Abstract
Introduction
We describe the case of two brothers diagnosed with autism who both carry a paracentic inversion of the short arm of chromosome 4 (46,XY, inv(4)(p12–p15.3)). We have determined that this inversion is inherited from an apparently unaffected mother and unaffected maternal grandfather.
Methods/Results
Using fluorescence in situ hybridisation analysis and Southern blot hybridisation we identified the breakpoints. The proximal breakpoint (4p12) maps to a region containing a cluster of gamma‐aminobutyric acid A (GABA(A)) receptor genes, and directly interrupts the GABRG1 gene, the distal‐most gene of the cluster. We also identified an insertion/deletion polymorphism for a ∼2 kb LINE1 (L1) element that occurs within intron 7 of GABRG1. Our genotype analysis amongst autism families indicated that the L1 deletion allele did not show increased transmission to affected individuals. No linkage disequilibrium was evident between the L1 and single nucleotide polymorphisms in adjacent GABA(A) receptor genes on 4p, where a recent study has identified significant association with autism.
Discussion
Despite this, the identification of an inversion breakpoint disrupting GABRG1 provides solid support for the genetic involvement of the short arm of chromosome 4 in the genetic aetiology of autism, and for the hypothesis of disrupted GABA neurotransmission in autism.
Keywords: autism, GABA receptor, GABRG1, inversion, 4p
Autism (MIM 209850) is a severe neurodevelopmental disorder, identified by deficits in social interaction and communication skills, coupled with unusual patterns of focus, along with stereotyped behaviours. It is believed there is a strong genetic component underlying the aetiology of autism, although the pattern of inheritance is not straightforward and is likely to involve genetic heterogeneity, gene‐gene interaction, and the added complication of possible environmental determinants. In a number of genetic diseases, structural chromosomal changes (deletions, duplications, translocations, or inversions) that segregate with the disease phenotype have served to narrow the search for disease determining genes to specific chromosome regions, and subsequently to specific candidate genes. Numerous chromosomal abnormalities among autistic individuals have been described, and include almost all chromosomes.1,2 In this study we describe the case of two autistic brothers who have inherited a paracentric inversion of the short arm of chromosome 4 from an apparently healthy mother and maternal grandfather.
The proband was born to a 39 year old woman and 40 year old father. The family history was negative for consanguinity, congenital anomalies, recurrent miscarriages, or other individuals with autism. The pregnancy and delivery were uncomplicated, and birth weight was 3178 g. The child began walking at 13 months and his first word was at 13 months. By 18 months he was using approximately 10 single words, which he subsequently lost. At 5 years 10 months he was using 5–10 words but articulating less well than at 18 months. His communication was primarily non‐verbal. He was non‐dysmorphic on physical examination at the age of 5 years 10 months. His height was at the 75th percentile, weight was at the 50th percentile, and head circumference was at the 75th percentile. Computed tomography (CT) brain scan at 3.5 years of age proved normal except for a deviated septum pellucidum. Urine organic acids, serum amino acids, and lactate and ammonia levels were all normal. Fragile X and 15q11–q13 methylation tests were both normal. Standard chromosome analysis revealed the karyotype 46,XY, inv(4)(p12p15.3)mat. At the age of 12 years 9 months, the Autism Diagnostic Interview‐Revised (ADI‐R)3 and the Autism Diagnostic Observation Schedule (ADOS)4 were administered, giving scores that were compatible with a diagnosis of autism. Tests were also administered for intellectual functioning (Leiter International Performance Scale), language (Peabody Picture Vocabulary Test–3rd Edition (PPVT IIIA)), and adaptive behaviour (Vineland Adaptive Behaviour Scales). While the proband was unable to complete the tests for intellectual functioning and language, the adaptive behaviour tests indicated profound developmental delay, with functioning below the 3 year age level.
The second male child was born 13 months after the first. The pregnancy and delivery were uncomplicated and birth weight was 3291 g. Neonatal seizures occurred within the first 24 h, but no further seizure activity was reported. The child was walking at 1 year of age. His speech was delayed and his first words were at 3.5 years of age. He was reported to have behavioural difficulties including temper tantrums. He did not appear dysmorphic on examination at the age of 5 years. A right sided ptosis, present since birth, was noted. His height was at the 50th percentile, weight was between the 25th and 50th percentiles, and head circumference was at the 50th percentile. CT brain scan proved normal. Urine organic acids, serum amino acids, and lactate and ammonia levels were all normal. ADOS and ADI were administered at the ages of 7 and 11, respectively, leading to a diagnosis of autism. Examination of intellectual functioning (Leiter) at the age of 11 years 8 months indicated mild developmental delay at the 6–6.5 year level. Language assessment (using Oral and Written Language Scales (OWLS)) indicated moderate developmental delay (at the 4 year 7 month level), while adaptive behaviour was in the severe developmental delay range (4.5–5 year level). Fluorescence in situ hybridisation (FISH) for 15q11–q13 was normal, however standard chromosome analysis revealed the karyotype 46,XY, inv(4)(p12p15.3)mat.
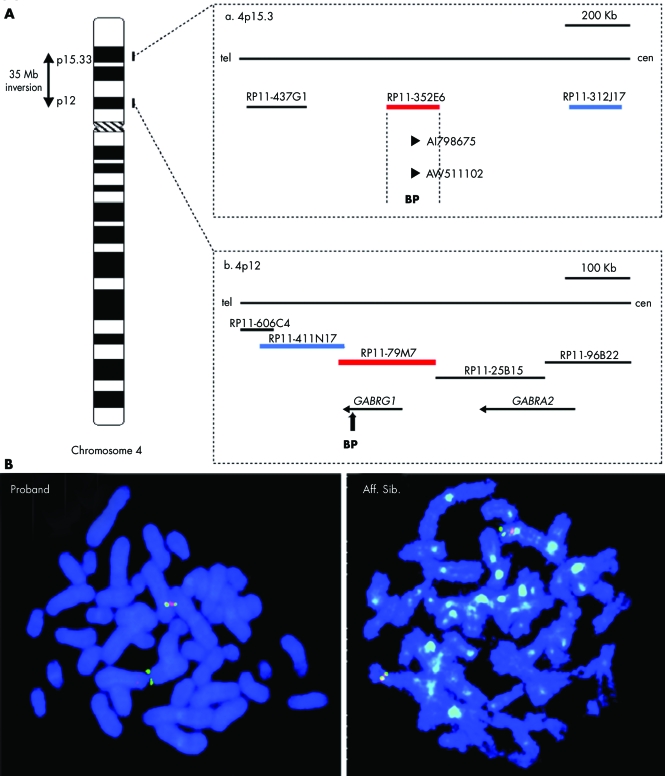
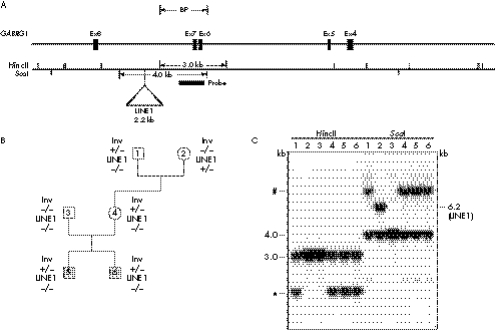
Bacterial artificial chromosome (BAC) clones from the RP11 human genomic library were used for FISH to narrow down the inversion breakpoints. BAC clone RP11‐775M18 (GenBank accession number AC096592) and RP11‐79M7 were shown to span the proximal breakpoint, and clone RP11‐352E06 (AC093809) the distal breakpoint (fig 1). The distal breakpoint at 4p15.3 maps to a region that lies close (∼12 Mb) to a region previously indicated to show linkage to autism (markers D4S2936 and D4S412)5. Genomic analysis of the sequence surrounding the 4p12 inversion breakpoint indicated the presence of a cluster of four gamma‐aminobutyric acid A (GABA(A)) receptor genes (fig 1). To refine the localisation of the proximal breakpoint, a number of probes across the GABRG1 gene were generated by long range PCR, radioactively labelled (α 32P dCTP) by random priming, and hybridised to Southern blots containing restriction digested genomic DNA from the proband, from family members, and from controls. The breakpoint was narrowed to within a ∼2.2 kb segment, lying between ScaI and HincII restriction sites, spanning from 267 bp upstream of the intron 5/exon 6 acceptor splice junction to within intron 7 (fig 2). The Southern hybridisation also revealed the presence of a polymorphic band present in the maternal grandmother and in some controls. This corresponds to a ∼2 kb LINE1 (L1) element (L1HS) that lies 461 bp downstream from the breakpoint critical region. This L1 element, which is present in the NCBI Build 35 human genome sequence (but not in the Pan troglodytes chimpanzee genome), was analysed by Southern hybridisation and by PCR, and shown to be polymorphic. All members of the family (proband, affected brother, parents, and maternal grandfather) are homozygous for the deletion, except for the maternal grandmother who is heterozygous for the polymorphism (fig 2).
Figure 1 (A) Physical map of the inv(4p) distal and proximal breakpoint regions, indicating BAC and fosmid clones used for FISH mapping of breakpoints. (a, b) Expressed sequence tags and known genes mapping to the distal and proximal breakpoint regions. (B) FISH studies of the inv(4p) regions in the proband, affected sibling, and mother, using two probes, one for the distal region, RP11‐312J17 which is inverted here. In a non‐rearranged genome it lies just proximal to the 4p15.3 breakpoint and the one for the proximal region, RP11‐79M7, which gives a signal for both 4p12 and 4p15, and thus spans the breakpoint. BP, breakpoint.
Figure 2 (A) Physical map of the GABRG1 gene on human chromosome 4p12. A precise breakpoint region is located in intron 7 of the GABRG1 gene. (B) Pedigree structure. 1: maternal grandfather, 2: maternal grandmother, 3: father, 4: mother, 5: proband, 6: affected sibling. The chromosome 4p inversion is inherited from the maternal grandfather. The L1 element insertion is only present, in heterozygous form, in the grandmother. (C) Southern blot analysis of the inversion region of GABRG1. Five PCR probes spanning the region were generated by PCR from human genomic DNA. The probe shown was generated using the primer sequences: 5′‐CCCTGTTTAGATGGCCAAGA‐3′ and 5′‐TCCGTAGAAGTGGCTGATCC‐3′; probes were labelled by random priming with α 32P dCTP, using standard protocols. Genomic DNA was digested with the indicated restriction enzymes. The locations of restriction sites for HincII and ScaI are shown in fig 2A. The resulting fragments were separated by electrophoresis on a 0.8% agarose gel, and transferred by Southern blotting onto Hybond‐N+ membranes (Amersham, Little Chalfont, UK). The Southern blots were hybridised with the PCR probe shown in the restriction maps. For HincII, the 3.0 kb fragments indicate the normal allele, and the asterisk (“*”) indicates the inverted allele fragments. For ScaI, the 4.0 kb fragments indicate the normal allele, and “#” indicates the inverted allele fragments, and the 6.0 kb fragment indicates the LINE1 polymorphism. Therefore, lanes 1, 4, 5, and 6 are positive for the 4p chromosome inversion. BP, breakpoint.
De novo insertions of retrotransposable elements, such as short interspersed nucleotide elements (SINEs) and long interspersed nucleotide elements (LINEs), have been reported in association with many genetic diseases (reviewed in Ostertag and Kazazian6). It is quite possible that the polymorphic L1 insertion within the GABRG1 gene attenuates its transcription, since it has been shown in vitro that inserting L1 sequences significantly reduces RNA expression, mainly through poor transcriptional elongation.7 Furthermore, it has been suggested that L1 elements act as molecular rheostats, fine tuning the transcriptome across evolution and acting as a medium for intra‐species differences, whereby heritable variations of L1 elements result in altered gene expression levels and hence phenotypic differences.7 This may have a direct impact on the way the brain develops and functions, since it has been shown recently, using in vitro and in vivo models, that L1 elements are active in neuronal progenitor cells.8 Hence, we hypothesised that the L1 insertion would be preferentially transmitted to affected offspring in autism families. To test this theory, we genotyped 262 autism families (1105 individuals) as well 40 reference Centre d'Etude Polymorphisme Humaine CEU families, using PCR primers either flanking the insertion or with one primer nested within the L1 sequence (table 1). All probands and siblings were diagnosed using ADI‐R and ADOS, and had a DSM‐IV diagnosis of autism. Using the family‐based association test,9 there was no association between the L1 polymorphism and autism (using an additive model, p = 0.59; 150 informative families).
Table 1 PCR primers and amplicon size for mutation screening and for LINE1 polymorphism genotyping.
| Exon 1 | Forward primer 5′‐3′ | Reverse primer 5′‐3′ | Product size (bp) |
|---|---|---|---|
| 1 | AGAGCAGAAGGGGAGAAAGG | GGGGAAAGGGGTAGATAGCA | 550 |
| 2 | TGTAATACCTTTTTGTCTGCCAT | TGGTCATCAAAATCCCAGAAT | 307 |
| 3 | AAGGCTACATTTCTGCATTTCA | CACATGCGAATTCTATTTTGGA | 294 |
| 4 | TCCGAAAAGACTGAGTTTAACT | TGGAAAATAAATGAGAGCAACT | 388 |
| 5 | CCTAGTTGACTGAAGAGGAAAT | ACAGCTTCTAAGGTTAAAATTC | 352 |
| 6 | AAATATTCTATGTTTGGCATATC | TGCTAGCATCATTTCACTATTTG | 307 |
| 7 | AAGAATACTCAAATAGTGAAAT | TGTGACTATGAACTTTTATCAC | 341 |
| 8 | TGCCAAATCAGATGGAAGAA | CTTTGGTTATGGGTCTCAAGC | 401 |
| 9 | CCTGTCAGTCTCCTAGAGTTTGC | TTTAACCTACTGGATTTTGCAAC | 580 |
| LINE1 | |||
| Flanking | CTTCCCCTACTCAAGATCCTACAAT | GCTAGAGTGTCATGAAGAGAAAAGC | LINE1+: 3 kb; LINE1−: 1 kb |
| Internal | CACATTGTTCCCATCGTCAG | TGACAAACCCACAGCCAATA | LINE1+: 1.4 kb |
To determine if the L1 indel was in linkage disequilibrium with single nucleotide polymorphisms (SNPs) within the GABA gene cluster (including some of those genotyped by Ma et al10), we genotyped it in the CEU samples from the HapMap data.11 Using Haploview v3.212 and release 18 of the HapMap data, we determined that the L1 indel was in strong linkage disequilibrium (r2>0.8) with 28 SNPs (rs11944736, rs6824076, rs2089156, rs6820046, rs1394345, rs970836, rs12502104, rs1604872, rs2882421, rs1504491, rs10938421, rs1566854, rs1504493, rs982461, rs7690876, rs13147984, rs1391173, rs10014198, rs7661204, rs1566856, rs7683876, rs1497569, rs1497570, rs1540741, rs1497571, rs1032850, rs1948609, rs6838525). The only SNP in GABRG1 that is both in strong linkage disequilibrium with the L1 indel and also genotyped by Ma et al10 is rs1497571, which produced a single marker (p = 0.90) in their study for association with autism. Therefore, neither the L1 indel (from our data) nor the Ma et al10 data on rs1497571 (which is in strong linkage disequilibrium with the L1 indel) shows association with autism.
Genomic analysis of the distal (4p15) breakpoint has identified an expressed sequence tag (EST) cluster. Sequences were obtained for the EST clones IMAGE #2348462 (AI798675), #2912255 (AW51110), and #2977939 (AW663909). Comparison of the EST clone sequences with the genomic sequence for BAC clone RP11‐352E06 (AC093809) revealed the presence of a gene. This gene spans 35 kb and contains five exons, with alternative splicing of exons 3, 4, and 5. However, neither the long nor the short isoform appears to contain an open reading frame. The genomic region surrounding the distal breakpoint is a gene‐poor region. Apart from the EST cluster within RP11‐352E06, the nearest other genes are EST clusters BC042433 ∼244 kb distal to RP11‐352E06, and BE501997 ∼636 kb proximal to RP11‐352E06, again with neither open reading frame nor homology to any known genes. The nearest known coding genes are HS3ST1 (a heparan sulphate biosynthetic enzyme) at 1.07 Mb distal to RP11‐352E06. HSP90BB (a heat shock protein) and RAB28 (a member of the RAS oncogene family) at 676 kb and 710 kb proximal to RP11‐352E06. The nearest known gene that could be considered a functional candidate is the neurotransmitter receptor gene DRD5, which is over 2.7 Mb distal to the breakpoint, however, it is unlikely that the translocation could affect a gene this far away by position effect.
Array comparative genomic hybridisation analysis was performed using a 1 Mb resolution BAC microarray (Spectral Genomics, Houston, TX) in order to test for the presence of other, cryptic genomic rearrangements in DNA from the inv(4p) proband and sibling, and revealed no additional changes. Deletions at 13q31.3 (RP11‐80B16) and 15q25.3 (RP11‐80J8) as well as a duplication at 14q32.2 (RP11‐90G22) were all noted but appear to be known genomic polymorphisms (see http://projects.tcag.ca/variation/).13
In order to assess whether GABRG1 frequently harbours coding or splice site mutations involved in autism, the coding region of GABRG1 was screened by direct sequencing using genomic DNA from 32 unrelated autistic patients from multiplex families, which were collected at The Hospital for Sick Children, were screened for sequence variants. PCR primers were selected from the intronic sequences flanking each exon using Primer3. Nine amplicons were designed for GABRG1. Primer sequences are shown in table 1. Sequencing was performed using BigDye cycle sequencing and analysed on an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). We identified a total of six sequence variants in GABRG1, none of which results in an amino acid substitution or is close enough to a splice site to result in disruption of splicing. Comparison with the SNP database, dbSNP, indicated that two of these variants are known SNPs. The frequency and positions of the changes identified through our screening are shown in table 2.
Table 2 Mutation screening in 32 autism probands.
| Location in gene | Variant | Comments | Frequency of variant in 32 autism probands: allele count (total number of chromosomes tested) |
|---|---|---|---|
| 5′UTR | −36T>G | 4 (64) | |
| 5′UTR | −65C>G | 4 (64) | |
| Exon 3 | A>G; Thr 88 Thr | 263 bp from START codon | Known SNP: rs976156 |
| Intron 6 | IVS6 −26 [C‐A] | 2 (64) | |
| Intron 7 | IVS7 +27 [T‐A] | 4 (64) | |
| 3′UTR | T>C | 59 bp downstream from STOP codon | Known SNP: rs6447493 |
It is possible that the proximal breakpoint may be disrupting one or more of the other GABA receptor genes within the 4p cluster by position effect and that the other 4p GABA receptor genes are involved in autism. It is plausible that disruption of any of the four GABA receptor genes in the 4p cluster may have a similar effect, since it would potentially affect the assembly or the integrity of the assembled heteropentameric GABA receptor. Ideally, we would have liked to use transcription analysis to demonstrate the effect of the translocation on adjacent genes, however, the low level of gene transcription for the 4p GABA receptor genes in the tissues available to us (lymphoblast cells) prevented us exploring this avenue of research.
We have described two autistic brothers carrying a paracentric inversion of chromosome 4p. Our molecular genetic analysis of the proximal breakpoint on 4p12 in the family described here highlights specific candidate genes for autism. These genes are of particular interest since they encode receptors for gamma‐aminobutyric acid (GABRG1, GABRA2, GABRA4, and GABRB1). These GABA type A receptors are heteropentameric ligand gated chloride channels that mediate fast synaptic inhibition in the mammalian brain.14 The receptors are assembled from a number of different subunits including α1–6, β1–3, γ1–3, δ, ε, π, and θ.15,16 Stoichiometric studies have suggested that the most common subunit composition is likely to be two α and two β subunits plus one γ subunit.17
GABA receptor genes are of particular interest in studies of autism for a number of reasons. Elevated plasma GABA levels have been found in infants with autism,18 thus suggesting a possible role for GABA as a biochemical marker for this disorder. Also, a quantitative receptor autoradiographic study showed significantly reduced hippocampal GABA receptor binding in autism.19 From the genetic point of view, the 15q11–q13 region which harbours another cluster of the GABA(A) receptors has been a focus of autism studies for a number of years. The most common chromosomal aberration among autism patients occurs within this region.1 Additionally, a number of genetic studies have implicated the GABA receptor cluster within this region.20,21,22,23,24 A recent family based association and linkage disequilibrium study of GABA(A) receptor genes has indicated this 4p cluster as showing the strongest evidence (of the GABA(A) gene clusters included in the study) of involvement in autism, and in particular the GABRA4 gene.10 Six additional autism cases with chromosomal rearrangements involving the short arm of chromosome 4 have been reported in The Autism Chromosome Rearrangement Database (available at http://projects.tcag.ca/autism/) suggesting that the short arm of chromosome 4 could harbour one or more genes implicated in the aetiology of autism. Two of these, a duplication of 4p12–p1325 and an unbalanced translocation, der(4) t(4;4) (p12q28),26 may also involve the 4p GABA receptor cluster, however, the boundaries of the duplication have not yet been established, hence it is not known whether the same locus is involved.
Our discovery of an inversion breakpoint disrupting one of the genes within the same GABA(A) gene cluster in two autistic brothers adds further evidence in support of a role for GABA genes, and the 4p GABA(A) receptor cluster in particular, in the aetiology of autism.
Electronic‐database information
The following web sites have been mentioned in this report: Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/; Primer3: http://www‐genome.wi.mit.edu/cgi‐bin/primer/primer3_www.cgi; dbSNP: http://www.ncbi.nlm.nih.gov/SNP/; Database of Genomic Variants: http://projects.tcag.ca/variation/; Autism Chromosome Rearrangement Database: http://projects.tcag.ca/autism/
Abbreviations
ADI‐R - Autism Diagnostic Interview‐Revised
ADOS - Autism Diagnostic Observation Schedule
BAC - bacterial artificial chromosome
CT - computed tomography
EST - expressed sequence tag
FISH - fluorescence in situ hybridisation
GABA(A) - gamma‐aminobutyric acid A
L1 - LINE1
LINE - long interspersed nucleotide element
PPVT IIIA - Peabody Picture Vocabulary Test–3rd Edition
SINE - short interspersed nucleotide element
SNP - single nucleotide polymorphism
Footnotes
This work was supported by Genome Canada/Ontario Genomics Institute, the Howard Hughes Medical Institute (HHMI), the McLaughlin Centre for Molecular Medicine, and The Centre for Applied Genomics. SC received a Postdoctoral Fellowship, in part, through the Hospital for Sick Children Research Training Centre. JBV is a Seaver Foundation and National Alliance for Research into Schizophrenia and Depression (NARSAD) Young Investigator. ADP holds a Canada Research Chair in Genetics of Complex Diseases. SWS is a Scientist of the Canadian Institutes of Health Research and an International Scholar of the HHMI.
Competing interests: none declared
Ethics statement: the study was approved by the Research Ethics Board of The Hospital for Sick Children and CAMH, and informed, written consent was obtained from the participating families
The following web sites have been mentioned in this report: Online Mendelian Inheritance in Man (OMIM): http://www.ncbi.nlm.nih.gov/Omim/; Primer3: http://www‐genome.wi.mit.edu/cgi‐bin/primer/primer3_www.cgi; dbSNP: http://www.ncbi.nlm.nih.gov/SNP/; Database of Genomic Variants: http://projects.tcag.ca/variation/; Autism Chromosome Rearrangement Database: http://projects.tcag.ca/autism/
References
- 1.Gillberg C. Chromosomal disorders and autism. J Autism Dev Disord 199828415–425. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Zwaigenbaum L, Szatmari P, Scherer S W. Molecular cytogenetics of autism. Curr Genomics 20044347–364. [Google Scholar]
- 3.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview‐Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 199424659–685. [DOI] [PubMed] [Google Scholar]
- 4.Lord C, Rutter M, Goode S, Heemsbergen J, Jordan H, Mawhood L, Schopler E. Autism diagnostic observation schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord 198919185–212. [DOI] [PubMed] [Google Scholar]
- 5. A full genome screen for autism with evidence for linkage to a region on chromosome 7q. International Molecular Genetic Study of Autism Consortium. Hum Mol Genet 19987571–578. [DOI] [PubMed] [Google Scholar]
- 6.Ostertag E M, Kazazian H H., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet 200135501–538. [DOI] [PubMed] [Google Scholar]
- 7.Han J S, Szak S T, Boeke J D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 2004429268–274. [DOI] [PubMed] [Google Scholar]
- 8.Muotri A R, Chu V T, Marchetto M C, Deng W, Moran J V, Gage F H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 2005435903–910. [DOI] [PubMed] [Google Scholar]
- 9.Horvath S, Xu X, Lake S L, Silverman E K, Weiss S T, Laird N M. Family‐based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol 20042661–69. [DOI] [PubMed] [Google Scholar]
- 10.Ma D Q, Whitehead P L, Menold M M, Martin E R, Ashley‐Koch A E, Mei H, Ritchie M D, Delong G R, Abramson R K, Wright H H, Cuccaro M L, Hussman J P, Gilbert J R, Pericak‐Vance M A. Identification of significant association and gene‐gene interaction of GABA receptor subunit genes in autism. Am J Hum Genet 200577377–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The International HapMap Consortium The International HapMap Project. Nature 2003426789–796. [DOI] [PubMed] [Google Scholar]
- 12.Barrett J C, Fry B, Maller J, Daly M J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 200521263–265. [DOI] [PubMed] [Google Scholar]
- 13.Iafrate A J, Feuk L, Rivera M N, Listewnik M L, Donahoe P K, Qi Y, Scherer S W, Lee C. Detection of large‐scale variation in the human genome. Nat Genet 200436949–951. [DOI] [PubMed] [Google Scholar]
- 14.Schofield P R, Darlison M G, Fujita N, Burt D R, Stephenson F A, Rodriguez H, Rhee L M, Ramachandran J, Reale V, Glencorse T A, Seeburg P H, Barnard E A. Sequence and functional expression of the GABA A receptor shows a ligand‐gated receptor super‐family. Nature 1987328221–227. [DOI] [PubMed] [Google Scholar]
- 15.Barnard E A, Skolnick P, Olsen R W, Mohler H, Sieghart W, Biggio G, Braestrup C, Bateson A N, Langer S Z. International Union of Pharmacology. XV. Subtypes of gamma‐aminobutyric acid A receptors: classification on the basis of subunit structure and receptor function, Pharmacol Rev 199850291–313. [PubMed] [Google Scholar]
- 16.Nayeem N, Green T P, Martin I L, Barnard E A. Quaternary structure of the native GABAA receptor determined by electron microscopic image analysis. J Neurochem 199462815–818. [DOI] [PubMed] [Google Scholar]
- 17.Chang Y, Wang R, Barot S, Weiss D S. Stoichiometry of a recombinant GABAA receptor. J Neurosci 1996165415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhossche D, Applegate H, Abraham A, Maertens P, Bland L, Bencsath A, Martinez J. Elevated plasma gamma‐aminobutyric acid (GABA) levels in autistic youngsters: stimulus for a GABA hypothesis of autism. Med Sci Monit 20028PR1–PR6. [PubMed] [Google Scholar]
- 19.Blatt G J, Fitzgerald C M, Guptill J T, Booker A B, Kemper T L, Bauman M L. Density and distribution of hippocampal neurotransmitter receptors in autism: an autoradiographic study. J Autism Dev Disord 200131537–543. [DOI] [PubMed] [Google Scholar]
- 20.Buxbaum J D, Silverman J M, Smith C J, Greenberg D A, Kilifarski M, Reichert J, Cook E H, Jr, Fang Y, Song C Y, Vitale R. Association between a GABRB3 polymorphism and autism. Mol Psychiatry 20027311–316. [DOI] [PubMed] [Google Scholar]
- 21.Cook E H, Jr, Courchesne R Y, Cox N J, Lord C, Gonen D, Guter S J, Lincoln A, Nix K, Haas R, Leventhal B L, Courchesne E. Linkage‐disequilibrium mapping of autistic disorder, with 15q11–13 markers. Am J Hum Genet 1998621077–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCauley J L, Olson L M, Delahanty R, Amin T, Nurmi E L, Organ E L, Jacobs M M, Folstein S E, Haines J L, Sutcliffe J S. A linkage disequilibrium map of the 1‐Mb 15q12 GABA(A) receptor subunit cluster and association to autism. Am J Med Genet B Neuropsychiatr Genet 200413151–59. [DOI] [PubMed] [Google Scholar]
- 23.Menold M M, Shao Y, Wolpert C M, Donnelly S L, Raiford K L, Martin E R, Ravan S A, Abramson R K, Wright H H, Delong G R, Cuccaro M L, Pericak‐Vance M A, Gilbert J R. Association analysis of chromosome 15 GABA(A) receptor subunit genes in autistic disorder. J Neurogenet 200115245–259. [DOI] [PubMed] [Google Scholar]
- 24.Nurmi E L, Amin T, Olson L M, Jacobs M M, McCauley J L, Lam A Y, Organ E L, Folstein S E, Haines J L, Sutcliffe J S. Dense linkage disequilibrium mapping in the 15q11–q13 maternal expression domain yields evidence for association in autism. Mol Psychiatry 20038624–634. [DOI] [PubMed] [Google Scholar]
- 25.Sabaratnam M, Turk J, Vroegop P. Case report: autistic disorder and chromosomal abnormality 46, XX duplication (4) p12–p13. Eur Child Adolesc Psychiatry 20009307–311. [DOI] [PubMed] [Google Scholar]
- 26.Medne L, Russell K, Ming J, Krantz I D, Souders M, Levy S, Gupta A, Spinner N B, Zackai E H, Morrissette J J D. Subtelomeric FISH analysis in 108 autistic patients as adjunct to chromosome analysis and fragile X testing. Am J Hum Genet 200373A847 [Google Scholar]