Abstract
We recently identified a novel non-synonymous variant, rs1143679, at exon 3 of the ITGAM gene associated with systemic lupus erythematosus (SLE) susceptibility in European-Americans (EAs) and African-Americans. Using genome-wide association approach, three other studies also independently reported an association between SLE susceptibility and ITGAM or ITGAM-ITGAX region. The primary objectives of this study are to assess whether single or multiple causal variants from the same gene or any nearby gene(s) are involved in SLE susceptibility and to confirm a robust ITGAM association across nine independent data sets (n = 8211). First, we confirmed our previously reported association of rs1143679 (risk allele ‘A’) with SLE in EAs (P = 1.0 × 10−8) and Hispanic-Americans (P = 2.9 × 10−5). Secondly, using a comprehensive imputation-based association test, we found that ITGAM is one of the major non-human leukocyte antigen susceptibility genes for SLE, and the strongest association for EA is the same coding variant rs1143679 (log10Bayes factor=20, P = 6.17 × 10−24). Thirdly, we determined the robustness of rs1143679 association with SLE across three additional case–control samples, including UK (P = 6.2 × 10−8), Colombian (P = 3.6 × 10−7), Mexican (P = 0.002), as well as two independent sets of trios from UK (PTDT = 1.4 × 10−5) and Mexico (PTDT = 0.015). A meta-analysis combing all independent data sets greatly reinforces the association (Pmeta = 7.1 × 10−50, odds ratio = 1.83, 95% confidence interval = 1.69–1.98, n = 10 046). However, this ITGAM association was not observed in the Korean or Japanese samples, in which rs1143679 is monomorphic for the non-risk allele (G). Taken together along with our earlier findings, these results demonstrate that the coding variant, rs1143679, best explains the ITGAM-SLE association, especially in European- and African-derived populations, but not in Asian populations.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a clinically heterogeneous autoimmune disease characterized by autoantibody production, complement activation, and organ-specific tissue destruction. SLE primarily affects women (women: men = 9:1) of childbearing age. It affects a higher proportion of people of African or Hispanic descent relative to those of European descent (1,2). Genetic predisposition has been implicated in the pathogenesis of SLE. In fact, SLE has a relatively strong genetic component (sibling risk ratio, λs, ∼30), compared with many other autoimmune diseases. Numerous candidate gene loci have been reported by genome-wide linkage studies in multiplex SLE families and by case–control studies (3).
Recently, applying a trans-ethnic approach, we identified and replicated an association occurring between a variant at exon 3 (rs1143679) of integrin-α-M (ITGAM) and SLE susceptibility in individuals with European- and African-descent (4). Additionally, using a genome-wide association study approach, three other studies independently reported an association between SLE susceptibility and intronic SNPs rs9888739 (5), rs11150610 (6) within ITGAM and with rs11574637 located between the ITGAM-ITGAX regions (7), respectively. The objectives of the present study are to (i) assess our previously reported genetic association within four independent case–control samples including European-Americans (EAs), Hispanic-Americans (HAs), Korean and Japanese; (ii) assess whether a single SNP or multiple SNPs from the ITGAM gene or any nearby gene(s) are involved in SLE susceptibility using a comprehensive imputation-based association analysis in a combined cohort of EAs (n = 5609) that includes current EA and published EA samples (4); and (c) assess the robustness of the genetic association across case–control and trio samples from multiple ethnic and geographic origins, including UK, Colombian and Mexican.
RESULTS
Allelic association test in EA, HA, Korean and Japanese samples
In EA, HA, and Korean samples, we genotyped 34 SNPs from TRIM2, ITGAM and ITGAX (Supplementary Material, Table S1). Twenty-four SNPs that passed quality control (QC) in EAs and HAs were included for further analysis. Twelve SNPs, not including rs1143679, which were polymorphic in Korean samples, were also genotyped in Japanese samples. Allelic association between individual SNPs and SLE for EA, HA, Korean and Japanese samples is shown in Table 1. In the EA group, odds ratios (ORs) were in the same direction, and the minor allele frequencies (MAFs) were similar at each SNP between our current EA analysis and previously published EA results (4). The strongest association was observed at rs1143679 (P = 1.0 × 10−8, OR = 1.73, 95% confidence interval (CI) = 1.43–2.10), with a risk allele ‘A’ frequency of 17.9% in cases and 11.1% in controls. The most significant association in the HA group was identified with the same SNP, rs1143679 (P = 2.85 × 10−5, OR = 2.09, 95% CI = 1.47–2.98). However, with the exception of borderline significance in rs7206295 and rs4597342, none of the SNPs was associated with SLE in Korean samples, in which most of the SNPs including rs1143679 were monomorphic for the non-risk allele ‘G’. In Japanese samples, rs1143679 was also monomorphic for ‘G’ allele, and none of the other genotyped SNPs yielded any significant association with SLE.
Table 1.
Results of allelic association for QC-checked SNPs in EAs, HAs, Korean and Japanese populations
| Gene | SNP | Allele† | European-American |
Hispanic-American |
Korean |
Japanese |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF |
MAF |
MAF |
MAF |
|||||||||||||||
| Case | Control | P-value | OR (95% CI) | Case | Control | P-value | OR (95% CI) | Case | Control | P-value | OR (95% CI) | Case | Control | P-value | OR (95% CI) | |||
| TRIM72 | rs8056505 | GA | 0.3 | 0.32 | 0.16 | 0.9 (0.78–1.04) | 0.42 | 0.39 | 0.33 | 1.11 (0.89–1.37) | 0.17 | 0.19 | 0.19 | 0.88 (0.73–1.06) | 0.16 | 0.17 | 0.93 | 0.47 (0.11,1.48) |
| ITGAM | rs8057320 | GA | 0.29 | 0.31 | 0.14 | 0.89 (0.77–1.03) | 0.39 | 0.37 | 0.64 | 1.05 (0.84–1.3) | 0.18 | 0.20 | 0.14 | 0.87 (0.72–1.04) | 0.17 | 0.18 | 0.68 | 0.93 (0.66–1.29) |
| ITGAM | rs7193943 | GA | 0.28 | 0.30 | 0.16 | 0.9 (0.77–1.04) | 0.36 | 0.30 | 0.37 | 1.1 (0.88–1.37) | 0.18 | 0.20 | 0.14 | 0.87 (0.72–1.04) | 0.16 | 0.18 | 0.61 | 0.918 (0.65–1.27) |
| ITGAM | rs11865830 | GA | 0.28 | 0.31 | 0.17 | 0.9 (0.78–1.04) | 0.37 | 0.36 | 0.77 | 1.03 (0.83–1.28) | 0.18 | 0.20 | 0.13 | 0.86 (0.72–1.04) | 0.17 | 0.18 | 0.52 | 1.07 (0.72–1.61) |
| ITGAM | rs1143679 | AG | 0.18 | 0.11 | 1.04 × 10−8 | 1.73 (1.43–2.10) | 0.16 | 0.09 | 2.85 × 10−5 | 2.09 (1.47–2.98) | 0 | 0 | – | – | 0 | 0 | – | – |
| ITGAM | rs8048583 | AG | 0.3 | 0.32 | 0.16 | 0.9 (0.78–1.04) | 0.41 | 0.41 | 0.86 | 1.01 (0.82–1.26) | 0.19 | 0.22 | 0.06 | 0.84 (0.7–1.01) | 0.17 | 0.19 | 0.54 | 1.11 (0.79–1.54) |
| ITGAM | rs9936831 | TA | 0.18 | 0.12 | 1.41 × 10−7 | 1.63 (1.35–1.96) | 0.19 | 0.11 | 0.0001 | 1.81 (1.32–2.47) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs11861251 | GA | 0.13 | 0.14 | 0.52 | 0.93 (0.77–1.13) | 0.07 | 0.07 | 0.82 | 0.95 (0.63–1.42) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs9888879 | GA | 0.19 | 0.12 | 6.92 × 10−8 | 1.64 (1.36–1.96) | 0.21 | 0.15 | 0.001 | 1.57 (1.18–2.09) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs12928810 | AG | 0.19 | 0.12 | 6.92 × 10−8 | 1.64 (1.36–1.96) | 0.15 | 0.11 | 0.02 | 1.44 (1.04–1.99) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs9888739 | AG | 0.19 | 0.12 | 3.90 × 10−8 | 1.65 (1.38–1.98) | 0.21 | 0.15 | 0.001 | 1.58 (1.18–2.1) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs7499077 | AG | 0.33 | 0.27 | 8.56 × 10−5 | 1.33 (1.15–1.54) | 0.29 | 0.22 | 0.008 | 1.39 (1.08–1.77) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs11860650 | AG | 0.18 | 0.12 | 1.34 × 10−7 | 1.64 (1.36–1.97) | 0.17 | 0.10 | 0.0001 | 1.91 (1.37–2.67) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs6565227 | TA | 0.19 | 0.12 | 8.76 × 10−8 | 1.63 (1.36–1.95) | 0.21 | 0.15 | 0.001 | 1.57 (1.18–2.09) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs7206295 | AG | 0.29 | 0.31 | 0.11 | 0.88 (0.76–1.02) | 0.40 | 0.39 | 0.72 | 1.04 (0.83–1.28) | 0.19 | 0.22 | 0.042 | 0.82 (0.69–0.99) | 0.16 | 0.18 | 0.51 | 1.11 (0.79–1.56) |
| ITGAM | rs1143683 | AG | 0.22 | 0.15 | 7.98 × 10−7 | 1.52 (1.29–1.81) | 0.19 | 0.12 | 0.0001 | 1.79 (1.31–2.43) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs1143678 | AG | 0.22 | 0.15 | 7.98 × 10−7 | 1.52 (1.29–1.81) | 0.20 | 0.12 | 0.0004 | 1.71 (1.26–2.31) | 0 | 0 | – | – | – | – | – | – |
| ITGAM | rs4597342 | AG | 0.29 | 0.31 | 0.15 | 0.9 (0.78–1.04) | 0.40 | 0.39 | 0.85 | 1.02 (0.82–1.26) | 0.19 | 0.23 | 0.045 | 0.83 (0.69–0.99) | 0.17 | 0.18 | 0.65 | 1.08 (0.77–1.5) |
| Outer | rs4506917 | CA | 0.47 | 0.51 | 0.01 | 0.83 (0.73–0.95) | 0.35 | 0.42 | 0.01 | 0.75 (0.61–0.93) | 0.21 | 0.24 | 0.09 | 0.86 (0.72–1.02) | – | – | – | – |
| Outer | rs4075052 | AC | 0.29 | 0.31 | 0.14 | 0.89 (0.77–1.03) | 0.40 | 0.40 | 0.92 | 1.01 (0.81–1.25) | 0.21 | 0.24 | 0.07 | 0.85 (0.71–1.01) | – | – | – | – |
| Outer | rs4261553 | AG | 0.29 | 0.31 | 0.16 | 0.90 (0.78–1.04) | 0.40 | 0.40 | 0.94 | 1 (0.81–1.24) | 0.21 | 0.24 | 0.07 | 0.85 (0.71–1.01) | 0.18 | 0.26 | 0.68 | 0.68 (0.22–1.82) |
| Outer | rs11150613 | GA | 0.30 | 0.32 | 0.21 | 0.91 (0.79–1.05) | 0.40 | 0.40 | 0.96 | 1 (0.81–1.24) | 0.21 | 0.24 | 0.09 | 0.85 (0.72–1.02) | – | – | – | – |
| ITGAX | rs2230429 | CG | 0.31 | 0.34 | 0.05 | 0.87 (0.75–1) | 0.28 | 0.35 | 0.004 | 0.72 (0.57–0.9) | 0.21* | 0.22 | 0.24 | 0.90 (0.75–1.08) | 0.16 | 0.18 | 0.23 | 0.35 (0.07–1.25) |
| ITGAX | rs9929832 | GA | 0.50 | 0.47 | 0.09 | 1.12 (0.98–1.28) | 0.48* | 0.49 | 0.61 | 0.94 (0.76–1.17) | 0.23 | 0.22 | 0.37 | 1.08 (0.91–1.29) | 0.16 | 0.17 | 0.96 | 1.05 (0.72–1.61) |
MAF, minor allele frequency.
*Minor allele flipped.
†Minor allele/major allele.
Haplotype and conditional analysis
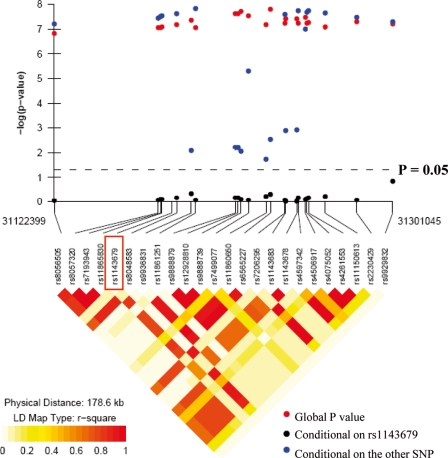
In EAs and HAs, multiple SNPs show highly significant associations (Table 1). This could be due either to the high correlation structure between SNPs or to involvement of multiple independently associated SNPs. A linkage disequilibrium (LD) plot, created for control samples in each population, revealed a highly correlated LD structure in EA and HA and an extremely strong correlation structure in Korean and Japanese populations (Supplementary Material, Fig. S1). We performed two-SNP haplotype analysis including rs1143679 paired with any other SNP. To exclude the possibility that multiple observed effects are caused by LD with a single true effect, pairs of SNPs were conditioned on each other, one at a time. If the global haplotype association disappeared, then the conditioned SNP explained the observed association and vice versa. As expected, all of the two-SNP combinations with rs1143679 showed highly significant global association. Conditional analyses demonstrated that the two-SNP global association disappeared for all sets if conditional on rs1143679, but remained significant when conditioned on the other SNP (Fig. 1). A similar pattern was seen in HA (data not shown). This further strengthens our hypothesis that all significant associations surrounding rs1143679 arise from the high correlation between themselves and rs1143679.
Figure 1.
Two-SNP conditional haplotype analysis plots for EAs. A two-SNP haplotype analysis including rs1143679 and any one of the other SNPs in order, in which both SNPs were conditional on each other, one at a time. Conditional analyses demonstrated that the two-SNP global association disappeared if conditional on rs1143679, but remained significant when conditional on the other SNP for all sets.
Structured association test in HAs
Hidden population substructure may lead to spurious associations, especially in HA samples in our current study. A structured association test (SAT) (8) was applied to HA samples using 76 Hispanic-specific ancestry informative markers (AIMs) (9). Information on AIMs is shown in Supplementary Material, Table S2. We first used STRUCTURE program (10) to estimate the admixture proportion for each individual. SAT was then applied for each of the 24 SNPs. As expected, a three-subpopulation model best fit the data with the overall admixture proportion of 48.7, 9 and 42.3%. The individual admixture proportion is presented in Supplementary Material, Figure S2. A structured-based association also yielded the highest significance at rs1143679 (PSAT = 2 × 10−5), and the covariate (individual admixture proportion) adjusted OR using logistic regression (2.26, 95% CI = 1.88–2.72) is comparable to the main analysis.
LD structure in and around ITGAM gene (from TRIM72 to ITGAX) across populations
To obtain a broad view of the LD structure across the region from TRIM72 to ITGAX, we used Hapmap data for Han Chinese (CHB), Japanese (JPT), European (CEU) and Yoruba Africans (YRI). As expected, the pair-wise LD structure was weaker in YRI samples and became much stronger in CEU samples. LD structure among common SNPs was most highly correlated in CHB and JPT (Supplementary Material, Fig. S3). This region in CHB and JPT samples showed many monomorphic SNPs or very low MAF compared with the CEU and YRI samples.
Imputation-based association testing
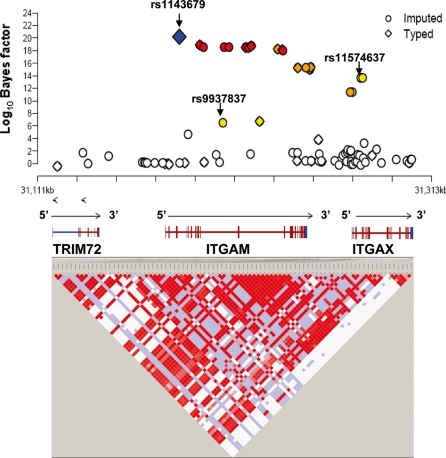
To create a more comprehensive fine map of the SNPs in and around the ITGAM region from TRIM72 to ITAGX, we imputed SNPs that were not genotyped in our study, but were available from the Hapmap database. We extracted the CEU Hapmap genotype data for 81 SNPs with MAFs greater than 0.01 from TRIM72 to ITGAX region (Hapmap-II). Among the 24 SNPs we used in EAs of our current study, 21 SNPs were common with Hapmap genotype data. Sixty SNPs were imputed and tested for association together with the genotyped SNPs, for a total of 84 SNPs. Both imputed and genotyped SNPs were assessed for association with the phenotype using a ‘Bayesian IMputation Based Association Mapping’ method implemented in BIMBAM (11). Bayesian factors (BFs) associated with each SNP were used to measure the strength of an association between genotype and phenotype.
The strongest evidence for association came from a highly correlated region starting from ITGAM and extended to ITGAX, where a total of 22 SNPs demonstrate convincing evidence of association (log10BFs>6) (Fig. 2). Interestingly, among these 22 highly associated SNPs, 11 SNPs were genotyped in our study and 11 SNPs were imputed, demonstrating the power of the imputation strategy for finding additional disease-related SNPs. The strongest evidence for association remained with rs1143679, and the other 21 SNPs showing convincing evidence for association were in high LD with rs1143679. This supports the hypothesis that rs1143679 might be the most promising candidate for a causal SNP and that significant associations from nearby SNPs arise due to the high correlation with rs1143679. It is also worth noting that the two most significant SNPs associated with SLE (7), rs9937837 and rs11574637, were also imputed out in our current study and had log10BFs of 6.5 and 13.6, respectively, compared with a log10BF of ∼20 at rs1143679. In addition, to assess the evidence for multiple SNPs affecting the phenotype, we used BIMBAM to compute multi-SNP BFs for all subsets of up to three SNPs. However, no improvement was found compared with the single SNP analysis, suggesting the absence of multi-SNP effects in ITGAM.
Figure 2.
Bayesian association assessment testing in and around ITGAM. All genotyped (diamond) and imputed (circle) SNPs are plotted with their log10 Bayes factor along with their physical position (NCBI build 35). The blue diamond is the proposed causal SNP in this study. The colors of white, yellow, orange and red represent the r2 correlations with rs1143679 (red: r2 ≥ 0.8; orange: 0.5 ≤r2 < 0.8; yellow: 0.2 ≤r2 < 0.5; white: r2 < 0.2). Blocks connecting pairs of SNPs are shaded according to the strength of the LD between the SNPs, from 0 (white) to 1.0 (bright red), as measured by the disequilibrium coefficient r2.
As a second approach for imputation-based association test, we performed imputing in the same region from TRIM72 to ITGAX region by IMPUTE/SNPTEST program (12,13) and tested the genotype–phenotype association in a frequentist framework completely taking into account the uncertainty of the genotypes. Significantly, a major association signal came from the same region detected by BIMBAM, starting from ITGAM and extending to ITGAX. Consistently, 22 SNPs with a log10BF greater than 6 showed a P-value less than 1 × 10−7, and the smallest P-value came from rs1143679 (P = 6.2 × 10−24). The other 21 SNPs that showed strong evidence for association (P < 1 × 10−7) were in moderate-to-high LD with rs1143679. For rs9937837 and rs11574637, the P-value was 1.76 × 10−9 and 1.86 × 10−17, respectively. A detailed result is shown in Supplementary Material, Table S4 and Figure S4.
To further explore the relationship of two previously reported (7) associated SNPs (rs9937837 and rs11574637) with rs1143679 in the association with SLE, we employed Hapmap CEU genotype data (60 unrelated parents) to impute rs9937837 and rs11574637 genotype data in combined EA samples by fastPhase program (14). Again, we used haplotype conditional analysis to dissect the correlation structure and determine which SNP was the best candidate to be the causal SNP. We performed a haplotype analysis using the three SNPs: rs1143679, rs9937837 and rs11574637. The global association was very significant (P = 2.2 × 10−20). However, conditional analysis demonstrated that the global association disappeared if conditioned on rs1143679 (P = 0.74), but remained significant if conditioned on rs9937837 (P = 7.6 × 10−14) or rs11574637 (P = 0.00012). This analysis further supports that rs1143679 explains the association of rs9937837 or rs11574637 with SLE.
Additional replication of rs1143679 association in Mexican, Colombian and UK samples
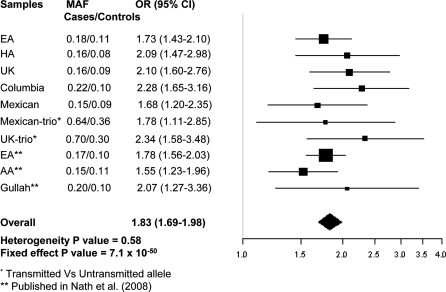
To assess the robustness of the genetic association across multiple populations, we genotyped rs1143679 in three additional independent case–control samples from Mexico, Colombia and UK, as well as two independent sets of trios from UK and Mexico. Association (same risk allele, ‘A’) in all three case–control samples was replicated: P = 3.6 × 10−7, OR = 2.28, 95% CI = 1.65–3.16 for Colombian; P = 0.002, OR = 1.68, 95% CI = 1.20–2.35 for Mexican and P = 6.2 × 10−8, OR = 2.10, 95% CI = 1.60–2.76 for UK. Replication was also demonstrated in both of the trio samples: PTDT = 1.4 × 10−5, OR = 2.34, 95% CI = 1.58–3.48 for UK and PTDT = 0.015, OR = 1.78, 95% CI = 1.11–2.85 for Mexican (Fig. 3), where the same risk allele ‘A’ is over-transmitted in probands. As allele frequencies were similar and the ORs are in the same direction, we combined the case-control and trio data together to get overall region-specific association. The combined P and OR (95% CI) for UK and Mexican are 1.18 × 10−11, 2.17 (1.73–2.72) and 1.1 × 10−4, 1.71 (1.30–2.25), respectively.
Figure 3.
Meta-analysis of rs1143679 combining all independent data sets in current study and our previously reported (4) data sets.
Meta-analysis
To obtain an overall strength of genetic association at rs1143679, we performed a meta-analysis (15,16) by obtaining an overall OR using all independent case–control data sets, including our previously reported data sets (4) and trio data sets. As rs1143679 was monomorphic in both Korean and Japanese populations, these populations were excluded. The effect of rs1143679 on SLE risk was consistent across all independent samples (heterogeneity P = 0.58). The magnitude of the overall P-value under the fixed-effect model was Pmeta = 7.1 × 10−50 (OR = 1.83, 95% CI = 1.69–1.98), again reinforcing the genetic association with SLE (Fig. 3).
Additional analysis
We performed additional analyses on rs1143679 to understand some relevant epidemiological characteristics. First, analysis of the data stratifying by gender yielded no significant differences between genders. Secondly, we compared different genetic models to identify a parsimonious genetic model for rs1143679 in different ethnic population samples. The best genetic model for each independent group was selected using both P-values and AIC. Our analysis revealed that the multiplicative model best explained the data in all independent populations (Supplementary Material, Table S3). Thirdly, using the overall risk allele frequency and OR, we analytically (17) estimated the sibling relative risk (λs) attributable to a given SNP. Assuming overall λs = 30, the locus-specific λs due to rs1143679 varies from 1.04 (Mexican) to 1.18 (Colombian), which explained ∼3–4% of the overall excess risk for SLE under a multiplicative model. Fourth, using the OR and risk allele frequency, we estimated the population attributable risk (PAR) due to rs1143679 in each population. We have estimated the lowest and highest PARs to be 12% (Mexican) and 24% (Colombian), respectively.
DISCUSSION
In the present study, we assessed ITGAM and SLE association in nine independent samples from ethnically diverged populations. We used both case–control and trio data sets for testing association. We confirmed and replicated our previous results that the minor allele ‘A’ of the non-synonymous coding SNP, rs1143679, in exon 3 of ITGAM is strongly associated with increased SLE risks. Imputation-based association results further extended our hypothesis that rs1143679 is the most promising candidate for causal association in EA, and hence, our intent was to replicate association with SLE in other ethnically and geographically diverged populations. We confirmed this association in independent EA, HA, UK, Mexican and Colombian samples; however, this SNP is monomorphic in both Korean and Japanese samples. LD structure from both our genotyped data (Korean and Japanese) and Hapmap data (CHB and JPT) surrounding the ITGAM region shows highly correlated LD structure in Asian populations (Supplementary Material, Figs S1 and S3), which might reflect some selection pressure existing to maintain this striking LD across this region in Asian populations.
As we reported elsewhere (4), we used a computer generated model to predict the position of the associated variant, rs1143679, how it might lie within the protein and how it might modulate the tertiary and quaternary structures of the protein. T-Coffee (18) was used to align the amino acid sequences corresponding to six known integrin α-chains (αM, αX, αD, αL, αIIb and αV). The position of the R77H polymorphism in this model suggests that it is not likely to affect the C3bi binding site. This polymorphism is also not likely to influence interactions between the β-propeller domain and the βI domain. However, from these models, it cannot be determined whether this polymorphism might alter binding of other αMβ2 ligands.
ITGAM encodes the α-chain of the αMβ2-integrin (Mac-1, CR3, CD11b/CD). αMβ2 regulates leukocyte activation and adhesion as well as migration from the bloodstream via interactions with a range of structurally unrelated ligands ICAM-1, C3bi, fibrinogen, glycoprotein Ibα (19,20). αMβ2 levels increase on neutrophils in lupus patients with active disease and may contribute to endothelial injury in SLE (21). The polymorphism rs1143679 associated with SLE causes the conversion of the normal arginine at amino acid position 77 to a histidine (R77H). The associated amino acid change has been shown to alter the tertiary and quaternary structures of the ligand-binding domain of αMβ2, changing its binding affinity (22). This polymorphism has been shown to be the target of the alloantibodies present in the mothers of neonates affected with neonatal autoimmune neutropenia, suggesting that the polymorphism indeed causes biologically and clinically significant structural changes in the αMβ2 molecule. Alloantibodies reactive against the polymorphic αMβ2 molecule block the αMβ2-dependent adhesion of neutrophils and monocytic U937 cells to fibrinogen, intracellular adhesion molecule-1 (ICAM-1) and glycoprotein Ibα; however, alloantibody does not block other αMβ2 ligands (22).
Our comprehensive imputation-based analysis indubitably identified that rs1143679 is the only variant that can best explain the observed ITGAM-SLE association. Although extremely promising as the causal polymorphism, rs1143679 could be in LD with other as yet unobserved independent causal variant(s) that may be rare or common. After imputation-based analysis, there was no strong evidence to support this possibility. However, as imputation was based on the common Hapmap (MAF>1%) SNPs, the possibility of other independent rare causal variant(s) within ITGAM cannot be ruled out with certainty. This issue raises possibilities that can be resolved by resequencing the entire ITGAM gene. Each new polymorphism with potential to confer functional consequences would be evaluated for a mechanism that could increase SLE risk.
In this context, we could add two pieces of information in favor of rs1143679 as the most promising variant (most likely a sole causal variant). First, we have resequenced entire exon 3 in 171 samples (85 cases and 86 controls), but we could not identify any novel variant (data not shown). Second, Genentech recently resequenced the entire ITGAM (all exons and UTRs) and shows that rs1143679 is the best variant that explains the ITGAM-SLE association, reconfirming our results (23).
It is worth noting that the model used in BIMBAM or IMPUTE is based on prospective likelihood and we applied this in our retrospective case–control data. Therefore, one can argue that this may not be an appropriate approach. However, Guan and Stephens (24) discussed and suggested that the use of the Bayesian prospective model remains appropriate for retrospective analysis and has already been applied to other case–control studies (13), where use of a retrospective likelihood would be more appropriate. For typed SNPs, results from Seaman and Richardson (25) provide conditions for the equivalence of prospective and retrospective Bayesian analysis. Hence, using BFs, from applying prospective methods to case–control data, will not be generally misleading.
In summary, our comprehensive imputation-based association analysis clearly demonstrates that rs1143679 is the only known non-synonymous exon 3 coding SNP of ITGAM that strongly associates with increased SLE risk. This genetic association is replicated in independent samples across multiple populations. Taken together with our earlier findings, these results show that the coding variant, rs1143679 of ITGAM, is robustly associated with SLE pathogenesis, especially in the European-derived, African-derived and Hispanic populations, but most likely not in Asian populations.
MATERIALS AND METHODS
Study populations
Samples were gathered from a variety of sources, including the LFRR (Lupus Family Registry and Repository) as well as collaborators, as part of other ongoing SLE-related research. All SLE patients met the revised SLE classification criteria of the American College of Rheumatology (26,27). A single patient was randomly selected when more than one affected individual was available from a pedigree multiplex for lupus, and all samples used in current study were independent. Genotypes were compared using PLINK to remove duplicate samples. This study was approved by the Institutional Review Boards or Ethics Committees at the Oklahoma Medical Research Foundation and University of Oklahoma Health Sciences Center, or the location at which subjects were recruited.
For the initial phase of our study, participants included 5378 unrelated SLE cases and controls (3818 EA and 1560 African-derived); the results of this analysis were reported elsewhere (4). The current study incorporated data from additional, independent cases and controls including 2130 unrelated SLE cases (738 independent EAs, 731 HAs and 661 Koreans) and 2063 unrelated controls (1053 independent EAs, 229 HAs and 781 Koreans). For the imputation-based association test in EAs, we used data from both the current EA (1791) and previously published EA samples (3818). Additionally, four additional independent cases–controls including Japanese (541), UK (973), Mexican (673) and Colombian (586) as well as two complete sets of trios from UK (243) and Mexico (172) populations were used for testing robust association (Table 2).
Table 2.
Demographics and sample sizes for independent data sets
| Study design | Population | Affection status | Sex (male/female) | Sample size | Total sample |
|---|---|---|---|---|---|
| Case–control | EA | Case | 59/679 | 738 | 1791 |
| Control | 405/648 | 1053 | |||
| Korean | Case | 40/621 | 661 | 1442 | |
| Control | 51/730 | 781 | |||
| Hispanic | Case | 73/658 | 731 | 960 | |
| Control | 36/193 | 229 | |||
| Japanese | Case | 13/163 | 176 | 541 | |
| Control | 299/66 | 365 | |||
| UK | Case | 28/417 | 445 | 973 | |
| Control | 0/528 | 528 | |||
| Mexican | Case | 37/352 | 389 | 673 | |
| Control | 14/270 | 284 | |||
| Colombian | Case | 7/198 | 205 | 586 | |
| Control | 125/256 | 381 | |||
| Total | Case | 257/3088 | 3345 | 6966 | |
| Control | 930/2691 | 3621 | |||
| Trio | UK | Parents | 243/243 | 486 | 729 |
| Proband | 23/220 | 243 | |||
| Mexican | Parents | 172/172 | 344 | 516 | |
| Proband | 27/145 | 172 | |||
| Total | Parents | 415/415 | 830 | 1245 | |
| Proband | 50/365 | 415 |
Genotyping
Most de-identified genomic DNA samples from SLE patients and control (EA, HA and Korean) subjects were genotyped at either the Oklahoma Medical Research Foundation or the University of Texas Southwestern Microarray Core Facility (Dallas, TX, USA). Samples were genotyped by a custom-designed, highly multiplexed, Illumina bead-based array method, and a detailed data cleaning and QC procedure was applied as we described (4). Thirty-four SNPs spanning three genes (TRIM2, ITGAM and ITGAX) were genotyped (Supplementary Material, Table S1) in the EA, HA and Korean populations. Japanese samples were genotyped for rs1143679, and 11 other SNPs that are polymorphic in Korean samples using the Invader assay combined with multiplex-PCR using ABI7700 and 7900 (Applied Biosystems). A 3-base extension method (28) and TaqMan assay were used to genotype the SNP, rs1143679, in the Mexican and Colombian samples. For UK samples, genotyping was performed by using an Illumina-based array, as described earlier.
QC of genotyping
Genotype data were only used from samples with a call rate greater than 90% of the SNPs screened (98.05% of the samples). The average call rate for all samples was 97.18%. Only genotype data from SNPs with a call frequency >90% in the samples tested and an Illumina GenTrain score greater than 0.7 (96.74% of all SNPs screened) were used for analysis. To verify sample identity, 91 SNPs that had been previously genotyped on 42.12% of the samples were retyped. In addition, at least one sample previously genotyped was randomly placed on each Illumina Infinium bead chip and used to track samples throughout the genotyping process.
STATISTICAL ANALYSIS
Single SNP analysis
Allele and genotype frequencies were calculated for each locus and tested for Hardy–Weinberg equilibrium (HWE) in controls. For single SNP QC, we employed a predetermined QC inclusion criterion (MAF > 1%, SNP call rate > 90%, control HWE P > 0.01). Case–control association studies were analyzed by χ2 test using 2 × 3 and 2 × 2 contingency tables of genotype and allele frequencies, respectively. Allelic OR and 95% CIs were calculated using PLINK 1.01 (29).
Haplotype and conditional analysis
LD plots were drawn with Haploview (30) using unrelated control populations. We used the squared correlation statistic (r2) as measures of LD strength. To disentangle the correlation structure in highly correlated regions, conditional haplotype analysis was performed using WHAP (31). WHAP directly calculates likelihood estimates, likelihood ratios and P-values and takes into account the loss of information due to haplotype phase uncertainty and missing genotypes. To test the hypothesis that all significant associations surrounding rs1143679 arise from the high correlation between themselves and rs1143679, we performed a two-SNP haplotype analysis including rs1143679 paired with any other SNP, and each SNP was conditional on singly, one at a time. If the global haplotype association disappeared, then the conditioned SNP explains the entire association and vice versa.
Admixture analysis and SAT in Hispanic samples
Spurious association between a marker and a phenotype can arise from population stratification, especially in admixed populations such as the HAs in our current study. To control for possible confounding due to population stratification, we performed an SAT in HA samples (8,9). We first used STRUCTURE to estimate the admixture proportion for each individual (10). The log likelihood of each analysis for varying numbers of population groups (k) was estimated from the average of three independent runs (20 000 burn in followed by 30 000 iterations). SAT was then applied for SNPs. SAT takes advantage of STRUCTURE to gain information about the population structure and to perform association tests conditional on ancestry proportion to exclude admixture effects. One hundred thousand simulated data sets under the null hypothesis were generated to evaluate the significance of the likelihood ratio test.
Imputation-based association test
We imputed all the SNPs that were not typed in our study but were available from the Hapmap database (Hapmap-II, MAF > 0.01) in order to create a more comprehensive fine map of the SNPs in and around the ITGAM region from TRIM72 to ITAGX. We first used BIMBAM (11) to impute SNPs that were not genotyped in our samples and to test for association with SLE. The imputation in BIMBAM was performed using the fastPHASE algorithm, which implements methods based on a cluster for haplotypes to estimate missing genotypes and reconstruct haplotypes from unphased SNP genotype data of unrelated individuals. Association between imputed and genotyped SNPs was assessed with the phenotype in a Bayesian regression framework, providing a natural means to consider uncertainty in estimated genotypes. BFs associated with each SNP were used to measure the strength of an association between genotype and phenotype. We used the default setting of the BIMBAM program, and BFs were computed under logistic regression of phenotypes on genotypes with additive (i) and dominance (ii) genetic effect incorporated in the model. The priors were obtained from prior D2, averaging over σa = 0.05, 0.1, 0.2, 0.4 and σd=σa/4. The authors of BIMBAM suggested in their latest work (24), when computing BFs based on imputed genotypes, that simply replacing the imputed genotypes with their posterior mean produces a good approximation to a full analysis, and the number of imputations (parameter ‘i’ in BIMBAM command line) was set to 1. Moreover, we also employed a sampling-based approach to calculate the BFs by setting ‘i’ to be 10 000, and the result was nearly identical to ‘i’ equal to 1 (Supplementary Material, Table S4 and Fig. S4). To assess the evidence for multiple SNPs affecting phenotype, we used BIMBAM to compute multi-SNP BFs for all subsets of up to three SNPs.
As a second and a complementary imputation-based association approach, we used the IMPUTE and SNPTEST programs (12,13) to impute SNPs in the same region as BIMBAM did and performed a genotype–phenotype association test in a frequentist framework. IMPUTE determined the probability distribution of missing genotype conditional on a set of known haplotypes and an estimated fine-scale recombination map. In our study, we used haplotypes and recombination rate files based on NCBI build 36. SNPTEST implements a statistical test based on missing data likelihood, taking account of the uncertainty of the genotypes completely. A score test P-value based on an additive genetic model was calculated and reported by SNPTEST.
Meta-analysis
We employed a meta-analysis (15,16) to obtain an overall OR using all independent case–control data sets, including previously reported data sets (4) and trio data sets. We performed meta-analysis for the case–control and trio data using CATMAP (R package), which implements methods proposed by Kazeem and Farrall (15), to obtain OR and its standard error from a standard transmission/disequilibrium test and integrating these with the results from the case–control study. A total of 10 046 individual samples were used in the meta-analysis. Korean and Japanese samples were excluded as rs1143679 appears to be monomorphic in these populations. Significance for combined result was assessed by the χ2 test. Cochran's Q statistic was used to test for heterogeneity between different data sets. In the absence of significant heterogeneity across the studies, fixed-effect models were used to combine the ORs.
SUPPLEMENTARY MATERIAL
FUNDING
The authors wish to thankfully acknowledge support from the National Institutes of Health (AI063622, RR020143, AR053483, AR049084, AI24717, AR42460, AR048940, AR445650, AR043274), the Alliance for Lupus Research, the US Department of Veterans Affairs, Wellcome Trust Senior Clinical Fellowship, ARC Project Grant (ref 17761), the Swedish Research Council, the Torsten and Ragnar Söderbergs Foundation, the SIDA/SAREC Foundation, the USC FCE, a grant of the Korea Health 21 R&D Project, Ministry of Health and Welfare, Republic of Korea (01-PJ3-PG6-01GN11-0002), Colciencias, Bogota, Colombia (2213-04-16484) and the Lupus Foundation of Minnesota.
Supplementary Material
ACKNOWLEDGEMENTS
These authors are grateful to the patients and their families for their cooperation and blood samples. We would like to thank the PROFILE Study Group (Drs Michelle Petri, John Reveille, Rosalind Ramsey-Goldman, Jeffrey Edberg and Graciela Alarcón) for contributing samples. We also acknowledge the support and help from all the LFRR staff, especially Gail Bruner and Jennifer Kelly. The authors also thankfully acknowledge the help and thoughtful discussion with Drs Rod McEver and Cheng Zhu and Wei Chen for providing the multiple sequence alignment and modeling of cd11b. Finally, the authors would like to thank Dr Matthew Stephens for clearing and helping in understanding some analytical issues regarding BIMBAM.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Danchenko N., Satia J.A., Anthony M.S. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus. 2006;15:308–318. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 2.Alarcón-Segovia D., Alarcón-Riquelme M.E., Cardiel M.H., Caeiro F., Massardo L., Villa A.R., Pons-Estel B.A. Grupo Latinoamericano de Estudio del Lupus Eritematoso (GLADEL) Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52:1138–1147. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 3.Sestak A.L., Nath S.K., Sawalha A.H., Harley J.B. Current status of lupus genetics. Arthritis Res. Ther. 2007;9:210. doi: 10.1186/ar2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nath S.K., Han S., Kim–Howard X., Kelly J.A., Viswanathan P., Gilkeson G.S., Chen W., Zhu C., McEver R.P., Kimberly R.P., et al. A nonsynonymous functional variant in integrin-alpha (M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat. Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 5.Harley J.B., Alarcón–Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham R.R., Cotsapas C., Davies L., Hackett R., Lessard C.J., Leon J.M., Burtt N.P., Guiducci C., Parkin M., Gates C., et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hom G., Graham R.R., Modrek B., Taylor K.E., Ortmann W., Garnier S., Lee A.T., Chung S.A., Ferreira R.C., Pant P.V., et al. Association of systemic lupus erythematosus with C8orf13–BLK and ITGAM–ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 8.Pritchard J.K., Stephens M., Rosenberg N.A., Donnelly P. Association mapping in structured populations. Am. J. Hum. Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian C., Hinds D.A., Shigeta R., Adler S.G., Lee A., Pahl M.V., Silva G., Belmont J.W., Hanson R.L., Knowler W.C., et al. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am. J. Hum. Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Servin B., Stephens M. Imputation–based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. doi:10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies via imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 13.The Wellcome Trust Case Control Consortium. Genomewide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheet P., Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazeem G.R., Farrall M. Integrating case–control and TDT studies. Ann. Hum. Genet. 2005;69:329–335. doi: 10.1046/j.1529-8817.2005.00156.x. [DOI] [PubMed] [Google Scholar]
- 16.Nicodemus K.K. Catmap: case–control and TDT meta-analysis package. BMC Bioinformatics. 2008;9:130. doi: 10.1186/1471-2105-9-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 18.Poirot O., O'Toole E., Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L., Li Z. The fourth blade within the β-propeller is involved specifically in C3bi recognition by integrin αMβ2. J. Biol. Chem. 2003;278:34395–34402. doi: 10.1074/jbc.M304190200. [DOI] [PubMed] [Google Scholar]
- 20.Fagerholm S.C., Varis M., Stefanidakis M., Hilden T.J., Gahmberg C.J. α-Chain phosphorylation of the human leukocyte CD11b/CD18 (Mac-1) integrin is pivotal for integrin activation to bind ICAMs and leukocyte extravasation. Blood. 2006;108:3379–3386. doi: 10.1182/blood-2006-03-013557. [DOI] [PubMed] [Google Scholar]
- 21.Buyon J.P., Shadick N., Berkman R., Hopkins P., Dalton J., Weissmann G., Winchester R., Abramson S.B. Surface expression of Gp 165/95, the complement receptor CR3, as a marker of disease activity in systemic Lupus erythematosus. Clin. Immunol. Immunopathol. 1988;46:141–149. doi: 10.1016/0090-1229(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 22.Sachs U.J.H., Chavakis T., Fung L., Lohrenz A., Bux J., Reil A., Ruf A., Santoso S. Human alloantibody anti-Mart interferes with Mac1-dependent leukocyte adhesion. Blood. 2004;104:727–734. doi: 10.1182/blood-2003-11-3809. [DOI] [PubMed] [Google Scholar]
- 23.Ferreira R.C., Hom G., Ortmann W., Petri M., Manzi S., Criswell L.A., Gregersen P.K., Graham R.R., Behrens T.W. A12, Genomics of Common Diseases, Nature Genetics and Welcome Trust Joint Conference. Cambridge, MA: The Broad Institute of MIT and Harvard; 2008. Deep re-sequencing of the ITGAM-ITGAX locus confirms the R77H allele as the functional variant associated with systemic lupus erythematosus. [Google Scholar]
- 24.Guan Y., Stephens M. Practical issues in imputation-based association mapping. PLoS Genet. 2008;4:e1000279. doi: 10.1371/journal.pgen.1000279. doi:10.1371/journal.pgen.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seaman S.R., Richardson S. Equivalence of prospective and retrospective models in the bayesian analysis of case–control studies. Biometrika. 2004;91:15–25. [Google Scholar]
- 26.Tan E.M., Cohen A.S., Fries J.F., Masi A.T., McShane D.J., Rothfield N.F., Schaller J.G., Talal N., Winchester R.J. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 27.Hochberg M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman K.M., Kelly J.A., Herring B.J., Adler A.J., Glenn S.B., Namjou B., Frank S.G., Dawson S.L., Bruner G.R., James J.A., Harley J.B. Evaluation of the genetic association of the PTPN22 R620W polymorphism in familial and sporadic systemic lupus erythematosus. Arthritis Rheum. 2006;54:2533–2540. doi: 10.1002/art.21963. [DOI] [PubMed] [Google Scholar]
- 29.Purcell S., Neale B., Todd–Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett F., Maller D. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 31.Purcell S., Daly M.J., Sham P.C. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.