Abstract
Colicins are plasmid-encoded toxic proteins produced by Escherichia coli strains to kill other E. coli strains that lack the corresponding immunity protein. Colicins intrude into the host cell by exploiting existing transport, diffusion, or efflux systems. We have traced the way colicin S4 takes to execute its function and show that it interacts specifically with OmpW, OmpF, and the Tol system before it inserts its pore-forming domain into the cytoplasmic membrane. The common structural architecture of colicins comprises a translocation, a receptor-binding, and an activity domain. We have solved the crystal structure of colicin S4 to a resolution of 2.5 Å, which shows a remarkably compact domain arrangement of four independent domains, including a unique domain duplication of the receptor-binding domain. Finally, we have determined the residues responsible for binding to the receptor OmpW by mutating exposed charged residues in one or both receptor-binding domains.
Colicins are plasmid-encoded toxic proteins produced by up to 50% of Escherichia coli strains in natural populations (1). They are produced to kill competing E. coli strains under stress conditions and are regulated by the SOS response (2, 3). Among the toxic functions of colicins, pore formation in the cytoplasmic membrane followed by a breakdown of the electrochemical gradients is the most abundant killing mechanism (4). Other colicins kill by exerting DNase, RNase, or phosphatase activity in the target cell (2). Colicin producer strains are protected from the toxic action by specific immunity proteins encoded on the same plasmid (5-7). In some cases, an additional protein is encoded on the plasmid, which ensures an efficient lysis of dying producer cells and, as a consequence, the maximal release of colicin into the environment (2, 8). The ubiquitous presence of non-colicinogenic strains in the same environments with colicin producers indicates that there has to be a narrow tradeoff between the costs and the benefits of colicin production (9).
Colicins exhibit a modular structure, which suggests that their domains are frequently recombined to create new toxic functions in highly competitive environments (1, 10). The modules comprise the N-terminal translocation domain, followed by a specific receptor-binding domain and the activity domain at the C terminus. The most pronounced sequence conservation can be observed among the activity domains of pore-forming colicins. Accordingly, the structures of colicin pore-forming domains solved so far are highly similar (2, 11). The highest variability in sequence and structure can be seen in the central receptor-binding domains that are adapted to bind to very different receptors with high affinity (2, 12).
The initial contact between colicin and the target cell is established between the receptor-binding domain and a specific receptor of the target cell, which in all cases is an outer membrane protein. Colicins recognize their receptor proteins with high specificity (12), and few point mutations in the receptor, or its entire deletion, can render the cell fully resistant to the colicin. This also explains the narrow range of strains that are sensitive to a certain colicin or groups of colicins.
The N-terminal translocation domain is subsequently used either to penetrate through the receptor pore itself into the periplasm or to recruit an additional translocation pore, typically the general diffusion pore OmpF, to do so (13, 14). It is typically not resolved in crystal structures, and NMR spectroscopy indicates a high flexibility in this domain (15). For some colicins, it has been shown that a whole cascade of interactions between different periplasmic proteins of the target cell and the N terminus of the colicin is necessary for the successful translocation. Colicins have been classified into two groups according to the energized uptake system that they abuse; group A colicins exploit the Tol system for translocation, whereas group B colicins exploit the Ton system (2, 10, 16). The Tol system comprises the lipoprotein Pal, the β-propeller protein TolB, the inner membrane protein TolQ, and TolA and TolR, which span the cytoplasmic membrane but also have extended periplasmic domains. The function of the Tol system is still not fully understood (17). The Ton system comprises TonB in complex with ExbB and ExbD; this complex uses the protonmotive force to interact with outer membrane transporters to energize the import of bound substrates, e.g. iron or cobalamin, and of colicins (18, 19). In the final step of colicin import, the C-terminal activity domain is translocated through the outer membrane to reach its place of action, which is the cytoplasmic membrane for pore-forming colicins, the periplasm for colicin phosphatases, or the cytosol for colicins with DNase or RNase activity.
In contrast to the common domain architecture, the three-dimensional structures known so far differ remarkably in their shape. The first colicin structures (e.g. colicins Ia and E3) solved by x-ray crystallography have been summarized as elongated, mainly α-helical molecules (20, 21). Compact structures are observed in colicins M, N, and B (22-24). Intrigued by the unusual receptor-binding domain duplication in colicin S4 and by its binding to the small outer membrane protein OmpW (25), we set out to investigate the pathway that colicin S4 takes through the outer membrane and the periplasm to insert into the cytoplasmic membrane. To this end, we have tested a series of E. coli knock-out strains with deletions of outer membrane proteins and proteins of the Tol and Ton systems for their resistance to colicin S4 and show that it is a Tol-dependent group A colicin. Using single channel conductance measurements, we were able to characterize the pore-forming activity. We have solved the structure of colicin S4 using x-ray crystallography, which displays an unusual, Y-shaped structure with two almost identical receptor-binding domains that form the arms. The sequence similarity of the two receptor-binding domains and their arrangement in the colicin structure raised the question whether colicin S4 would indeed need two functional receptor-binding domains to bind to target cells. On the basis of the structure, we have mapped the crucial charged residues for receptor binding and mutated them to alanines in one or both domains.
EXPERIMENTAL PROCEDURES
Strains, Constructs, and Primers—All strains and constructs used in this study are listed in supplemental Table SI, and all primers used can be found in supplemental Table SII.
Sequence Analysis—Homology searches were done using PSI-BLAST and the non-redundant data base (26). Alignments were computed with ClustalW (27) and were further edited manually. Secondary structure elements in the pore alignment were assigned using Protein Data Bank codes 2i88 (colicin E1) and 1cii (colicin Ia). All relevant GenBank™ identifiers are listed in supplemental Table SIII.
Construction of the 5KΔompW Strain—The ompW deletion strain was produced using the pKO3 vector for homologous recombination (28); to be able to compare it with the spontaneous ompW mutation generated in a mutagenesis screen (25), the same background strain (E. coli 5K) was chosen. The genome regions upstream and downstream of the OmpW coding region were amplified by PCR and fused in a second PCR using primers KOvfwd, KOvrev, KOhfwd, and KOhrev, which contain compatible ends. The resulting PCR product was subcloned into pCR-BLUNT (Invitrogen), digested with BamHI, and ligated into the pKO3 vector. Selection for deletion clones was done as described (28) and was verified by PCR of the genomic DNA.
Cloning, Expression, and Purification of OmpW—OmpW was cloned, expressed, and purified as described (29).
Cloning and Expression of Colicins S4 and S4His—The DNA sequence was amplified from the pColS4 plasmid using primers S4-fwd and S4-rev or S4-His-rev, respectively. The PCR product was digested with Eco91I and cloned into an Eco91I-digested pASK-IBA33+ vector (IBA GmbH, Göttingen, Germany). The E. coli strain 5KΔompW was transformed with the resulting plasmids, p33-S4 and p33-S4His. Expression of colicin S4 or S4His was induced by adding 0.2 μg/ml anhydrotetracycline (IBA GmbH).
Generation of Mutant Colicin Variants—To introduce mutations into the receptor-binding domain, the sequence was first divided between the receptor-binding domains to avoid misannealing of primers in the almost identical sequence parts. The first part was engineered by fusing the PCR product obtained from the wild-type colicin S4 sequence with primers S4-fwd-all and Colmut-2-rev with the PCR product obtained by fusion of primers Colmut-3-fwd, Colmut-4-rev, Colmut-5-fwd, and Colmut-6-rev. To generate the first part without amino acid mutation in receptor-binding domain R1, the primer Colmut-4wt-rev was used instead of Colmut-4-rev. The second part was generated from the wild-type colicin S4 sequence using primers S4-fwd-R2P and ColS4-rev-long. The mutations in the second part were introduced by fusing the PCR products obtained with primers S4-fwd-R2P and S4mut-R2rev and primers Colmut-R2fwd and ColS4-rev-long using the previously generated second part fragment as a template. The two parts were fused by adding primers S4-fwd-all and ColS4-rev-long using parts with and without amino acid changes in every combination to yield S4m1, S4m2, and S4m12, respectively. The PCR products were digested with Eco91I and cloned into Eco91I-digested pASK-IBA33+ vector (IBA GmbH).
Purification of Colicin S4 and Its Mutant Variants—4 h post-induction, cells were pelleted at 7000 × g. After resuspension of the pellet in buffer containing 20 mm Tris (pH 8.5), 1 mm MgSO4, 1 mm MnSO4, and a pinch of DNase, cells were passed three times through a French pressure cell. The lysate was centrifuged in an ultracentrifuge at 60,000 × g for 30 min. The supernatant was applied to a Mono Q column (GE Healthcare). The protein was eluted with a gradient to 1 m NaCl. The fractions containing colicin S4 were pooled, concentrated using an Amicon concentrator (Millipore), and applied to a preparative Superdex 200 column (GE Healthcare) equilibrated with 150 mm NaCl and 20 mm MOPS3 (pH 7.0).
Purification of Colicin S4His—Cell lysis was performed as described for colicin S4. After centrifugation in an ultracentrifuge for 30 min at 60,000 × g, the supernatant was applied to a nickel-nitrilotriacetic acid column (GE Healthcare). The protein was eluted with a gradient to 0.5 m imidazole. The colicin S4-containing fractions were pooled, concentrated using an Amicon concentrator, and applied to a preparative Superdex 200 column equilibrated with 150 mm NaCl and 20 mm MOPS (pH 7.0).
Colicin S4 Sensitivity Assay—The E. coli strains used in this assay (supplemental Table SI) were obtained from the Keio Collection (30). 10 μl of an overnight culture was mixed with 3 ml of melted LB top agar and poured on an LB agar plate. After cooling to room temperature, round filter platelets (Schleicher & Schüll) were placed on the plates. The filter platelets were subsequently soaked with colicin S4 dilutions, with total colicin S4 amounts ranging from 10 to 10-7 μg. The so-treated LB agar plates were incubated overnight at 37 °C. Colicin S4 sensitivities of the different strains were evaluated by checking for clear inhibition zones around the filter platelets.
Single Channel Conductance Measurements—Single channel conductance values were recorded using a BLM workstation (Warner Instruments, Hamden, CT) with a BC-535 amplifier and an LPF-8 Bessel filter connected to an Axxon Digidata 1440A digitizer. Data were recorded and evaluated using pCLAMP 10.0 software (Molecular Devices, Sunnyvale, CA) supplied with the digitizer. First of all, 3 μl of a 1% (w/v) solution of ultrapure hen egg phosphatidylcholine (a gift from Lipoid GmbH, Ludwigshafen, Germany) in 1:1 (v/v) methanol/chloroform was applied to a 150-μm aperture in a 4-ml polysulfone cuvette (Warner Instruments). After evaporation of the solving agents, the cuvette was filled with the measurement buffer, which was 1 m KCl and 20 mm MES (pH 6.0). 3 μl of a 1% (w/v) solution of ultrapure hen egg phosphatidylcholine in 9:1 (v/v) n-decane/butanol was painted onto the 150-μm aperture of the cuvette. 1 μl colicin of S4 solution (3-4 mg/ml) was added to the cuvette, which contained the ground electrode of the setup. Colicin S4 inserted readily into the membrane at a potential of approximately -100 mV.
Protein Crystallization—Crystals of colicin S4 were obtained at 293 K by the vapor diffusion hanging drop method against 200 μl of a reservoir solution. Crystal drops were prepared by mixing 2.5 μl of protein at 30 mg/ml concentration with 2.5 μl of reservoir solution. Crystals were obtained with 0.16 m MgCl2, 24% polyethylene glycol 4000, and 20% glycerol in 80 mm Tris-HCl (pH 8.5) at a size of up to 1000 × 100 × 100 μm. Platinum derivatives were obtained by soaking the crystal in the mother liquid containing 2 mm of K2PtCl4, (NH4)2PtCl4, K2Pt(NO2)4, or K2Pt(CN)4.
X-ray Data Collection—Single crystals were flash-frozen in their mother liquid, and data collection was performed at 100 K. The crystal system is R32 with cell constants of a = b = 240.34 Å, c = 80.03 Å, α = β = 90°, and γ = 120°. The crystals contained one monomer in the asymmetric unit, diffracted to a resolution limit of 2.45 Å, and showed a solvent content of 70%. All data sets were collected at Swiss Light Source beamline PXII on a MarCCD225 detector at 90 K. Data were indexed, integrated, and scaled with the XDS program package (31).
Structure Determination and Refinement—Data sets were collected at the theoretical platinum edge + 10 eV, and 180 images (1°/image) were recorded. All data of native and derivative crystals were processed identically and were transferred to the SHARP program package (32). The electron density provided by SHARP and DM was used in Buccaneer for initial model building (33, 34). This initial model was partially improved by using the pore-forming domain structure of colicin A (Protein Data Bank code 1col) together with model bits derived from Buccaneer. This model was then refined in REF-MAC5 (35), and both maps including experimental and model phases were used for model building. To improve this model, Coot (36) and iterative REFMAC rebuilding were used to rebuild side chains and to add missing residues. A random set of 5% of the data was neglected during the refinement process and marked as the test set for cross-validation. Atoms were refined and TLS parameters determined using the TLS server and REF-MAC (37). ARP/wARP was used to build the solvent structure (38). Together, this procedure returned a final model consisting of 3530 non-hydrogen atoms and 199 water molecules (corresponding to Val68-His499). Together with the hydrogen atoms generated for all amino acid residues, a crystallographic R/Rfree factor of 0.22/0.25 was achieved.
Secondary structure elements were defined according to DSSP criteria (molbio.info.nih.gov/structbio/basic.html). Figures were prepared using the programs DINO, RasMol, the Swiss-PdbViewer (39), and POV-Ray. The atomic coordinates of colicin S4 have been deposited in the Protein Data Bank (code 3few).
RESULTS
Colicin S4 Sequence Features and Classification of Pore-forming Domains—The colicin S4 sequence shares regions of high homology with other colicins but also comprises sequence parts that are unique (25). As in other colicins, the N-terminal translocation domain of colicin S4 (residues 1-118) is very rich in glycines (22%). The N-terminal 43 residues of colicins K and S4 are 98% identical, but a canonical TolA or TolB box (2) is missing. The receptor-binding domain of colicin S4 is unique in two ways: it has no sequence homology to other colicin domains (presumably because it is the only colicin that binds to OmpW), and it arose from a recent domain duplication event (25). PSI-BLAST searches using only the sequence of this domain against the non-redundant data base did not yield significant hits. The two 86-residue repeats display a sequence identity of 61% (25). Colicin S4 belongs to the group of pore-forming colicins, as the C-terminal 201 residues of colicin S4 (residues 299-499) and colicin A (residues 392-592) are 77% identical (see alignment in Fig. 1C) (25).
FIGURE 1.
A, structural features of the different groups of colicin pores. The pore-forming domains of colicins S4, E1, and Ia shown in the top and side views; the hydrophobic hairpin is drawn in red. The major differences between the colicin groups are the length of the hydrophobic hairpin (see also B), the angle between Pα1 and Pα2 (colicin S4, 5°; colicin E1, 34°; and colicin Ia, 37 °), and the position of the hairpin comprising Pα5 and the loop connection to Pα6. B, superimposition of hydrophobic hairpins. Left, colicin S4 (group PI, in red) versus colicin E1 (group PIIa, in pink); right, colicin E1 (group PIIa, in pink) versus colicin Ia (group PIIb, in purple). C, alignment of the pore-forming domains of colicins. Colicins are grouped according to sequence similarity. Groups PI and PIIa/b differ by several insertions and deletions, whereas groups PIIa and PIIb are highly similar and differ only in several conserved positions, affecting the overall charge and isoelectric point of the domains. Moreover, the structure of colicin Ia contains four very short β-strands where colicin E1 only has unstructured loops.
Based on the sequence alignment, the pore-forming domains of colicins can be divided into three groups. These differences can also be observed on the structural level (Fig. 1A). Based on these observations, we decided to term the groups (see alignment in Fig. 1C). Colicin S4 is more similar to colicins A, B, U, Y, and N (group PI) than to colicins E1, 5, 10, and K (group PIIa) and colicins Ia and Ib, pyocin S5 from Pseudomonas aeruginosa, and alveicin B from Hafnia alvei (group PIIb). The main difference between the group PI pore-forming domains and the other two groups is in the different length of helices Pα1, Pα3, Pα4, and Pα8; in an insertion in the turn region of the hydrophobic hairpin between helices Pα8 and Pα9; and in the angle between Pα1 and Pα2. The differences between groups PIIa and PIIb are more subtle and are based mostly on point mutations and on four short β-strands visible only in group PIIb.
Colicin S4 Expression and Purification—The DNA sequence of colicin S4 was obtained from plasmid pColS4 (25). To express the protein, the gene was cloned in a tet-inducible expression vector (pASK-IBA33+). Upon induction, colicin S4 accumulated in the cytosol of the expressing bacteria. However, the presence of the receptor protein OmpW rendered the cells sensitive to colicin S4, which was released into the growth medium at low concentrations, most likely after lysis of single cells. This led to the death of all cells shortly after induction, and the protein yield remained low. To overcome this problem, we used a ΔompW knock-out strain based on E. coli 5K, which was resistant to colicin S4.
Colicin S4 can be easily obtained in vast amounts and high purity using either ion exchange or, for His-tagged constructs, nickel affinity chromatography and preparative gel-sizing chromatography for polishing. In solution, colicin S4 is a monomer. The protein can be concentrated up to 80 mg/ml using spin concentrators. The protein remains active (i.e. kills wild-type E. coli cells and displays pore-forming activity in single channel conductance measurements) for several weeks when stored on ice.
Colicin S4 Forms Pores in Artificial Membranes—To show that the killing activity of colicin S4 is due to pore formation in the same fashion as in other colicins, we performed single channel conductance measurements. Black lipid membranes were produced from hen egg phosphatidylcholine, and colicin S4 was added to the bilayer setup on the ground side of the membrane. In the first assays, colicin S4 with a C-terminal His tag was used. This protein inserted spontaneously into the membrane but only with very low frequency and at high negative voltages (less than -100 mV). When colicin S4 without the His tag was used, the insertion frequency was much higher, and the voltage needed for insertion was lower (less than -80 mV). It seems that the C-terminal His tag hinders membrane insertion, but once the pore is formed, it does influence the conductance compared with the non-tagged protein (data not shown). Intriguingly, when very small amounts of purified OmpW were added to the bilayer setup with the His-tagged colicin, the insertion frequency increased dramatically. We assume that OmpW recruits the colicin to the proximity of the bilayer and thus increases the chances of insertion. The effect of OmpW on the insertion frequency seemed much less pronounced for the non-tagged colicin, albeit this is difficult to quantify. After these observations, we exclusively used non-tagged colicin (without addition of OmpW) in all single channel conductance measurements.
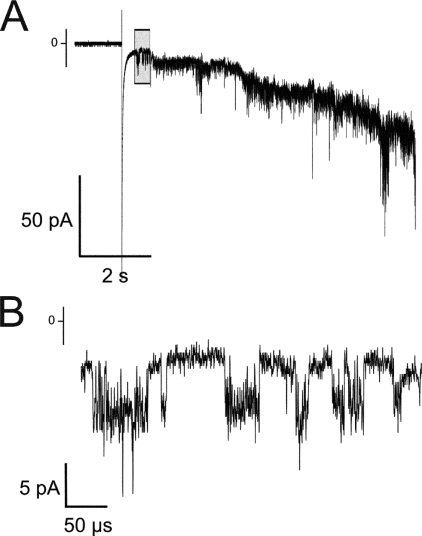
Colicin S4 inserted spontaneously into the lipid bilayer in 1 m KCl buffers at pH <7. At higher pH, no insertion events were observed. The recordings shown in Fig. 2 were taken in 20 mm MES-KOH (pH 6) and 1 m KCl. In Fig. 2A, numerous successive insertion events are visible. Fig. 2B shows a close-up of the same recording, where the signal of an individual colicin channel can be seen. As with other colicins, the single channel characteristics are very noisy, and the channels have different conductance states that make a quantitative evaluation of the conductance difficult (40). But the behavior of colicin S4 in these experiments seems similar to that of colicin A (41) as expected from the sequence alignments (Fig. 1C).
FIGURE 2.
Single channel conductance measurements. A shows the insertion of numerous channels just after injection of the protein. B shows the action of a single channel flickering at a high frequency, prior to permanent opening. The trace was recorded at -100 mV. The buffer conditions were 1 m KCl and 20 mm MES (pH 6.0).
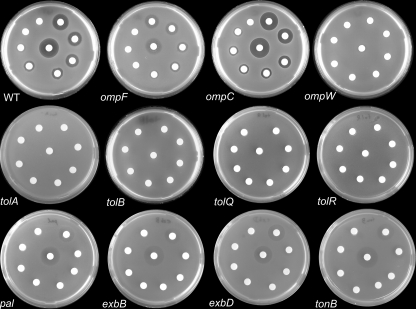
Colicin S4 Exploits OmpW as a Receptor and OmpF and Parts of the Tol System for Translocation—To examine the interaction partners of colicin S4 in the target cell, we performed a sensitivity screening using single knock-out strains of genes that were previously shown to be involved in colicin import and function, obtained from the Keio Collection (30). The putative interaction partners can be divided into two groups: possible receptors, i.e. outer membrane proteins, and proteins involved in translocation, i.e. proteins of the Tol and Ton systems. The target strains were plated on LB agar, and filter platelets soaked with decreasing amounts of colicin S4 were put on the plates. If the target strain was sensitive to colicin S4, a clear zone around the filter platelets indicated the killing of the cells by the colicin. Resistant strains were not affected by colicin S4, resulting in a homogeneous lawn. Some knock-outs rendered the cells only partially resistant, leading to inhibition zones that were not entirely clear (Fig. 3).
FIGURE 3.
Colicin S4 sensitivity of selected E. coli strains. The white filter platelets were soaked with colicin S4 (10 μg, middle platelet;1 μg, upper right platelet; continuing clockwise to 10-7 μg). The ΔompW strain shows full resistance. The ΔompF strain shows only partial resistance, indicating that other porins might be used alternatively for the translocation process. The ΔompC strain shows hypersensitivity, probably due to increased ompF expression levels. The ΔtolA, ΔtolB, ΔtolQ, and ΔtolR strains show full resistance, and Δpal shows partial resistance, indicating the Tol-dependent translocation of colicin S4. However, the ΔtonB, ΔexbB, and ΔexbD strains also show partial resistance. WT, wild-type.
OmpW is an eight-stranded monomeric β-barrel of unknown function that forms a hydrophobic channel that is not suited for the transport of soluble macromolecules (29, 42). ΔompW cells were absolutely resistant to colicin S4 treatment, confirming the finding of Pilsl et al. (25) that OmpW is the primary receptor. OmpF and OmpC are closely related 16-stranded trimeric β-barrels that form unspecific diffusion pores with a size cutoff of ∼600 Da (43). Nevertheless, different colicins are known to translocate using these pores or the outer membrane efflux protein TolC (13, 20, 44). The ΔompF strain showed almost complete resistance to colicin S4, whereas the ΔompC strain was hypersensitive compared with the wild type, probably because in a ΔompC background, the amount of OmpF protein in the outer membrane is increased. The double knock-out E. coli strain BL21omp7 (ΔompC ΔompF ΔlamB) (45) was fully resistant. The ΔtolC strain remained sensitive to colicin S4 (data not shown). The ΔompT strain was also hypersensitive (data not shown), which is in accordance with the findings of Cavard and Lazdunski (46), who showed that the outer membrane protease OmpT is an efficient defense mechanism that cleaves colicin molecules in the medium.
To investigate whether colicin S4 is translocated by the Tol or Ton system, we used the knock-out strains ΔtolA, ΔtolB, ΔtolQ, ΔtolR, and Δpal (Tol system) and ΔtonB, ΔexbB, and ΔexbD (Ton system). The ΔtonB, ΔexbB, and ΔexbD strains showed a decreased sensitivity to colicin S4 but were not fully resistant. The ΔtolB, ΔtolQ, and ΔtolR strains were fully resistant to colicin S4 treatment, and Δpal was at least partially resistant, but the ΔtolA strain was not. In control experiments using different strains lacking TolA, we found that the supposed ΔtolA strain from the Keio Collection still contains the tolA gene. The other ΔtolA strains, namely A592 and TPS94 (47), behaved as expected, i.e. they were fully resistant to colicin S4. In conclusion, colicin S4 import depends on the complete Tol system.
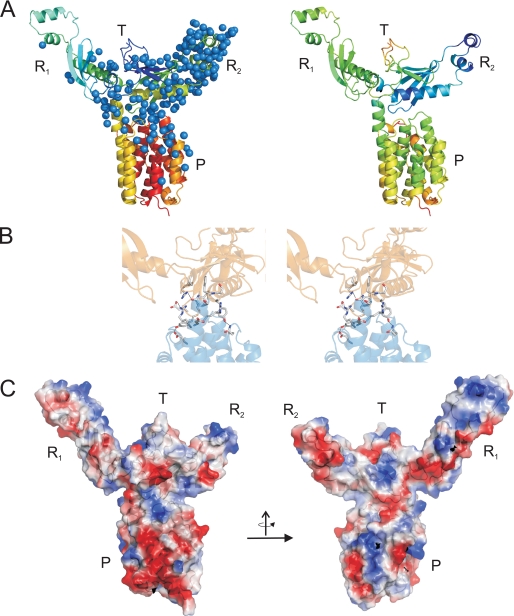
Crystal Structure of Colicin S4—In comparison with other colicins whose structure is typically dominated by coiled-coil domains and thus is extended and rod-like, colicin S4 is a compact molecule (Fig. 4A). However, as anticipated from sequence analysis, colicin S4 consists of four distinct domains, namely the N-terminal translocation domain, two receptor-binding domains, and the C-terminal pore-forming domain (Fig. 4B). The crystallographic statistics are summarized in Table 1. The N-terminal translocation domain is only partially resolved in the structure; residues 1-67 could not be modeled into the density maps, most probably because of the intrinsic flexibility of this domain type that has been described also for other colicins (21-23, 48-50). The visible part of the translocation domain in the electron density map consists of an antiparallel β-sheet formed by three strands (Tβ1-Tβ3), with an extended, glycine-rich loop structure between strands Tβ2 and Tβ3, followed by a short helix that connects the domain to the first receptor-binding domain (R1) (Fig. 4). Interestingly, the β-sheet of the translocation domain continues into the second receptor-binding domain (R2), as strand Tβ1 is connecting to the parallel R2β1(Fig. 4). It seems that this arrangement holds the translocation domain in place. Colicin S4 is the only colicin that has two nearly identical receptor-binding domains. Both domains consist of a three-stranded antiparallel β-sheet (R1, β1-β3; and R2, β1-β3), followed by three helices. The termini of the domains are in close proximity to each other. The small β-sheet covers the longest helix (R1α3 and R2α3, respectively) via a hydrophobic core and several residues forming salt bridges and hydrogen bonds between these elements. The two shorter helices (R1α1 and R1α2, R2α1, and R2α2) are extending outward from the overall structure, suggesting that they might play a direct role in receptor binding, as discussed below.
FIGURE 4.
Structure of colicin S4. A, stereo view of the complete structure. B, annotation of domains and secondary structure elements: translocation domain (T), receptor-binding domain 1 (R1), receptor-binding domain 2 (R2), and pore-forming domain (P).
TABLE 1.
Summary of data collection, phasing, and refinement statistics
FOM, figure of merit; r.m.s.d., root mean square deviation.
| Colicin S4 | |
|---|---|
| Data collectiona | |
| Wavelength (Å) | 0.976 |
| Space group | R32 |
| Resolution (Å) | 40-2.45 (2.60-2.45) |
| Cell constants (Å) | a = b = 240.4, c = 80.0 |
| Unique reflections | 32,353 (5185) |
| Redundancy | 6.2 (6.2) |
| Completeness (%) | 99.5 (99.7) |
| Rmerge (%) | 6.2 (43.8) |
| I/σ(I) | 16.4 (3.8) |
| Wilson B-factor | 59.5 |
| Phasing statistics | |
| FOM after SHARP (40-2.8) | 0.28 |
| FOM after DM (40-2.8) | 0.78 |
| Refinement statisticsa | |
| Space group | R32 |
| Resolution (Å) | 25-2.45 (2.51-2.45) |
| Rcryst (%) | 0.22 (0.37) |
| Rfree (%) | 0.25 (0.40) |
| Non-hydrogen atoms | 3530 |
| Waters | 199 |
| Ligand (sodium) | 1 |
| Mean B-value (Å2) | 52 |
| r.m.s.d. of bond length (Å2) | 0.008 |
| r.m.s.d. of angle | 1.3° |
| Model quality | |
| Residues in most favored region | 345 (91.8%) |
| Residues in most allowed region | 24 (6.4%) |
| Residues in generously allowed region | 5 (1.3%) |
| Residues in disallowed region | 2 (0.5%) |
Numbers in parentheses refer to the highest resolution shell.
Whereas domain R1 has a very incomplete hydration sphere, R2 is fully hydrated (Fig. 5). This observation is also supported by the B-factors of these domains, which are relatively high for the first domain but low for the second domain. The low hydration of R1 can be only partially attributed to crystal contacts. The two domains superimpose with a root mean square density of only 0.9 Å over 76 residues (Fig. 6A), which was expected from the high sequence identity.
FIGURE 5.
Structure of colicin S4. A: left, hydration of colicin S4 in the crystal arrangement. Water molecules are represented by blue balls. Note the significant difference in hydration between domains R1 and R2. Right, structure of colicin S4. B: network of hydrophobic and electrostatic interactions between domain R2 and the pore-forming domain (stereo view). C: electrostatic characterization of colicin S4. Blue, positively charged surface; red, negatively charged surface.
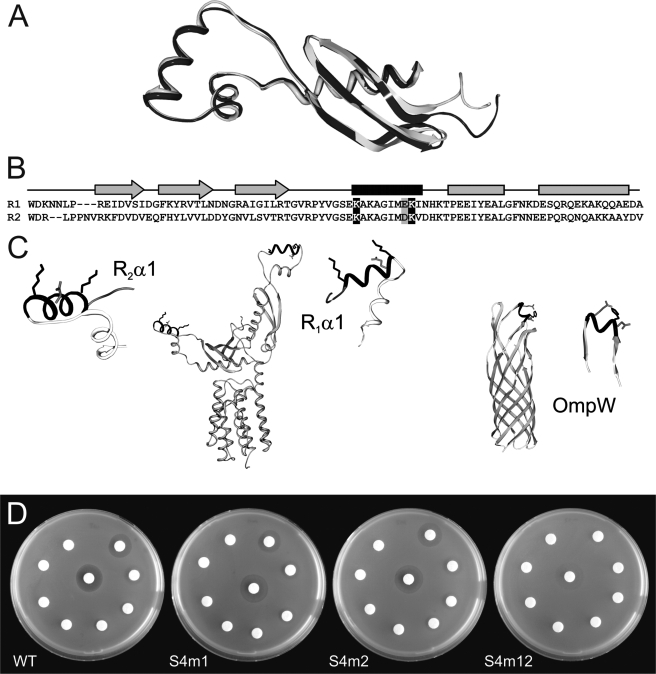
FIGURE 6.
Receptor-binding domain mutants. A, superimposition of the two receptor-binding domains. R1 is shown in black, and R2 is shown in light gray. B, alignment of the two receptor-binding domains. The mutated residues are highlighted in black (positive charge) and gray (negative charge). C, location of the α-helices (black) that were mutated to interfere with OmpW binding. The mutated residues Lys163, Glu170, and Lys171 in R1, and Lys250, Asp257, and Lys258 in R2 are shown in stick representation. D, sensitivity assay. Colicin S4 was spotted on filter platelets in decreasing concentrations (see Fig. 3). The wild-type (WT) E. coli strain is resistant to the colicin mutant S4m12, in which both domains are modified; it is partially resistant to the two colicin mutants S4m1 and S4m2, in which only one domain is changed, and is fully sensitive to control colicin S4.
The pore-forming domain is similar in structure (root mean square deviation of 1.5 Å over 193 residues) and sequence (77% identity) to colicin A (see also alignment Fig. 1C). As in all other pore-forming colicins, the domain consists of a bundle of amphipathic helices (Pα1-Pα7 and Pα10) that cover two hydrophobic helices (Pα8 and Pα9) in the core of the domain. These two hydrophobic helices are involved in pore formation by inserting into membranes (51-53).
Overall, the four domains of colicin S4 are interconnected by different modes. The translocation domain, together with parts of R2, forms an extended β-sheet. Domain R1 is rather loosely attached to the rest of the molecule, whereas R2 forms interactions with both the translocation and the pore-forming domain. The network of hydrophobic and electrostatic interactions between R2 and pore-forming domain is shown in detail in Fig. 5B. Together, this results in an asymmetric arrangement of the two otherwise similar domains.
Fig. 5C shows the surface charge distribution of colicin S4. The functional implications of the charges on the helices of the pore-forming domains of colicins have been discussed (11). We noticed that the two receptor-binding domains are rather positively charged at their tip-like structures and that this might be the locus of OmpW binding. To test this hypothesis, we produced a number of point mutation variants of colicin S4 (see below).
Both Receptor-binding Domains Recognize OmpW, but One Is Sufficient for Effective Killing of E. coli Cells—The presence of two almost identical receptor-binding domains raises the question whether both are necessary or if only one functional copy would be sufficient for activity. The first helix of both receptor-binding domains represents the most accessible secondary structure element within the whole molecule. Both helices display several charged residues. In R1α1, we found Lys163, Glu170, and Lys171, and in R2α1, we found Lys250, Asp257, and Lys258 protruding from the same side of either α-helix (Fig. 6C).
In the crystal structure of OmpW, one striking feature is a single α-helix, framed by two short β-strands, that is exposed to the extracellular space (42). Asp116, His117, and Glu120 of this helix are exposed to the extracellular space and form a charged patch with a negative-positive-negative charge pattern that is complementary to the positive-negative-positive charge pattern of α1 in either of the receptor-binding domains of colicin S4. To show that these residues are indeed involved in the interaction of colicin S4 with OmpW, we mutated the three charged residues in either or both receptor-binding domains of colicin S4 to alanines, yielding constructs S4m1 (K163A/E170A/K171A), S4m2 (K250A/D257A/K258A), and S4m12 with all six mutations. The mutant colicins were expressed and purified in the same way as the wild-type protein. The expression levels and yields of the purified proteins were comparable with those of the wild-type protein. The CD spectra of colicin S4, S4m1, and S4m12 were recorded to exclude improper folding, but the spectra of the three proteins showed no significant differences in shape and thus in secondary structure content (supplemental Fig. S1). The activity of S4m1, S4m2, and S4m12 in comparison with wild-type colicin S4 was assayed as described above using the wild-type E. coli BW25113 (K12) strain provided by the Keio Collection (Fig. 6D). S4m1 and S4m2, which have mutations in only one receptor-binding domain, displayed no significant changes in killing efficiency compared with wild-type colicin S4, whereas S4m12 showed a greatly reduced killing efficiency. It appears that both receptor-binding domains can bind to the target receptor OmpW and that only one functional domain is sufficient for binding and killing.
DISCUSSION
Structure and Mechanism of Colicin S4—Colicins have been studied intensely since their discovery in 1925 (54). Despite this, new members of this class of proteins with unique features can still be found, as only a relatively small number of colicin plasmids have been sequenced. Here, we present the structure and function of colicin S4, originally described by Pilsl et al. (25).
After binding to the receptor OmpW, the N-terminal glycine-rich translocation domain recruits OmpF to translocate through the outer membrane. Colicin S4 then exploits the Tol system to reach the inner membrane. The mostly disordered, flexible N terminus is well suited to interact with multiple binding partners as seen in other colicins (13, 55), yet canonical binding sequences for TolA and TolB are not present in the colicin S4 sequence. The deletion of the lipoprotein Pal renders the cells partially resistant. This is probably due to the dislocation of the periplasmic binding partner of colicin S4, TolB, which is kept in close proximity to the outer membrane by Pal (16). Alternatively, the distance between the inner and outer membranes might increase because of the disrupted coupling via Pal, making it harder for colicin S4 to reach the cytoplasmic membrane to exert its pore-forming activity (56).
Evolutionary Aspects—The pore-forming domain of colicin S4 is highly similar to that of other pore-forming colicins in sequence, structure, and behavior in artificial membranes. The sequence and structure comparison clearly shows the descent of all pore-forming domains from a common ancestor, now diverging into three different clades. In contrast to this common descent, colicin S4 contains a unique receptor-binding domain type. The most striking feature of this part of colicin S4 is the presence of two copies of the receptor-binding domain. In protein evolution, gene duplication is a first step in the evolution of new functions. This way, one part can keep the necessary function, whereas the duplicate is free to evolve, a process that is also seen on the domain and subdomain levels (57). In colicin S4, both duplicates retained their function so far: the sequence is highly conserved, and both domains are still able to bind to the receptor OmpW. This suggests that the duplication occurred very recently.
As all other pathogenicity factors, colicins have to evolve fast to overcome host defense mechanisms that evolve just as fast; in other words, co-evolving organisms must develop continuously to maintain their fitness. This evolutionary law has been termed the “red queen principle” (58). In the case of colicin S4, we observe such evolution in action. It would be tempting to construct a stable colicin S4 with only one receptor-binding domain or to search for such a “precursor” colicin S4 in natural isolates.
Supplementary Material
Acknowledgments
We thank Volkmar Braun and Klaus Hantke for the colicin clones and helpful discussions, the staff at Swiss Light Source beamline PXII, and Andrei Lupas for continuing support.
The atomic coordinates and structure factors (code 3few) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the Max Planck Society and by German Science Foundation Grant SFB766/B4. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables SI-SIII.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: MOPS, 4-morpholinepropanesulfonic acid; MES, 4-morpholineethanesulfonic acid.
References
- 1.Riley, M. A., and Wertz, J. E. (2002) Annu. Rev. Microbiol. 56 117-137 [DOI] [PubMed] [Google Scholar]
- 2.Cascales, E., Buchanan, S. K., Duche, D., Kleanthous, C., Lloubes, R., Postle, K., Riley, M., Slatin, S., and Cavard, D. (2007) Microbiol. Mol. Biol. Rev. 71 158-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillor, O., Vriezen, J. A., and Riley, M. A. (2008) Microbiology (Read.) 154 1783-1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lakey, J. H., and Slatin, S. L. (2001) Curr. Top. Microbiol. Immunol. 257 131-161 [DOI] [PubMed] [Google Scholar]
- 5.de Zamaroczy, M., and Buckingham, R. H. (2002) Biochimie (Paris) 84 423-432 [DOI] [PubMed] [Google Scholar]
- 6.Duche, D. (2002) Biochimie (Paris) 84 455-464 [DOI] [PubMed] [Google Scholar]
- 7.Kleanthous, C., Hemmings, A. M., Moore, G. R., and James, R. (1998) Mol. Microbiol. 28 227-233 [DOI] [PubMed] [Google Scholar]
- 8.Cavard, D. (1991) J. Bacteriol. 173 191-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riley, M. A. (1993) Mol. Biol. Evol. 10 1048-1059 [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., Patzer, S. I., and Hantke, K. (2002) Biochimie (Paris) 84 365-380 [DOI] [PubMed] [Google Scholar]
- 11.Zakharov, S. D., and Cramer, W. A. (2002) Biochim. Biophys. Acta 1565 333-346 [DOI] [PubMed] [Google Scholar]
- 12.Cao, Z., and Klebba, P. E. (2002) Biochimie (Paris) 84 399-412 [DOI] [PubMed] [Google Scholar]
- 13.Housden, N. G., Loftus, S. R., Moore, G. R., James, R., and Kleanthous, C. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13849-13854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zakharov, S. D., and Cramer, W. A. (2004) Front. Biosci. 9 1311-1317 [DOI] [PubMed] [Google Scholar]
- 15.Deprez, C., Blanchard, L., Guerlesquin, F., Gavioli, M., Simorre, J. P., Lazdunski, C., Marion, D., and Lloubes, R. (2002) Biochemistry 41 2589-2598 [DOI] [PubMed] [Google Scholar]
- 16.Lazzaroni, J. C., Dubuisson, J. F., and Vianney, A. (2002) Biochimie (Paris) 84 391-397 [DOI] [PubMed] [Google Scholar]
- 17.Lloubes, R., Cascales, E., Walburger, A., Bouveret, E., Lazdunski, C., Bernadac, A., and Journet, L. (2001) Res. Microbiol. 152 523-529 [DOI] [PubMed] [Google Scholar]
- 18.Braun, V. (2006) ACS Chem. Biol. 1 352-354 [DOI] [PubMed] [Google Scholar]
- 19.Postle, K., and Kadner, R. J. (2003) Mol. Microbiol. 49 869-882 [DOI] [PubMed] [Google Scholar]
- 20.Kurisu, G., Zakharov, S. D., Zhalnina, M. V., Bano, S., Eroukova, V. Y., Rokitskaya, T. I., Antonenko, Y. N., Wiener, M. C., and Cramer, W. A. (2003) Nat. Struct. Biol. 10 948-954 [DOI] [PubMed] [Google Scholar]
- 21.Wiener, M., Freymann, D., Ghosh, P., and Stroud, R. M. (1997) Nature 385 461-464 [DOI] [PubMed] [Google Scholar]
- 22.Hilsenbeck, J. L., Park, H., Chen, G., Youn, B., Postle, K., and Kang, C. (2004) Mol. Microbiol. 51 711-720 [DOI] [PubMed] [Google Scholar]
- 23.Vetter, I. R., Parker, M. W., Tucker, A. D., Lakey, J. H., Pattus, F., and Tsernoglou, D. (1998) Structure 6 863-874 [DOI] [PubMed] [Google Scholar]
- 24.Zeth, K., Römer, C., Patzer, S. I., and Braun, V. (2008) J. Biol. Chem. 283 25324-25331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilsl, H., Smajs, D., and Braun, V. (1999) J. Bacteriol. 181 3578-3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Link, A. J., Phillips, D., and Church, G. M. (1997) J. Bacteriol. 179 6228-6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albrecht, R., Zeth, K., Soding, J., Lupas, A., and Linke, D. (2006) Acta Crystallogr. F Struct. Biol. Crystalliz. Comm. 62 415-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L., and Mori, H. (2006) Mol. Systems Biol. 2 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795-800 [Google Scholar]
- 32.Vonrhein, C., Blanc, E., Roversi, P., and Bricogne, G. (2007) Methods Mol. Biol. 364 215-230 [DOI] [PubMed] [Google Scholar]
- 33.Cowtan, K. (2006) Acta Crystallogr. 62 1002-1011 [DOI] [PubMed] [Google Scholar]
- 34.Wang, B. C. (1985) Methods Enzymol. 115 90-112 [DOI] [PubMed] [Google Scholar]
- 35.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D 53 240-255 [DOI] [PubMed] [Google Scholar]
- 36.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 37.Winn, M. D., Murshudov, G. N., and Papiz, M. Z. (2003) Methods Enzymol. 374 300-321 [DOI] [PubMed] [Google Scholar]
- 38.Cohen, S. X., Ben Jelloul, M., Long, F., Vagin, A., Knipscheer, P., Lebbink, J., Sixma, T. K., Lamzin, V. S., Murshudov, G. N., and Perrakis, A. (2008) Acta Crystallogr. Sect. D 64 49-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guex, N., and Peitsch, M. C. (1997) Electrophoresis 18 2714-2723 [DOI] [PubMed] [Google Scholar]
- 40.Davidson, V. L., Brunden, K. R., Cramer, W. A., and Cohen, F. S. (1984) J. Membr. Biol. 79 105-118 [DOI] [PubMed] [Google Scholar]
- 41.Collarini, M., Amblard, G., Lazdunski, C., and Pattus, F. (1987) Eur. Biophys. J. 14 147-153 [DOI] [PubMed] [Google Scholar]
- 42.Hong, H., Patel, D. R., Tamm, L. K., and van den Berg, B. (2006) J. Biol. Chem. 281 7568-7577 [DOI] [PubMed] [Google Scholar]
- 43.Delcour, A. H. (2003) Front. Biosci. 8 1055-1071 [DOI] [PubMed] [Google Scholar]
- 44.Zakharov, S. D., Eroukova, V. Y., Rokitskaya, T. I., Zhalnina, M. V., Sharma, O., Loll, P. J., Zgurskaya, H. I., Antonenko, Y. N., and Cramer, W. A. (2004) Biophys. J. 87 3901-3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prilipov, A., Phale, P. S., Van Gelder, P., Rosenbusch, J. P., and Koebnik, R. (1998) FEMS Microbiol. Lett. 163 65-72 [DOI] [PubMed] [Google Scholar]
- 46.Cavard, D., and Lazdunski, C. (1990) J. Bacteriol. 172 648-652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, T. P., and Webster, R. E. (1986) J. Bacteriol. 165 107-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng, Y. S., Shi, Z., Doudeva, L. G., Yang, W. Z., Chak, K. F., and Yuan, H. S. (2006) J. Mol. Biol. 356 22-31 [DOI] [PubMed] [Google Scholar]
- 49.Collins, E. S., Whittaker, S. B., Tozawa, K., MacDonald, C., Boetzel, R., Penfold, C. N., Reilly, A., Clayden, N. J., Osborne, M. J., Hemmings, A. M., Kleanthous, C., James, R., and Moore, G. R. (2002) J. Mol. Biol. 318 787-804 [DOI] [PubMed] [Google Scholar]
- 50.Soelaiman, S., Jakes, K., Wu, N., Li, C., and Shoham, M. (2001) Mol. Cell 8 1053-1062 [DOI] [PubMed] [Google Scholar]
- 51.Kienker, P. K., Qiu, X., Slatin, S. L., Finkelstein, A., and Jakes, K. S. (1997) J. Membr. Biol. 157 27-37 [DOI] [PubMed] [Google Scholar]
- 52.Song, H. Y., Cohen, F. S., and Cramer, W. A. (1991) J. Bacteriol. 173 2927-2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zakharov, S. D., and Cramer, W. A. (2002) Biochimie (Paris) 84 465-475 [DOI] [PubMed] [Google Scholar]
- 54.Gratia, A. (1925) C. R. Soc. Biol. (Paris) 93 1040-1041 [Google Scholar]
- 55.Loftus, S. R., Walker, D., Mate, M. J., Bonsor, D. A., James, R., Moore, G. R., and Kleanthous, C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 12353-12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clavel, T., Germon, P., Vianney, A., Portalier, R., and Lazzaroni, J. C. (1998) Mol. Microbiol. 29 359-367 [DOI] [PubMed] [Google Scholar]
- 57.Soding, J., and Lupas, A. N. (2003) BioEssays 25 837-846 [DOI] [PubMed] [Google Scholar]
- 58.van Valen, L. (1973) Evol. Theory 1 1-30 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.