–8 consistent with linkage to AGS2.
Table 3 Two point LOD scores (θ = 0) for each family at specified markers.
| Family number | D13S325 | D13S291 | D13S623 | D13S284 | D13S768 | D13S176 | D13S1309 | D13S634 | D13S800 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | −0.75 | −0.73 | −1.47 | 2.46 | 1.19 | 1.34 | 0.96 | −1.1 | 1.34 |
| 2 | −0.6 | −1.20 | 1.11 | 1.25 | 1.67 | 1.13 | −0.61 | −0.81 | 0.52 |
| 3 | −1.6 | −0.97 | −0.88 | 1.04 | 0.76 | 0.68 | 1.22 | 1.09 | −0.83 |
| 4 | 0.8 | 0.71 | ? | 0.89 | 0.99 | 0.57 | 0.29 | 0.94 | 0.81 |
| 5 | 0.23 | 0.40 | 0.88 | 0.79 | 0.34 | 0.49 | 0.29 | −1.63 | −1.58 |
| 6 | −∞ | −∞ | −∞ | 1.32 | 0.73 | 1.07 | −∞ | −∞ | 0.43 |
| 7 | 0.99 | 0.82 | 0.83 | 0.96 | 0.78 | 0.94 | 1.05 | −∞ | −∞ |
| 8 | 1.3 | −1.3 | −0.97 | −0.96 | 0.86 | 1.11 | −2.04 | −1.82 | −1.04 |
| 9 | 0.74 | −1.66 | −1.66 | −1.65 | −1.66 | −1.66 | −1.66 | −1.66 | −1.66 |
| 10 | −∞ | −∞ | −1.32 | −0.11 | −0.46 | −1.02 | 1.29 | −0.26 | 0.38 |
| 11 | −∞ | −∞ | −1.63 | −∞ | 0.27 | −1.55 | −1.63 | −0.96 | −∞ |
| 12 | −∞ | −∞ | −∞ | −∞ | −∞ | −∞ | −0.47 | −∞ | −∞ |
| 13 | −∞ | −1.33 | ? | −1.07 | −∞ | −∞ | −∞ | −∞ | −∞ |
| 14 | −∞ | −1.6 | ? | −1.58 | 0.23 | −∞ | −∞ | −1.78 | −∞ |
| 15 | −∞ | −∞ | −∞ | −∞ | −∞ | −∞ | −∞ | −∞ | −∞ |
| 16 | 0.63 | −1.61 | −1.61 | −1.76 | 0.40 | −∞ | −∞ | −1.61 | −1.66 |
| 17 | −∞ | −∞ | −∞ | −0.76 | −∞ | −∞ | −∞ | −∞ | −∞ |
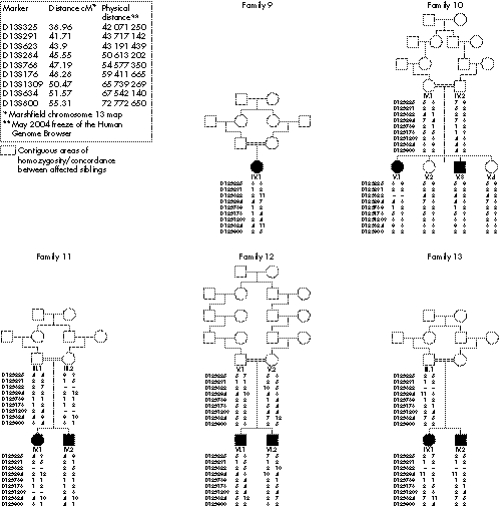
Figure 3 Genotype data for families 11–15 unlikely to be linked to the AGS2 locus.
Discussion
Our search for outstanding AGS disease loci was initiated by the previously observed genetic heterogeneity. We report here the mapping of the AGS2 locus to chromosome 13q14–21, with a maximum multipoint HLOD score of 5.75. This exceeds the accepted threshold value of 3.3 for significant linkage of a Mendelian disorder in the presence of locus heterogeneity.24 On this basis, our data establish the existence of a novel AGS locus, AGS2, on chromosome 13q14–21 and suggest that mutations in at least one other, as yet unmapped, gene produce a similar phenotype.
Based on the minimum region of shared homozygous markers between consanguineous families, the AGS2 critical interval covers a genetic distance of less than 5 cM (D13S284 to D13S1309) (fig 1). However, no single family shows independent linkage to AGS2 (that is, a LOD score >3) (table 3). Consequently, the AGS2 critical interval is most appropriately defined using a 1 LOD‐unit support interval (fig 3).25 On this basis, the AGS2 locus contains approximately 70 genes and unannotated transcripts. Of note, the majority of these genes are positioned towards the centromeric end of the AGS2 locus. Consequently, refinement of the region may significantly reduce the number of positional candidate genes. To this end, work to identify shared ancestral haplotypes within the critical region is ongoing.
We were unable to discern any obvious differences in the clinical characteristics of those families linking to either AGS1, AGS2, or other, as yet undefined, AGS loci (tables 1 and 2). However, phenotypic differences may become apparent when the genes for AGS are eventually identified. In particular, gene identification will enable the issue of phenotypic overlap of AGS with pseudo‐TORCH syndrome to be addressed.9 Some patients demonstrate elevated levels of CSF IFN‐α even when the number of white cells in the CSF is not raised. In the absence of CSF IFN‐α measurements, such cases might be inappropriately considered as pseudo‐TORCH syndrome. Furthermore, Blau et al recently described three children with intracranial calcification and a neurological phenotype reminiscent of AGS in whom both CSF white cells and IFN‐α were consistently normal.26 A molecular basis for classifying these disorders will therefore be of significant clinical utility.
The mechanism responsible for the elevated levels of CSF IFN‐α observed in AGS is not understood. In general terms, such a finding might result from the loss of a negative regulator of IFN‐α production or the presence of an IFN‐α inducer normally absent from the central nervous system. None of the genes within the AGS2 critical interval are known to have such a role and we have been unable to recognise any obvious paralogs or genes from the same cellular pathway within the AGS1 and AGS2 critical intervals.
AGS is a Mendelian mimic of congenital viral infection that continues to be overlooked in the clinical diagnosis of in utero infection.27 The concept of AGS as a primary genetic “interferon‐opathy” highlights a possible unifying theme in the neuropathogenesis of AGS, congenital viral infection, and cerebral systemic lupus erythematosus.9 Consequently, identification of the genes responsible for AGS may provide novel insights into a common neurodegenerative mechanism resulting from exposure of the developing human brain to abnormally high levels of IFN‐α. As in a number of other disorders such as Joubert syndrome, Walker‐Warburg syndrome, and Meckel‐Gruber syndrome, genetic heterogeneity poses a significant obstacle to defining the molecular basis of AGS. Localisation of the AGS2 locus represents an important step towards gene identification in AGS.
Acknowledgements
We wish to express our gratitude to the families for their participation in this study. We would like to acknowledge the National Heart, Lung and Blood Institute for genome scanning and Genethon for sample storage. We wish to acknowledge Dr Geoff Woods for his help in initiating this work. We would like to thank all the clinicians who have contributed samples and clinical data for patients not included in the current paper.
Electronic‐database information
Information on Marshfield linkage maps can be found at http://research.marshfieldclinic.org/genetics and the Human Genome Browser at http://genome. ucsc.edu/.
Abbreviations
AGS - Aicardi‐Goutières syndrome
HLOD - heterogeneity LOD
IFN‐α - interferon α
LOD - logarithm of the odds
Footnotes
Work in the authors' laboratories was funded by the Wellcome Trust and the Medical Research Council
Competing interests: none declared
Information on Marshfield linkage maps can be found at http://research.marshfieldclinic.org/genetics and the Human Genome Browser at http://genome. ucsc.edu/.
References
- 1.Aicardi J, Goutières F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol 19841549–54. [DOI] [PubMed] [Google Scholar]
- 2.Goutières F, Aicardi J, Barth P G, Lebon P. Aicardi‐Goutières syndrome: an update and results of interferon‐α studies. Ann Neurol 199844900–907. [DOI] [PubMed] [Google Scholar]
- 3.Garrec M L, Doret M, Pasquier J C, Till M, Lebon P, Buenerd A, Escalon J, Gaucherand P. Prenatal diagnosis of Aicardi‐Goutières syndrome. Prenat Diagn 20052528–30. [DOI] [PubMed] [Google Scholar]
- 4.Black D N, Watters G V, Andermann E, Dumont C, Kabay M, Kaplan P, Meagher‐Villemure K, Michaud J, O'Gorman G, Reece E. Encephalitis among Cree children in Northern Québec. Ann Neurol 198824483–489. [DOI] [PubMed] [Google Scholar]
- 5.Dale R C, Tang S P, Heckmatt J Z, Tatnall F M. Familial systemic lupus erythematosus and congenital infection‐like syndrome. Neuropediatrics 200031155–158. [DOI] [PubMed] [Google Scholar]
- 6.Aicardi J, Goutières F. Systemic lupus erythematosus or Aicardi‐Goutières syndrome? Neuropediatrics 200031113. [DOI] [PubMed] [Google Scholar]
- 7.Stephenson J B P. Aicardi‐Goutierès syndrome – observations of the Glasgow school. Eur J Paediatr Neurol 20026(Suppl A)A67–A70. [DOI] [PubMed] [Google Scholar]
- 8.Lebon P, Badoual J, Ponsot G, Goutières F, Hémeury‐Cukier F, Aicardi J. Intrathecal synthesis of interferon‐α in infants with progressive familial encephalopathy. J Neurol Sci 198884201–208. [DOI] [PubMed] [Google Scholar]
- 9.Crow Y J, Black D N, Ali M, Bond J, Jackson A P, Lefson M, Michaud J, Roberts E, Stephenson J B, Woods C G, Lebon P. Cree encephalitis is allelic with Aicardi‐Goutieres syndrome: implications for the pathogenesis of disorders of interferon alpha metabolism. J Med Genet 200340183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebon P, Meritet J F, Krivine A, Rozenberg F. Interferon and Aicardi‐Goutieres syndrome. Eur J Paediatr Neurol 20026(Suppl A)A47–A53. [DOI] [PubMed] [Google Scholar]
- 11.Lieberman A P, Pitha P M, Shin H S, Shin M L. Production of tumour necrosis factor and other cytokines by astrocytes stimulated with lipopolysaccharide or a neurotropic virus. Proc Natl Acad Sci U S A 1989866348–6352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiyama H, Ikeda K, Katoh M, McGeer E G, McGeer P L. Expression of MRP14, 27E10, interferon‐α and leukocyte common antigen by reactive microglia in postmortem human brain tissue. J Neuroimmunol 199450195–201. [DOI] [PubMed] [Google Scholar]
- 13.Barth P G, Walter A, van Gelderen I. Aicardi‐Goutières syndrome: a genetic microangiopathy? Acta Neuropathol 199998212–216. [DOI] [PubMed] [Google Scholar]
- 14.Indraccolo S. Undermining tumour angiogenesis by gene therapy: an emerging field. Curr Gene Ther 20044297–308. [DOI] [PubMed] [Google Scholar]
- 15.Cid M C, Hernández‐Rodríguez J, Robert J, del Río A, Casademont J, Coll‐Vinent B, Grau J M, Kleinman H K, Urbano‐Marquez A, Cardellach F. Interferon‐α may exacerbate cryoglobulinemia‐related ischaemic manifestations: an adverse effect potentially related to its anti‐angiogenic activity. Arthritis Rheum 1999421051–1055. [DOI] [PubMed] [Google Scholar]
- 16.Akwa Y, Hassett D E, Eloranta M L, Sandberg K, Masliah E, Powell H, Whitton J L, Bloom F E, Campbell I L. Transgenic expression of IFN‐α in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J Immunol 19981615016–5026. [PubMed] [Google Scholar]
- 17.Campbell I L, Krucker T, Steffensen S, Akwa Y, Powell H C, Lane T, Carr D J, Gold H G, Henriksen S J, Siggins G R. Structural and functional neuropathology in transgenic mice with CNS expression of IFN‐α. Brain Res 199983546–61. [DOI] [PubMed] [Google Scholar]
- 18.Crow Y J, Jackson A P, Roberts E, van Beusekom E, Barth P, Corry P, Ferrie C D, Hamel B C, Jayatunga R, Karbani G, Kalmanchey R, Kelemen A, King M, Kumar R, Livingstone J, Massey R, McWilliam R, Meager A, Rittey C, Stephenson J P B, Tolmie J L, Verrips A, Voit T, van Bokhoven H, Brunner H G, Woods C G. Aicardi‐Goutières syndrome displays genetic heterogeneity with one locus (AGS1) on chromosome 3p21. Am J Hum Genet 200067213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faure S, Bordelais I, Marquette C, Rittey C, Campos‐Castello J, Goutieres F, Ponsot G, Weissenbach J, Lebon P. Aicardi‐Goutieres syndrome: monogenic recessive disease, genetically heterogeneous disease, or multifactorial disease? Clin Genet 199956149–153. [DOI] [PubMed] [Google Scholar]
- 20.Weber J L, Broman K W. Genotyping for human whole‐genome scans: past, present, and future. Adv Genet 20014277–96. [DOI] [PubMed] [Google Scholar]
- 21.O'Connell J R, Weeks D E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 199863259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lathrop G M, Lalouel J ‐ M, Julier C, Ott J. Strategies for multilocus analysis in humans. Proc Natl Acad Sci U S A 1984813443–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruglyak L, Daly M J, Reeve‐Daly M P, Lander E S. Parametric and non‐parametric linkage analysis: a unified multipoint approach. Am J Hum Genet 1996581347–1363. [PMC free article] [PubMed] [Google Scholar]
- 24.Terwilleger J D, Ott J.Handbook of human genetic linkage. Baltimore, MD: Johns Hopkins University Press, 1994235–242.
- 25.Conneally P M, Edwards J H, Kidd K K, Lalouel J M, Morton N E, Ott J, White R. Report of the committee of linkage analysis and reporting. Cytogenet Cell Genet 198540356–359. [DOI] [PubMed] [Google Scholar]
- 26.Blau N, Bonafe L, Krageloh‐Mann I, Thony B, Kierat L, Hausler M, Raemakers V. Cerebrospinal fluid pterins and folates in Aicardi‐Goutierès syndrome: a new phenotype. Neurology 200361642–647. [DOI] [PubMed] [Google Scholar]
- 27.Modlin J F, Grant P E, Makar R S, Roberts D J, Krishnamoorthy K S. Case records of the Massachusetts General Hospital. Weekly clinicopathological exercises. Case 25‐2003. A newborn boy with petechiae and thrombocytopenia. N Engl J Med 2003349691–700. [DOI] [PubMed] [Google Scholar]