Abstract
In addition to the hallmark neurological manifestations of Huntington's disease (HD), weight loss with metabolic dysfunction is often observed in the later stages of disease progression and is associated with poor prognosis. The mechanism for weight loss in HD is unknown. Using two mouse models of HD, the R6/2 transgenic and CAG140 knock-in mouse strains, we demonstrate that adipose tissue dysfunction is detectable at early ages and becomes more pronounced as the disease progresses. Adipocytes acquire a ‘de-differentiated’ phenotype characterized by impaired expression of fat storage genes. In addition, HD mice exhibit reduced levels of leptin and adiponectin, adipose tissue-derived hormones that regulate food intake and glucose metabolism. Importantly, some of these changes occur prior to weight loss and development of some of the characteristic neurological symptoms. We demonstrate that impaired gene expression and lipid accumulation in adipocytes can be recapitulated by expression of an inducible mutant huntingtin transgene in an adipocyte cell line and that mutant huntingtin inhibits transcriptional activity of the PGC-1α co-activator in adipocytes, which may contribute to aberrant gene expression. Thus, our findings indicate that mutant huntingtin has direct detrimental effects in cell types other than neurons. The results also indicate that circulating adipose-tissue-derived hormones may be accessible markers for HD prognosis and progression and suggest that adipose tissue may be a useful therapeutic target to improve standard of life for HD patients.
INTRODUCTION
Huntington's disease (HD) is a fatal genetic disorder caused by a mutation in the huntingtin (HTT) gene (1). In addition to the hallmark neurological manifestations of HD, weight loss is often observed in the course of the disease, becoming malignant at the late stages (2–5). Patients exhibiting higher body mass index at presentation tend to experience slower disease progression (6). The weight loss occurs despite normal to very high caloric intake (2–4,7). It had been previously proposed that weight loss in HD patients is due to increased energy expenditure associated with hyperkinesia or dystonia, but this is unlikely to explain the loss, which occurs in HD patients with minimal chorea at early stages of the disease (8). One possibility is that weight loss and metabolic abnormalities are secondary to central degeneration that is known to occur in the cortex, striatum and other areas such as the hypothalamus, which in turn could affect food intake or energy expenditure (9). It has also been proposed that alterations in the diurnal cycle of hormones involved in the HPA axis could lead to weight loss; however, studies in HD mouse models suggest that the alterations in the HPA axis may reflect stress responses (10). Another possibility is that metabolic irregularities result from direct effects of the huntingtin gene defect in peripheral tissues that influence metabolic homeostasis. Since weight loss and metabolic perturbations may contribute to the overall decline in health of HD patients, an understanding of the mechanisms involved may be valuable for designing therapeutic interventions. In addition, identifying predictive (subclinical) factors or markers may help to identify HD patients who are more susceptible to weight loss.
Evidence from studies in mouse models suggests that HD progression is indeed characterized by changes in peripheral tissues in addition to the brain. For example, the R6/2 mouse strain, which expresses a transgene encoding the N-terminal 3% of the human huntingtin protein with an expanded glutamine tract (11), exhibits alterations in key metabolic tissues such as adipose tissue (12,13), skeletal muscle (14–16) and pancreas (17–19). Abnormalities have been described in both white and brown adipose tissue of the R6/2 strain, suggesting alterations in both energy storage (white adipose tissue) and energy dissipation (brown adipose tissue). Specifically, the R6/2 mouse fails to induce thermogenesis in response to cold exposure, indicating impaired brown adipose tissue function (13). In addition, R6/2 mice exhibit an increase in white adipose tissue mass at 8–9 weeks of age, associated with reduced lipolysis of stored fat (12). This is followed by loss of fat tissue and wasting by 12 weeks, which has been associated with increased energy expenditure (20,21). These observations suggest that adipose tissue alterations may contribute to the progression of HD.
In recent years, white adipose tissue has been shown to be a key modulator of metabolism in humans and other animals. Conditions in which white adipose tissue mass is abnormally increased (obesity) or decreased (lipodystrophy) lead to impaired metabolic homeostasis manifested as glucose intolerance and insulin resistance, which may progress to diabetes (22,23). Weight loss in human HD patients and mouse models is also associated with the development of glucose intolerance and insulin resistance, which may progress to pancreatic beta cell failure and diabetes (17,18,24,25). A primary mechanism by which adipose tissue influences glucose homeostasis is through the production of adipokine hormones such as leptin and adiponectin, which are secreted into the circulation and act in tissues with key roles in energy metabolism. In general, leptin levels increase with obesity, whereas adiponectin levels decrease with obesity, and the two have opposing effects on glucose metabolism, insulin sensitivity and energy expenditure in tissues such as skeletal muscle and liver. Thus, leptin acts in the hypothalamus to influence food intake and energy expenditure, and adiponectin acts on skeletal muscle and liver to promote glucose uptake and inhibit glucose production (26–28). Abnormal levels of these adipokines have been used as a marker for insulin resistance and atherosclerotic risk (29–31), but they have not been evaluated during the progression of HD.
Here we determine that disease progression in two independent HD mouse models—R6/2 transgenic (Tg) and CAG140 knock-in (KI) mice—is characterized by alterations in the expression of key adipose tissue genes and in adipokine secretion. We also demonstrate that the mechanism for altered adipocyte gene expression is a cell-intrinsic defect in adipocytes, as it can be replicated by expression of an inducible transgene in cultured adipocytes. These findings indicate that metabolic disturbances in HD may result from direct effects in adipose tissue and raise the possibility that treatment of peripheral abnormalities in HD patients may be a useful therapeutic intervention to improve some symptoms. They also suggest that circulating adipose tissue-derived hormone levels may serve as accessible subclinical markers to monitor HD progression and therapeutic efficacy.
RESULTS
Altered body weight and fat mass in R6/2 Tg and CAG140 KI mice
We studied the potential role of white adipose tissue abnormalities during the progression of HD in two independent mouse models. The R6/2 Tg mouse is a model of rapid disease progression that is fatal within 12–15 weeks due to the expression of a transgene consisting of exon 1 of the human huntingtin gene containing an expanded CAG repeat (reviewed in 32). The CAG140 KI mouse, which carries a chimeric mouse/human exon 1 containing an expanded CAG repeat inserted into the endogenous mouse Hdh gene, exhibits a slower disease course with a lifespan of about 2 years (33,34).
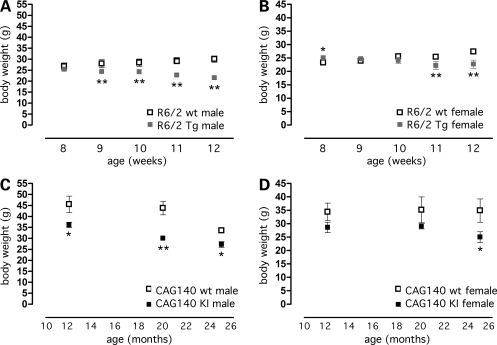
Both R6/2 Tg and CAG140 KI mice exhibited alterations in body weight during disease progression, although with different timing due to the distinct disease time course in the two models. Thus, compared with wild-type animals, R6/2 mice had normal or slightly increased body weight at 8 weeks of age and reduced body weight by 9 weeks (male) or 11 weeks (female) [Fig. 1A and B; males: genotype × age interaction F(4,32) = 11.6, P < 0.0001; females: genotype × age interaction F(4,44) = 12.6, P < 0.0001], in agreement with previous reports (12). CAG140 KI mice had normal body weight through 7 months of age (data not shown), but reduced body weight compared with wild-type controls beginning at 12 (males) or 25 months (females) [Fig. 1C and D; males: effect of genotype F(1,45) = 28.7, P < 0.0001; females: effect of genotype F(1,27) = 7.46, P < 0.02]. The weight loss late in the disease course in both models is reminiscent of that observed in HD patients (2–5).
Figure 1.
Body weight curves for R6/2 transgenic and CAG140 KI mice. Body weight was determined in groups of transgenic (Tg) and KI mice and their corresponding wild-type (wt) littermates at the indicated time points. (A) Male R6/2 Tg (n = 5) and wt (n = 5) mice weighed repeatedly. (B) Female R6/2 Tg (n = 4) and wt (n = 9) mice weighed repeatedly. (C) Male CAG140 KI (n = 8–13) and wt (n = 4–11) mice, separate groups used for each age. (D) Female CAG140 KI (n = 3–6) and wt (n = 5–6) mice, separate groups used for each age. Body weight of CAG140 KI mice aged 1–7 months was indistinguishable from wt mice (data not shown). *P < 0.05; **P < 0.01 versus corresponding wt group. Values represent mean ± SEM.
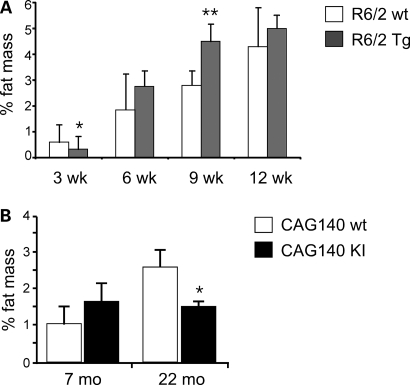
Although male mice showed more pronounced body weight differences, subsequent studies were performed with females due to the availability of greater numbers of female mice. Both the R6/2 and CAG140 strains exhibited age-dependent alterations in white adipose tissue mass. R6/2 mice were analyzed at four ages: 3 weeks (prior to the onset of motor deficits), 6 weeks (commensurate with the onset of motor deficits), 9 weeks (commensurate with the appearance of overt behavioral abnormalities) and 12 weeks (advanced disease stage with reduced body weight). When expressed as % body weight, R6/2 mice had reduced visceral fat pad mass at 3 weeks and increased fat mass at 9 and 12 weeks (Fig. 2A). Subcutaneous fat mass was not altered in R6/2 mice at any age (Supplementary Material, Fig. S1A). The increased body fat beginning at 9 weeks of age in combination with reduced body weight (Fig. 1) suggests a reduction in lean body mass. CAG140 mice did not differ from wild-type mice at 7 months, but had 40–60% reductions in gonadal and subcutaneous fat mass at 22 months of age (Fig. 2B and Supplementary Material, Fig. S1B). Thus, the expression of mutant huntingtin as a transgene or as a gene KI is associated with reduced body weight at advanced stages of the disease and either increased (R6/2) or decreased (CAG140) visceral adipose tissue mass. These findings in the R6/2 strain are similar to those originally reported by Bjorkqvist et al. (17).
Figure 2.
Age-dependent alterations in adipose tissue mass in R6/2 Tg and CAG140 KI mice. Visceral adipose tissue from the gonadal fat depot was weighed and expressed as % body weight in female (A) R6/2 Tg and (B) CAG140 KI mice compared with corresponding wt mice at ages indicated (n = 5–6 mice for each genotype). Values represent mean ± SD. *P < 0.05; **P < 0.01 versus corresponding wt group.
Altered adipokine levels in R6/2 and CAG140 mice
It is well established that white adipose tissue has an important endocrine function in metabolic homeostasis through the action of adipokines such as leptin and adiponectin, which are secreted from adipocytes and act in the brain and peripheral tissues to influence glucose metabolism, appetite and energy expenditure (35,36). To test whether the altered white adipose tissue phenotype of the R6/2 and CAG140 mice might be associated with abnormal adipokine levels, we quantitated leptin and adiponectin in blood of R6/2 Tg and CAG140 KI mice.
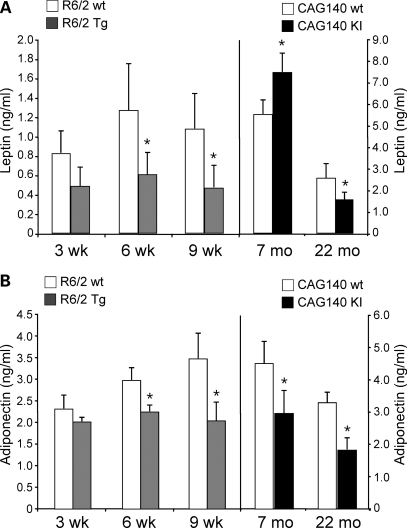
In normal animals, circulating leptin levels are typically proportional to body fat. To avoid confounding effects due to weight reductions at advanced stages of the disease, here we examined leptin levels at ages prior to these changes, i.e. at 3–9 weeks in R/62 mice and at 7–22 months in CAG140 mice (Fig. 1B and D). At ages 3, 6 and 9 weeks, R6/2 mice had ∼50% lower leptin levels than wild-type mice, an effect that became significant at 6 and 9 weeks (Fig. 3A). This is in spite of the fact that at 6 and 9 weeks the transgenic animals have body weight and % body fat equal to or greater than wild-type mice (Figs 1B and 2A). In CAG140 mice, absolute leptin levels were higher than those in R6/2 mice as would be expected (notice different scales on left and right panels of Fig. 3A) given their greater adipose tissue accumulation due to older age (7–22 months versus 3–9 weeks for R6/2 mice). Although 7-month-old CAG140 mice showed significantly elevated leptin levels compared with wild-type, by 22 months leptin levels dropped by 80% and were significantly lower than in wild-type mice. The drop in leptin levels of CAG140 KI mice between 7 and 22 months of age occurred despite the fact that KI mice maintained constant % fat mass (Fig. 2B) and body weight (29.8 ± 4.1 versus 28.6 ± 2.1 g, for 7 and 22 months, respectively). Thus, HD progression in the two mouse models is characterized by a drop in leptin levels that is disproportionately low for their body weight and adiposity.
Figure 3.
Age-dependent reductions in circulating leptin and adiponectin levels in R6/2 Tg and CAG140 KI mice. (A) Leptin and (B) adiponectin levels were determined in fasting plasma samples from female R6/2 Tg and CAG140 KI mice and wt littermates at the ages indicated (n = 4–5 mice for each genotype). Values represent mean ± SD. *P < 0.05 versus the corresponding wt group.
A second adipokine, adiponectin, also showed reduced levels in HD mouse models. In healthy animals and humans, adiponectin levels are typically highest in lean individuals and are reduced in conditions with impaired adipose tissue function, such as obesity (37). Both the R6/2 Tg and CAG140 KI mice exhibited reduced adiponectin levels compared with wild-type mice at nearly all ages (Fig. 3B). The reduced adipokine levels in HD mice prior to the weight loss that occurs in late stages of the disease suggest compromised adipose tissue function. Together, these results suggest that adipose tissue from HD mice may have reduced functional capacity.
Impaired expression of mature adipocyte genes in HD mouse models
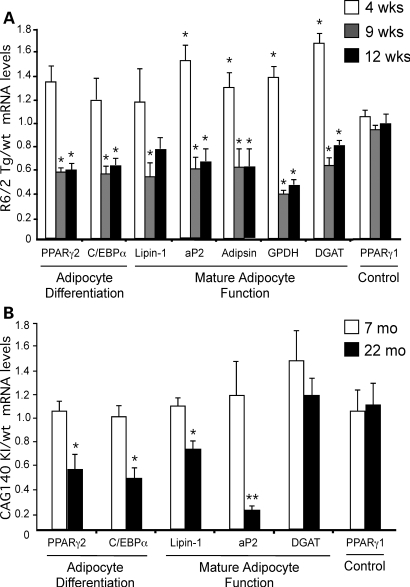
We investigated whether HD progression is marked by alterations in white adipose tissue gene expression. Mature adipocytes are formed from precursor cells that are induced to differentiate in response to hormonal signals (38,39). During early stages of adipocyte differentiation, the transcription factors peroxisome proliferator-activated receptor γ2 (PPARγ2) and CAAT enhancer binding protein α (C/EBPα) are induced, and these subsequently induce target genes for triglyceride biosynthesis [e.g. diacylglycerol acyltransferase (DGAT) and lipin-1] and other mature adipocyte factors [e.g. the adipocyte fatty acid binding protein (aP2) and adipsin]. Using qPCR, we quantitated the expression of adipogenic and lipogenic genes in adipose tissue of R6/2 mice at 4, 9 and 12 weeks and in CAG140 mice at 7 and 22 months of age. All values were normalized to control genes that did not vary between wild-type and mutant mouse tissues (described in Materials and Methods) and are expressed as a ratio of the levels in the mutant compared with the average value for wild-type mice.
Both HD mouse models exhibited substantial alterations in white adipose tissue gene expression during disease progression. In R6/2 mice, we detected a dramatic change in adipose tissue gene expression between 4 and 9 weeks of age. At 4 weeks of age, R6/2 mice exhibited 20–60% elevations in adipocyte gene expression compared with wild-type mice (Fig. 4A). However, by 9 weeks of age, the expression levels of key adipogenic and lipogenic genes such as PPARγ2, C/EBPα, lipin-1 and DGAT were reduced by more than 50% and remained low at 12 weeks of age. Levels of the constitutively expressed PPARγ1 isoform were similar in wild-type and R6/2 Tg mice throughout the lifetime of the animals, indicating that these effects were specific to genes involved in adipocyte maturation or function. The gene expression profile in adipose tissue from 9- and 12-week-old R6/2 mice suggests a ‘de-differentiated’ phenotype characterized by reduced expression of mature adipocyte genes.
Figure 4.
Age-dependent impairment in adipose tissue gene expression in R6/2 Tg and CAG140 KI mice. Adipose tissue mRNA levels were determined by qRT–PCR in R6/2 Tg (A) and CAG140 KI mice (B), and their corresponding controls at the ages indicated. Expression levels were normalized to HPRT and TBP genes, and Tg or KI levels expressed relative to the average value for respective wild-type animals. n = 4–5 mice for each group. Values represent mean ± SD. *P < 0.05 and **P < 0.01 versus the corresponding wt group (e.g. a value of 1.0).
As observed for R6/2 mice, CAG140 mice exhibited a reduction in the level of adipose tissue gene expression commensurate with disease progression. Gene expression levels were normal at 7 months of age, but by 22 months, CAG140 KI mice exhibited 30–80% reductions in expression of adipocyte differentiation factors and aP2, although DGAT expression was not significantly reduced (Fig. 4B). Thus, even though the R6/2 Tg and CAG140 KI mouse strains exhibit substantially different disease time courses and lifespan, they have remarkably similar gene expression changes in adipose tissue as the disease progresses.
Mutant huntingtin alters adipocyte gene expression in a cell-intrinsic manner
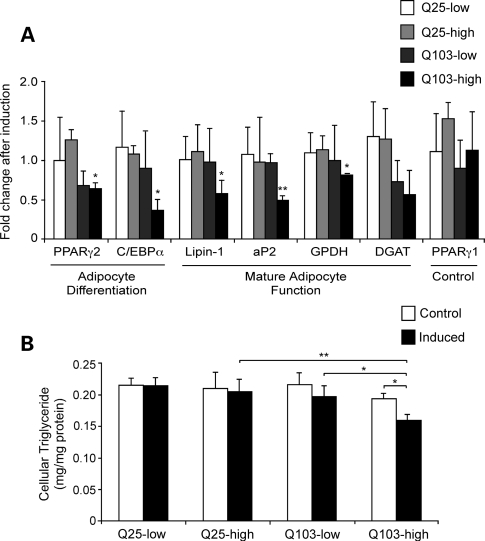
The impaired adipose tissue gene expression observed in the two HD mouse models might arise from a direct effect of mutant huntingtin in adipocytes or as a secondary effect due to systemic alterations. Consistent with a potential direct effect in adipocytes, endogenous huntingtin is expressed ubiquitously in tissues throughout the body, including different adipose tissue depots (gonadal and retroperitoneal white adipose tissue, and interscapular brown adipose tissue; see Supplementary Material, Fig. S2). Both the R6/2 Tg and CAG140 KI mice express mutant huntingtin under the control of native huntingtin gene regulatory elements (11,40). To test whether mutant huntingtin can affect adipocyte gene expression in a cell-intrinsic manner, we generated 3T3-L1 pre-adipocyte cell lines that express human huntingtin exon 1 containing either wild-type (Htt-Q25) or expanded (Htt-Q103) glutamine repeat number under the control of a hormone-inducible promoter. The stable 3T3-L1 cell lines were differentiated to mature adipocytes, treated with either vehicle or with hormone to induce Htt transgene expression and adipocyte gene expression was measured. We selected cell lines with similar low and high copy numbers of the Htt-Q25 and -Q103 transgenes as determined by qPCR to investigate potential dosage effects. Each cell line served as its own control, with gene expression values presented as the ratio of levels in cells after hormonal induction of the transgene versus treatment with vehicle control. All expression levels were normalized to 18S ribosomal RNA.
Expression of Htt-Q25 had no effect on expression of adipogenic or mature adipocyte genes (Fig. 5A). This was true in cells expressing both low and high copy numbers of the Htt-Q25 plasmid. Expression of the PPARγ1 control did not change in response to induction of either Htt-Q25 or Htt-Q103 expression. However, induction of Htt-Q103 did have a deleterious effect on expression of key adipogenic genes (PPARγ2, C/EBPα) and mature adipocyte markers and lipogenic genes, including aP2, lipin-1 and glycerol-3-phosphate dehydrogenase (GPDH) (Fig. 5A). The effect of Htt-Q103 on gene expression appeared to be dose-dependent, becoming statistically significant in cells expressing high levels of the mutant huntingtin construct. The expression of mutant huntingtin also resulted in reduced triglyceride storage within the adipocytes (Fig. 5B). This effect was observed in adipocytes expressing either low or high levels of the mutant huntingtin transgene, but not in cells expressing the wild-type version of the transgene. These results indicate that expression of mutant Htt within adipocytes has a direct effect on gene expression and triglyceride storage capacity in these cells.
Figure 5.
Acute expression of mutant huntingtin exon 1 impairs gene expression in 3T3-L1 adipocytes. 3T3-L1 stable cell lines containing Htt-Q25 and Htt-Q103 at low and high copy numbers were differentiated into mature adipocytes as described in Materials and Methods. On day 6, parallel groups of adipocytes were continued in basal conditions or induced for 48 h with tebufenozide to induced Htt transgene expression. (A) Gene expression levels were determined by qRT–PCR and normalized to 18S rRNA for four independent culture dishes for each construct and treatment. Data are expressed as mRNA levels after tebufenozide induction relative to the average value for the respective cell line under basal conditions. *P < 0.05 and **P < 0.01 versus Q25-low and Q25-high samples. (B) Triglyceride levels in cells treated as in (A) and expressed relative to total cell protein. Determinations are from four independent culture dishes for each cell line and treatment; values represent mean ± SD. *P < 0.05 and **P < 0.01 between samples indicated.
Mutant huntingtin inhibits PGC-1α co-activation of the PPARγ response element in adipocytes
A potential mechanism by which mutant huntingtin may alter gene expression is through dysregulation of transcriptional co-activator activity. One co-activator recently implicated in HD pathogenesis in brain is PPARγ co-activator 1α (PGC-1α), which exhibits impaired function through reduced expression and/or binding to mutant huntingtin (13,41,42). Known partners for PGC-1α in metabolic tissues include PPARγ and lipin-1 (43,44) and known targets include PPARγ-regulated genes such as aP2. Here we have shown that several of these genes exhibit altered expression in white adipose tissue of HD mouse models and in adipocytes expressing mutant huntingtin in vitro, raising the question of whether reduced PGC-1α levels and/or activity may result from expression of mutant huntingtin in adipocytes.
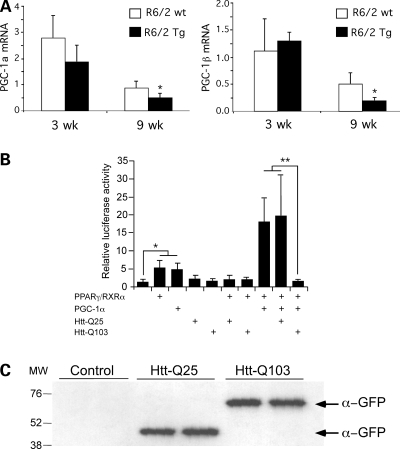
We first assessed the expression levels of PGC-1 in adipose tissue from R6/2 Tg mice at early (3 weeks) and late (9 weeks) points during disease progression. PGC-1α mRNA levels were not different between wild-type and transgenic mice at 3 weeks. Both showed a decline with age at 9 weeks, but levels in the transgenic mice were significantly lower than wild-type animals (Fig. 6A). The related co-activator, PGC-1β, which exhibits overlapping function with PGC-1α including the ability to co-activate PPARγ (44), also exhibited reduced expression levels in adipose tissue of 9-week-old R6/2 Tg compared with wild-type mice (Fig. 6A). Similar studies performed in adipose tissue from 7- and 22-month-old CAG140 KI mice showed extremely low levels of PGC-1 expression in wild-type and mutant mice, due to age-dependent decline in PGC-1 expression in adipose tissue, making it impossible to accurately determine the expression levels (data not shown). Thus, as has been reported in human brain, progression of HD in R6/2 Tg mice is associated with reduced adipose tissue expression of PGC-1 co-activators.
Figure 6.
Reduced PGC-1α expression and activity adipocytes expressing mutant huntingtin. (A) mRNA levels for PGC-1α and PGC-1β were determined by qRT–PCR in adipose tissue from R6/2 Tg mice at the ages indicated for five mice of each genotype. Expression levels were normalized to HPRT and TBP. (B) PCG-1α co-activator activity for PPARγ was assessed in the presence of Htt-Q25 and Htt-Q103. 3T3-L1 pre-adipocytes were co-transfected with a luciferase reporter construct containing the multimerized (3×) PPAR-responsive element upstream of the viral thymidine kinase minimal promoter in the indicated combinations with PPARγ/RXRα, PGC-1α and Htt-Q25 or Htt-Q103 huntingtin expression vectors. All samples also received a Renilla luciferase expression vector as a normalization control. The luciferase activity produced from the PPAR-responsive reporter gene are shown as the average ± SD of four replicates from a representative experiment. *P < 0.05; **P < 0.01 for comparisons indicated.
To investigate whether mutant huntingtin may interfere with PGC-1α transcriptional co-activation in adipocytes, we tested the effects of Htt-Q25 and Htt-Q103 on PGC-1α activated gene expression in 3T3-L1 pre-adipocytes. Cells were transfected with PGC-1α together with a luciferase reporter gene driven by PPAR response elements (PPRE×3-Luc), and expression vectors for PPARγ and RXRα. Compared with the control samples containing only the reporter plasmid, addition of PGC-1α or PPARγ/RXRα-stimulated reporter activity more than 5-fold and more than 18-fold when added in combination (Fig. 6B). To test the effect of huntingtin, cells were also transfected with Htt-Q25 or Htt-Q103 and expression was induced for 48 h. The addition of Htt Q-25 had no effect on reporter gene activation, but addition of Htt-Q103 interfered with PGC-1α co-activation, reducing the luciferase reporter levels back to baseline (Fig. 6B). Htt-Q25 and Htt-Q103 proteins were expressed at similar levels, as ascertained by western blot analysis (Fig. 6C). Thus, mutant huntingtin inhibits PGC-1α activation of the PPARγ gene regulatory element. A similar mechanism may be in effect in adipose tissue of R6/2 Tg or CAG140 KI mice, leading to the observed reductions in expression of PPARγ and PPARγ target genes.
DISCUSSION
In addition to neurological impairment, metabolic changes are an important clinical problem during the progression of HD in human patients. Here we have characterized alterations in a key metabolic tissue, white adipose tissue, in two mouse models of HD. Both the R6/2 Tg and CAG140 KI mouse strains exhibited evidence of adipose tissue dysfunction with disease progression. Despite the fact that the time course of disease in the two models ranges from 12–15 weeks for R6/2 Tg to more than 2 years for CAG140 KI mice, both strains had progressive reductions in body weight, circulating adipokine levels and alterations in gene expression in a set of key adipogenic and lipogenic genes. These findings have implications for understanding the pathogenesis, monitoring the progression and treating the symptoms of HD, as discussed below.
The changes in gene expression in white adipose tissue may contribute to several of the metabolic symptoms that have been noted in late stages of HD in both mice and humans, including weight loss and insulin resistance (22,23). Normal white adipose tissue plays an essential role in maintaining metabolic homeostasis, as demonstrated by the fact that genetic deficiency of mature adipose tissue in lipodystrophy leads to insulin resistance, diabetes, increased food intake and hypermetabolic rate (45). We purposely selected a set of genes that specifically reflects adipocyte differentiation and important functional features of mature adipocytes. Indeed, we found that the gene expression patterns in adipose tissue from HD mice tend to be biphasic; expression of some genes increase at early stages of the disease, but decrease as the disease progresses, in a manner similar to, but less severe than, that observed in mouse models of congenital lipodystrophy (23). Our gene expression changes were extremely sensitive to disease progression, particularly up to overt spontaneous phenotype development in the fast progressing R6/2 mice. Specifically, there are substantial reductions in the expression of key adipogenic transcription factors such as PPARγ, triglyceride synthesis and storage genes (lipin-1, DGAT, GPDH) and in genes that are characteristic of mature adipocytes (aP2, adipsin). In lipodystrophy, similar alterations in gene expression leads to adipocytes that are less efficient at lipid synthesis and storage, exhibit impaired glucose uptake and have reduced secretion of adipose tissue-derived hormones (22,23,45). Such changes are therefore likely to contribute to the weight loss and insulin resistance that develop in HD. White adipose tissue is uniquely useful for biopsy and it is possible that the changes observed here could be used to track disease progression in HD.
Impaired adipocyte function would also lead to the reduced leptin and adiponectin levels observed in the HD mice. As with adipocyte gene expression, circulating adiponectin and leptin levels are dynamic during the course of the disease, with normal or elevated levels at early time points, and become diminished compared with wild-type mice at later ages. The reduction in adipokine levels could not be accounted for by changes in body weight or fat mass and were most likely related to impaired adipose tissue function and corresponding reduced synthesis of mature adipocyte proteins. The altered function, rather than altered amount, of white adipose could make for more accurate, useful and accessible measurements in the clinical environment. Particularly noteworthy is the 4-fold reduction in leptin levels in CAG140 KI mice from 7 to 22 months of age. Because of their important roles in regulating food intake, energy expenditure and glucose homeostasis, the reduced adipokine levels may contribute to the metabolic dysregulation in HD. Reduced plasma leptin levels have also been reported in human HD patients, although no data exist as to whether the levels diminish within an individual over the course of the disease (46). These results suggest that circulating levels of leptin and other secreted adipocyte factors normalized to body weight may be a gauge of disease severity and stage, and thus a useful biomarker for monitoring disease progression and/or effects of therapeutic interventions.
Transcriptional dysregulation has been proposed as a key mechanism by which the mutant huntingtin protein exerts its toxic effects in the cell (reviewed in 47,48). Transcriptional dysregulation in the brain is an early event in disease pathogenesis and has been shown across different models, including humans and the R6/2 and N171–82Q Tg mouse strains (14,49–51). Altered gene expression levels in peripheral tissues, such as peripheral blood lymphocytes, muscle and pancreatic islets, have also been observed in HD patients and mouse models (14,15,18,52). Here, we used a set of key genes to determine specifically whether white adipocytes showed dysfunctional gene expression patterns that would alter their function and indeed we found that this was so in two in vivo models of HD. However, it is not clear whether abnormal gene expression in peripheral tissues is a primary effect of mutant huntingtin expression in those tissues or secondary to systemic changes. Here, we addressed this key question by examining the effect of mutant huntingtin expression in cultured adipocytes. We found that high-level expression of Htt-Q103 in mature adipocytes potently reduced the levels of several genes found to be reduced in vivo, including PPARγ, lipin-1, aP2 and GPDH. These results strongly indicate that mutant huntingtin within adipocytes has a deleterious effect on gene expression.
Various mechanisms have been proposed for the effects of mutant huntingtin on gene expression in neurons, including interaction of protein fragments/aggregates with components of the core transcription apparatus (53), with components of the RNA-mediated gene silencing machinery (54) and with the transcriptional co-activator PGC-1α. PGC-1α has a key role in transcriptional activation of genes in adipose tissue, heart and skeletal muscle and in protection of neurons against oxidative damage (42,44). In HD, PGC-1α function is impaired through reduced expression and/or binding to mutant huntingtin (13,41,42), and PGC-1α-deficient mice exhibit spongiform lesions in the striatum, impaired thermogenesis and hyperactivity (32,33,55,56). Since PGC-1α is known to interact with proteins such as PPARγ2 and lipin-1 (43) and to co-activate PPARγ2 target genes such as aP2, we investigated whether mutant huntingtin expression in adipocytes affects PGC-1 expression levels and/or activity. We found evidence for reduced PGC-1α and PGC-1β expression levels in adipose tissue of R6/2 Tg mice specifically at a late stage of the disease, and also found that mutant huntingtin interferes with PGC-1α co-activation in cultured adipocytes. These findings identify one potential mechanism by which mutant huntingtin may impair adipocyte gene expression.
HD is a debilitating and fatal disease that exacts its toll through neurodegeneration in the striatum, cerebral cortex and hypothalamus. Our studies demonstrate that expression of mutant huntingtin in white adipose tissue is directly detrimental to tissue function and may influence severity or course of the disease. These observations open the door for novel approaches to study and treat HD, and a detailed study of human patient samples is now warranted. The ability to isolate and culture pre-adipocytes from mouse models provides an additional tool to characterize the molecular pathogenesis of HD. More importantly, the accessibility of small amounts of subcutaneous adipose tissue, and the ability to detect adipose tissue-derived hormones in small volumes of blood, may prove useful in designing and monitoring drug therapies. An important aspect of the current findings is that impaired adipose tissue function is detectable in HD mouse strains at early stages of disease progression. For example, reduced leptin levels were detected in the R6/2 Tg mice at 6 weeks of age, where as hypoactivity, limb clasping, changes in gait and involuntary movements typically do not become evident until 8–12 weeks of age (32,57). This raises the possibility that analysis of leptin levels may have diagnostic or prognostic applications, serving as a readily accessible marker to identify patients at risk for poor prognosis or to monitor the progression of disease or response to experimental treatments at early stages of disease progression.
White adipose tissue function is extremely important for glucose homeostasis and energy balance. This suggests that restoring its function in HD patients through peripheral inhibition of mutant huntingtin via RNAi or antisense strategies would have therapeutic value. Furthermore, the activation of adipose tissue PPARγ with thiazolidinedione drugs, which are currently in use for treatment of non-insulin-dependent diabetes, may have beneficial effects on promoting adipocyte maturation, as well as recently reported central (i.e. striatal) benefits (58,59). Together, these results support the idea that treatment of peripheral tissues in HD may be valuable in retarding progression of the disease and improving quality of life.
MATERIALS AND METHODS
R6/2 and CAG140 mouse strains
All procedures were carried out in accordance with the USPHS Guide for the Care and Use of Laboratory Animals and were approved by the UCLA Animal Research Committee. R6/2 mice were generated by breeding female mice with ovarian transplants from female R6/2 mice (Jackson Laboratory, Bar Harbor, ME, USA) to C57BL/6J × CBA F1 males. CAG140 KI mice were generated by breeding heterozygous mice in-house. Homozygous KI mice were used. Because repeat lengths of expanded CAG repeats are unstable, CAG repeat lengths for the cohorts used in this study were determined by Laragen, Inc. (Culver City, CA, USA) to be 146 ± 4 (mean ± SEM) for R6/2 transgenic mice and 123.3 ± 0.6 for CAG140 KI mice. Wild-type littermates were used as controls. Mice were housed in a temperature- and humidity-controlled room. Food and water were available ad libitum and mice were kept on a 12:12 h light:dark cycle.
Blood and tissue collection
Blood was collected from mice after a 14 h overnight fast under isoflurane anesthesia and were euthanized immediately afterwards for tissue collection. Leptin and adiponectin levels were determined in 50 and 1 µl plasma samples, respectively, using a Mouse Leptin ELISA Kit (Crystal Chem, Inc., Downers Grove, IL, USA) or Quantikine Adiponectin Immunoassay Kit (R&D, Minneapolis, MN, USA). Fat pads from the gonadal and inguinal subcutaneous depots were dissected and weighed and weights expressed as a percentage of total body weight.
RNA quantitation
Total RNA was isolated from adipose tissue or cultured cells with Trizol (Invitrogen, Carlsbad, CA, USA) and treated with RNase-free DNase (Ambion, Austin, TX, USA) to remove any contaminating genomic DNA. First-strand cDNA synthesis was performed using oligo-dT primers (Invitrogen). Real-time PCR reactions were performed on the iCycler iQ real-time detection system (BioRad, Hercules, CA, USA) using SYBR Green PCR QuantiTect reagent kit (Qiagen, Valencia, CA, USA) as described previously (60). Each assay included (in triplicate): a standard curve of four serial dilution points of control cDNA (ranging from 100 ng to 100 pg), a no template control and 25–50 ng of each sample cDNA. The relative concentration of the endogenous controls (TBP, TATA box-binding protein; HPRT, hypoxanthine phosphoribosyltransferase; 18S ribosomal RNA) and genes of interest were determined by plotting the threshold cycle (Ct) versus the log of the serial dilution points, and relative expression of the gene of interest was determined after normalizing to endogenous controls. Primers used for real-time PCR were designed to span an intron and are as follows: TBP (acccttcaccaatgactcctatg, atgatgactgcagcaaatcgc); HPRT (cacaggactagaacacctgc, gctggtgaaaaggacctct); 18S RNA (accgcagctaggaataatgga, gcctcagttccgaaaacca); PPARγ1 (cacgttctgacaggactgtgt, cagcaaccattgggtcagctc); PPARγ2 (ccagagcatggtgccttcgct, cagcaaccattgggtcagctc); C/EBPα (gaacagcaacgagtaccgggta, gccatggccttgaccaaggag); lipin-1 (ggtcccccagccccagtcctt, gcagcctgtggcaattca); DGAT (tgctacgacgagttcttgag, ctctgccacagcattgagac); GPDH (gtggtaccccatcagttcattg, gtccttcaggagctgtccctg); aP2 (gaacctggaagcttgtcttcg, accagcttgtcaccatctcg); adipsin (actccctgtccgcccctgaacc, cgagagccccacgtaaccacacct); PGC-1α (ctcacagagacactggacagt, tgtagctgagctgagtgttgg); PGC-1β (ccgatgactccgagctcttcca, tgcagataagggggcaggtga).
Adipocyte cell lines expressing inducible wild-type and mutant huntingtin constructs
The 3T3-L1 pre-adipocyte cell line was transfected with plasmids generously provided by Dr Erik Schweitzer containing exon 1 of human huntingtin (Htt) fused to enhanced green fluorescent protein (EGFP) under the direction of an ecdysone-responsive promoter (61). Two versions of the plasmid were used: Htt-Q25, which contains a polyglutamine tract of 25 residues and served as the control, and Htt-Q103, which contains an expanded polyglutamine tract of 103 residues. Stably transfected 3T3-L1 cell lines were obtained by selection in G418, and cell lines that had integrated the plasmid at ‘low’ and ‘high’ copy numbers were characterized by quantitation of EGFP sequences in genomic DNA by real-time PCR (primers, EGFP-f, acgtaaacggccacaagttc; EGFP-r, aagtcgtgctgcttcatgtg). Inducible expression of Htt-EGFP was confirmed by treating cells with 1 µm tebufenozide in DMSO (62) and monitoring the induction of Htt-EGFP mRNA by qRT–PCR. To distinguish between Htt–EGFP sequences present in genomic DNA and in mRNA transcripts, the PCR was performed after treatment of RNA samples with RNase-free DNase to remove contaminating genomic DNA and in the presence and absence of reverse transcriptase.
For studies of adipocyte gene expression, the stable 3T3-L1 cell lines were induced to differentiate into mature adipocytes by treating confluent cultures for 2 days with Dulbecco's modified Eagle's medium containing 10% bovine serum, 10 µg/ml insulin, 5 µm dexamethasone and 0.5 mm 3-isobutyl-1-methylxanthine. Incubation was continued for an additional 4 days in the same medium containing 10% fetal bovine serum and 10 µg/ml insulin to produce mature adipocytes with lipid accumulation. At day 6 after initiation of differentiation, adipocytes were treated with DMSO vehicle or 1 µm tebufenozide for the determination of gene expression in basal conditions or after induction of the Htt constructs, respectively. RNA was prepared and gene expression determined by real-time RT–PCR as described above. Cellular triglyceride levels were quantitated using an acetyl-acetone-based colorimetric assay (Wako Chemicals, Richmond, VA, USA) and normalized to protein levels determined by Bradford assay.
PGC-1α co-activator assays
To determine the effects of mutant huntingtin on gene activation by PGC-1α in adipocytes, the 3T3-L1 pre-adipocyte cell line was transfected with expression plasmids for PGC-1α, PPARγ and RXRα (kindly provided by Dr Peter Tontonoz), and a reporter plasmid containing three copies of a PPAR response element upstream of a viral thymidine kinase minimal promoter firefly luciferase vector (kindly provided by Dr. Ronald Evans; (63). In addition, a plasmid expressing Renilla luciferase (phRL-TK Renilla, Promega Corporation, Madison, WI, USA) was transfected to normalize for variations in transfection efficiency. Duplicate sets of samples included an expression plasmid for exon 1 of the huntingtin gene containing either Htt-Q25 or Htt-Q103, described above. Using Effectene (Qiagen) 80 000 cells were transfected, and 36 h later, firefly and Renilla luciferase activities were quantitated using the Dual-Luciferase Reporter System (Promega) and a Glomax microplate luminometer (Promega). In each experiment, samples were analyzed in quadruplicate, and each experiment was repeated at least twice. To assess Htt-Q25 and Htt-Q103 protein levels in transfected cells, parallel samples were harvested, lyzed and equivalent amounts of cellular protein were resolved by PAGE in 12% Tris-glycine gels. Htt proteins were detected by virtue of the EGFP fusion using monoclonal anti-GFP antibody diluted 1:500 (MAB2510, Chemicon).
Statistics
Body weights were compared using repeated measures ANOVA (R6/2) or completely randomized ANOVA (CAG140), using GB-Stat v8.0. Post hoc comparisons were made using Fisher's LSD test. Student's t-test was used for comparisons in gene expression and adipokine levels between Tg or KI mice and their respective controls. A critical value of P < 0.05 was used as a significance threshold for all comparisons.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by The Hereditary Disease Foundation and the National Institutes of Health (HL28481 to K.R., GM08042 to J.P.).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Erik Schweitzer for providing the plasmid containing Htt exon 1-GFP, Dr Peter Tontonoz for PGC-1α, PPARγ and RXRα expression plasmids and Dr Ronald Evans for the PPRE-luciferase reporter plasmid.
Conflict of Interest statement. The authors have no conflicts to declare.
REFERENCES
- 1.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Morales L.M., Estevez J., Suarez H., Villalobos R., Chacin de Bonilla L., Bonilla E. Nutritional evaluation of Huntington disease patients. Am. J. Clin. Nutr. 1989;50:145–150. doi: 10.1093/ajcn/50.1.145. [DOI] [PubMed] [Google Scholar]
- 3.Trejo A., Tarrats R.M., Alonso M.E., Boll M.C., Ochoa A., Velasquez L. Assessment of the nutrition status of patients with Huntington's disease. Nutrition. 2004;20:192–196. doi: 10.1016/j.nut.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Farrer L.A., Meaney F.J. An anthropometric assessment of Huntington's disease patients and families. Am. J. Phys. Anthropol. 1985;67:185–194. doi: 10.1002/ajpa.1330670304. [DOI] [PubMed] [Google Scholar]
- 5.Sanberg P.R., Fibiger H.C., Mark R.F. Body weight and dietary factors in Huntington's disease patients compared with matched controls. Med. J. Aust. 1981;1:407–409. doi: 10.5694/j.1326-5377.1981.tb135681.x. [DOI] [PubMed] [Google Scholar]
- 6.Myers R.H., Sax D.S., Koroshetz W.J., Mastromauro C., Cupples L.A., Kiely D.K., Pettengill F.K., Bird E.D. Factors associated with slow progression in Huntington's disease. Arch. Neurol. 1991;48:800–804. doi: 10.1001/archneur.1991.00530200036015. [DOI] [PubMed] [Google Scholar]
- 7.Stoy N., McKay E. Weight loss in Huntington's disease. Ann. Neurol. 2000;48:130–131. [PubMed] [Google Scholar]
- 8.Djousse L., Knowlton B., Cupples L.A., Marder K., Shoulson I., Myers R.H. Weight loss in early stage of Huntington's disease. Neurol. 2002;59:1325–1330. doi: 10.1212/01.wnl.0000031791.10922.cf. [DOI] [PubMed] [Google Scholar]
- 9.Morton G.J., Cummings D.E., Baskin D.G., Barsh G.S., Schwartz M.W. Central nervous system control of food intake and body weight. Nature. 2006;443:289–295. doi: 10.1038/nature05026. [DOI] [PubMed] [Google Scholar]
- 10.Petersen A., Bjorkqvist M. Hypothalamic-endocrine aspects in Huntington's disease. Eur. J. Neurosci. 2006;24:961–967. doi: 10.1111/j.1460-9568.2006.04985.x. [DOI] [PubMed] [Google Scholar]
- 11.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 12.Fain J.N., Del Mar N.A., Meade C.A., Reiner A., Goldowitz D. Abnormalities in the functioning of adipocytes from R6/2 mice that are transgenic for the Huntington's disease mutation. Hum. Mol. Genet. 2001;10:145–152. doi: 10.1093/hmg/10.2.145. [DOI] [PubMed] [Google Scholar]
- 13.Weydt P., Pineda V.V., Torrence A.E., Libby R.T., Satterfield T.F., Lazarowski E.R., Gilbert M.L., Morton G.J., Bammler T.K., Strand A.D., et al. Thermoregulatory and metabolic defects in Huntington's disease transgenic mice implicate PGC-1alpha in Huntington's disease neurodegeneration. Cell Metab. 2006;4:349–362. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Luthi-Carter R., Hanson S.A., Strand A.D., Bergstrom D.A., Chun W., Peters N.L., Woods A.M., Chan E.Y., Kooperberg C., Krainc D., et al. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum. Mol. Genet. 2002;11:1911–1126. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- 15.Strand A.D., Aragaki A.K., Shaw D., Bird T., Holton J., Turner C., Tapscott S.J., Tabrizi S.J., Schapira A.H., Kooperberg C., et al. Gene expression in Huntington's disease skeletal muscle: a potential biomarker. Hum. Mol. Genet. 2005;14:1863–1876. doi: 10.1093/hmg/ddi192. [DOI] [PubMed] [Google Scholar]
- 16.Ribchester R.R., Thomson D., Wood N.I., Hinks T., Gillingwater T.H., Wishart T.M., Court F.A., Morton A.J. Progressive abnormalities in skeletal muscle and neuromuscular junctions of transgenic mice expressing the Huntington's disease mutation. Eur. J. Neurosci. 2004;20:3092–3114. doi: 10.1111/j.1460-9568.2004.03783.x. [DOI] [PubMed] [Google Scholar]
- 17.Bjorkqvist M., Fex M., Renstrom E., Wierup N., Petersen A., Gil J., Bacos K., Popovic N., Li J.Y., Sundler F., et al. The R6/2 transgenic mouse model of Huntington's disease develops diabetes due to deficient beta-cell mass and exocytosis. Hum. Mol. Genet. 2005;14:565–574. doi: 10.1093/hmg/ddi053. [DOI] [PubMed] [Google Scholar]
- 18.Andreassen O.A., Dedeoglu A., Stanojevic V., Hughes D.B., Browne S.E., Leech C.A., Ferrante R.J., Habener J.F., Beal M.F., Thomas M.K. Huntington's disease of the endocrine pancreas: insulin deficiency and diabetes mellitus due to impaired insulin gene expression. Neurobiol. Dis. 2002;11:410–424. doi: 10.1006/nbdi.2002.0562. [DOI] [PubMed] [Google Scholar]
- 19.Hurlbert M.S., Zhou W., Wasmeier C., Kaddis F.G., Hutton J.C., Freed C.R. Mice transgenic for an expanded CAG repeat in the Huntington's disease gene develop diabetes. Diabetes. 1999;48:649–651. doi: 10.2337/diabetes.48.3.649. [DOI] [PubMed] [Google Scholar]
- 20.Goodman A.O., Murgatroyd P.R., Medina-Gomez G., Wood N.I., Finer N., Vidal-Puig A.J., Morton A.J., Barker R.A. The metabolic profile of early Huntington's disease—a combined human and transgenic mouse study. Exp. Neurol. 2008;210:691–698. doi: 10.1016/j.expneurol.2007.12.026. [DOI] [PubMed] [Google Scholar]
- 21.van der Burg J.M., Bacos K., Wood N.I., Lindqvist A., Wierup N., Woodman B., Wamsteeker J.I., Smith R., Deierborg T., Kuhar M.J., et al. Increased metabolism in the R6/2 mouse model of Huntington's disease. Neurobiol. Dis. 2008;29:41–51. doi: 10.1016/j.nbd.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 22.Chehab F.F. Obesity and lipodystrophy—where do the circles intersect? Endocrinol. 2008;149:925–934. doi: 10.1210/en.2007-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reue K., Phan J. Metabolic consequences of lipodystrophy in mouse models. Curr. Opin. Clin. Nutr. Metabol. Care. 2006;9:436–441. doi: 10.1097/01.mco.0000232904.82038.db. [DOI] [PubMed] [Google Scholar]
- 24.Josefsen K., Nielsen M.D., Jorgensen K.H., Bock T., Norremolle A., Sorensen S.A., Naver B., Hasholt L. Impaired glucose tolerance in the R6/1 transgenic mouse model of Huntington's disease. J. Neuroendocrinol. 2008;20:165–172. doi: 10.1111/j.1365-2826.2007.01629.x. [DOI] [PubMed] [Google Scholar]
- 25.Lalic N.M., Maric J., Svetel M., Jotic A., Stefanova E., Lalic K., Dragasevic N., Milicic T., Lukic L., Kostic V.S. Glucose homeostasis in Huntington disease: abnormalities in insulin sensitivity and early-phase insulin secretion. Arch. Neurol. 2008;65:476–480. doi: 10.1001/archneur.65.4.476. [DOI] [PubMed] [Google Scholar]
- 26.Ahima R.S., Lazar M.A. Adipokines and the peripheral and neural control of energy balance. Mol. Endocrinol. (Baltimore, Md.) 2008;22:1023–1031. doi: 10.1210/me.2007-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry B.A., Clarke I.J. Adipose tissue hormones and the regulation of food intake. J. Neuroendocrinol. 2008;20:842–849. doi: 10.1111/j.1365-2826.2008.1730.x. [DOI] [PubMed] [Google Scholar]
- 28.Scherer P.E. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes. 2006;55:1537–1545. doi: 10.2337/db06-0263. [DOI] [PubMed] [Google Scholar]
- 29.Koebnick C., Shaibi G.Q., Kelly L.A., Roberts C.K., Lane C.J., Toledo-Corral C., Davis J.N., Byrd-Williams C., Weigensberg M.J., Goran M.I. Leptin-to-adiponectin ratio as independent predictor of insulin sensitivity during growth in overweight Hispanic youth. J. Endocrinol. Invest. 2007;30:RC13–RC16. doi: 10.1007/BF03346344. [DOI] [PubMed] [Google Scholar]
- 30.Norata G.D., Raselli S., Grigore L., Garlaschelli K., Dozio E., Magni P., Catapano A.L. Leptin:adiponectin ratio is an independent predictor of intima media thickness of the common carotid artery. Stroke. 2007;38:2844–2846. doi: 10.1161/STROKEAHA.107.485540. [DOI] [PubMed] [Google Scholar]
- 31.Oda N., Imamura S., Fujita T., Uchida Y., Inagaki K., Kakizawa H., Hayakawa N., Suzuki A., Takeda J., Horikawa Y., et al. The ratio of leptin to adiponectin can be used as an index of insulin resistance. Metab. Clin. Exptl. 2008;57:268–273. doi: 10.1016/j.metabol.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Li J.Y., Popovic N., Brundin P. The use of the R6 transgenic mouse models of Huntington's disease in attempts to develop novel therapeutic strategies. NeuroRx. 2005;2:447–464. doi: 10.1602/neurorx.2.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menalled L.B., Sison J.D., Dragatsis I., Zeitlin S., Chesselet M.F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J. Comp. Neurol. 2003;465:11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- 34.Hickey M.A., Kosmalska A., Enayati J., Cohen R., Zeitlin S., Levine M., Chesselet M.F. Extensive early motor and non-motor behavioral deficits are followed by striatal neuronal loss in knock-in Huntington's disease mice. Neuroscience. 2008;157:280–295. doi: 10.1016/j.neuroscience.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahima R.S. Adipose tissue as an endocrine organ. Obesity. 2006;14(Suppl. 5):242S–249S. doi: 10.1038/oby.2006.317. [DOI] [PubMed] [Google Scholar]
- 36.Trujillo M.E., Scherer P.E. Adipose tissue-derived factors: impact on health and disease. Endocrinol. Rev. 2006;27:762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 37.Berg A.H., Scherer P.E. Adipose tissue, inflammation, and cardiovascular disease. Circ. Res. 2004;96:939–949. doi: 10.1161/01.RES.0000163635.62927.34. [DOI] [PubMed] [Google Scholar]
- 38.Farmer S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosen E.D., MacDougald O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 40.Levine M.S., Klapstein G.J., Koppel A., Gruen E., Cepeda C., Vargas M.E., Jokel E.S., Carpenter E.M., Zanjani H., Hurst R.S., et al. Enhanced sensitivity to N-methyl-d-aspartate receptor activation in transgenic and knockin mouse models of Huntington's disease. J. Neurosci. Res. 1999;58:515–532. [PubMed] [Google Scholar]
- 41.Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1alpha by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 42.St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 43.Finck B.N., Gropler M.C., Chen Z., Leone T.C., Croce M.A., Harris T.E., Lawrence J.C., Jr, Kelly D.P. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 45.Garg A. Acquired and inherited lipodystrophies. N. Engl. J. Med. 2004;350:1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 46.Popovic V., Svetel M., Djurovic M., Petrovic S., Doknic M., Pekic S., Miljic D., Milic N., Glodic J., Dieguez C., et al. Circulating and cerebrospinal fluid ghrelin and leptin: potential role in altered body weight in Huntington's disease. Eur. J. Endocrinol. 2004;151:451–455. doi: 10.1530/eje.0.1510451. [DOI] [PubMed] [Google Scholar]
- 47.Sadri-Vakili G., Cha J.H. Mechanisms of disease: histone modifications in Huntington's disease. Nat. Clin. Pract. Neurol. 2006;2:330–338. doi: 10.1038/ncpneuro0199. [DOI] [PubMed] [Google Scholar]
- 48.Cha J.H. Transcriptional signatures in Huntington's disease. Prog. Neurobiol. 2007;83:228–248. doi: 10.1016/j.pneurobio.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu H., McCaw E.A., Hebb A.L., Gomez G.T., Denovan-Wright E.M. Mutant huntingtin affects the rate of transcription of striatum-specific isoforms of phosphodiesterase 10A. Eur. J. Neurosci. 2004;20:3351–3363. doi: 10.1111/j.1460-9568.2004.03796.x. [DOI] [PubMed] [Google Scholar]
- 50.Luthi-Carter R., Strand A., Peters N.L., Solano S.M., Hollingsworth Z.R., Menon A.S., Frey A.S., Spektor B.S., Penney E.B., Schilling G., et al. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum. Mol. Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- 51.McCaw E.A., Hu H., Gomez G.T., Hebb A.L., Kelly M.E., Denovan-Wright E.M. Structure, expression and regulation of the cannabinoid receptor gene (CB1) in Huntington's disease transgenic mice. Eur. J. Biochem. 2004;271:4909–4920. doi: 10.1111/j.1432-1033.2004.04460.x. [DOI] [PubMed] [Google Scholar]
- 52.Borovecki F., Lovrecic L., Zhou J., Jeong H., Then F., Rosas H.D., Hersch S.M., Hogarth P., Bouzou B., Jensen R.V., et al. Genome-wide expression profiling of human blood reveals biomarkers for Huntington's disease. Proc. Natl Acad. Sci. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhai W., Jeong H., Cui L., Krainc D., Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 54.Savas J.N., Makusky A., Ottosen S., Baillat D., Then F., Krainc D., Shiekhattar R., Markey S.P., Tanese N. Huntington's disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. PNAS. 2008;105:10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leone T.C., Lehman J.J., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 57.Hickey M.A., Gallant K., Gross G.G., Levine M.S., Chesselet M.F. Early behavioral deficits in R6/2 mice suitable for use in preclinical drug testing. Neurobiol. Dis. 2005;20:1–11. doi: 10.1016/j.nbd.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 58.Quintanilla R.A., Jin Y.N., Fuenzalida K., Bronfman M., Johnson G.V. Rosiglitazone treatment prevents mitochondrial dysfunction in mutant huntingtin expressing cells: Possible role of PPARgamma in the pathogenesis of huntington disease. J. Biol. Chem. 2008;283:25628–25637. doi: 10.1074/jbc.M804291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kiaei M. Peroxisome proliferator-activated receptor-gamma in amyotrophic lateral sclerosis and Huntington's disease. PPAR Res. 2008;2008:418765. doi: 10.1155/2008/418765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phan J., Peterfy M., Reue K. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 2004;279:29558–29564. doi: 10.1074/jbc.M403506200. [DOI] [PubMed] [Google Scholar]
- 61.Aiken C.T., Tobin A.J., Schweitzer E.S. A cell-based screen for drugs to treat Huntington's disease. Neurobiol. Dis. 2004;16:546–555. doi: 10.1016/j.nbd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 62.Addison J.A. Safety testing of tebufenozide, a new molt-inducing insecticide, for effects on nontarget forest soil invertebrates. Ecotoxicol. Environ. Saf. 1996;33:55–61. doi: 10.1006/eesa.1996.0006. [DOI] [PubMed] [Google Scholar]
- 63.Kliewer S.A., Umesono K., Noonan D.J., Heyman R.A., Evans R.M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.