Abstract
Thrombospondin (TSP) 1 is a trimeric multidomain protein that contains motifs that recognize distinct host cell receptors coupled to multiple signaling pathways. Selected TSP1-induced cellular responses are tyrosine kinase-dependent, and TSP1 contains epidermal growth factor (EGF)-like repeats. Specific receptor interactions or functions for the EGF-like repeats have not been identified. We asked whether one or more biological responses to TSP1 might be explained through EGF receptor (EGFR) activation. In A431 cells, TSP1 increased autophosphorylation of Tyr-1068 of EGFR in a dose- and time-dependent manner. The ability of TSP1 to activate EGFR was replicated by the tandem EGF-like repeats as a recombinant protein. The three EGF-like repeats alone produced a high level of Tyr-1068 phosphorylation. EGF-like repeats from TSP2 and TSP4 also activated EGFR. Tyr-1068 phosphorylation was less when individual EGF-like repeats were tested or flanking sequences were added to the three EGF-like repeats. TSP1 and its EGF-like repeats also increased phosphorylation of EGFR Tyr-845, Tyr-992, Tyr-1045, Tyr-1086, and Tyr-1173, activated phospholipase Cγ, and increased cell migration. No evidence was found for binding of the EGF-like repeats to EGFR. Instead, EGFR activation in response to TSP1 or its EGF-like repeats required matrix metalloprotease activity, including activity of matrix metalloprotease 9. Access to the ligand-binding portion of the EGFR ectodomain was also required. These findings suggest release of an endogenous EGFR ligand in response to ligation of a second unknown receptor by the TSPs.
Thrombospondin (TSP)2 1 is an ∼420-kDa trimeric glycoprotein composed of three identical 145-kDa polypeptide chains linked by disulfide bonds (1). TSP1 is one of five TSP family members (2). Each subunit of TSP1 and TSP2 contains the following structural elements: an NH2-terminal globular domain of the laminin G domain and concanavalin A-like lectin/glucanase superfamily; an α-helical region that presumably forms a parallel homotrimeric coiled coil as in matrilin-1; a von Willebrand factor type C module; three TSP type 1 (TSR) or properdin repeats, each of which is elongated and consists of a novel, antiparallel three-stranded fold; and the TSP “signature piece” that is made up of three EGF-like TSP type 2 repeats, 13 calcium-binding repeats, and a COOH-terminal domain that forms a lectin-like β-sandwich (2). The elements of the “signature piece” interact extensively to form three structural regions termed the stalk, wire, and globe, and are further stabilized by disulfide bonds and bound calcium (3). All five vertebrate TSPs contain the signature piece. TSP3, TSP4, and TSP5 have a pentameric coiled coil (4), lack the von Willebrand factor C domain and TSRs, and have an extra EGF-like repeat (5, 6).
TSP1 recognizes multiple cell surface receptors coupled to specific signaling pathways that evoke distinct, and sometimes opposing, biological responses (5, 7, 8). The NH2-terminal globular domain recognizes heparan sulfate proteoglycans (6), low density lipoprotein receptor-related protein 1 (9), sulfated glycolipids (10), calreticulin (9), and integrins (11). In the TSRs, the CSVTCG and GVQXR motifs recognize CD36 (12), and in the calcium-binding wire an RGD-containing sequence binds to αvβ3 and other integrins (13). β1 integrins also have been shown to interact with TSRs (14). Finally, two cell-binding motifs in the COOH-terminal lectin-like domains, RFYVVM and FIRVVM (15), interact with CD47 or integrin-associated protein (16). Therefore, the NH2-terminal globular domain, the TSRs and calcium-binding repeats, and the COOH-terminal domain of TSP1 all exhibit receptor binding activities that elicit distinct host cell responses. Aside from minimal recognition by integrins (14), no such receptor binding activity or biological function has been ascribed to the type 2 EGF-like repeats of TSP1. TSP1 is secreted by numerous cell types and is present in the extracellular matrix (17). TSP1 was first demonstrated in the releasate of thrombin-stimulated platelets (18). It is expressed by endothelial cells, smooth muscle cells, fibroblasts, keratinocytes, cells of monocytes/macrophage lineage, and tumor cells (17). Much of our understanding of TSP1 biology has been established in these cell systems. TSP1 is also expressed in epithelia, is abundant in the basement membranes underlying these cells (17), and participates in epithelial cell responses, including re-epithelialization during wound healing (8), bronchial epithelial cell morphogenesis and development (19), and migration of epithelium-derived tumor cells (20). In human epithelium-derived cancer cells, EGF increases TSP1 expression (21). TSP1 null mice display epithelial cell alterations (22, 23).
In previous studies, we demonstrated that TSP1 increases tyrosine phosphorylation of the zonula adherens proteins γ-catenin and p120ctn (24), an event that can occur downstream of the EGF receptor (EGFR) (25, 26), also referred to as HER1 or ErbB1 (27). EGFR contains an NH2-terminal, ligand-binding ectodomain that is coupled to an intracellular catalytic domain and its tyrosine autophosphorylation sites (27). Ligand binding to the EGFR ectodomain induces receptor homodimerization and heterodimerization with other ErbB family members, intrinsic kinase activity, and autophosphorylation of specific tyrosine residues which, in turn, serve as docking sites within the cytoplasmic domain for signaling molecules (27). High affinity EGFR ligands share a 45-55-aa EGF motif with six spatially conserved cysteine residues that form three intramolecular disulfide bonds that dictate their tertiary conformation (28). These ligands are synthesized as transmembrane precursor proteins that are cleaved by cell surface matrix metalloproteases (MMP) (28) and ADAMs (a disintegrin and metalloproteinases) (29-32) to release mature growth factors for auto-crine/paracrine stimulation. EGFR ligands that specifically activate EGFR include EGF, transforming growth factor α, amphiregulin, and others that activate both EGFR and ErbB4, including heparin-binding EGF, betacellulin, and epiregulin (28).
In addition to these “authentic” ErbB ligands, EGF-like sequences are present in many other proteins, including TSP1 (33-35). EGF-like repeats in the γ2 chain of laminin-5 (34) and in the counter-adhesive protein, tenascin-C (35), have been demonstrated to activate EGFR. EGFR not only responds to direct binding of EGF motif-containing ligands, but it can be transactivated by heterologous receptors, including G protein-coupled receptors (36) and integrins (37). Whether TSP1 elicits biological responses through EGFR and/or other ErbB receptors, either through direct binding or transactivation, is not known. Here, we provide evidence that the EGF-like repeats of TSP1 and other TSP family members, likely through an MMP-mediated indirect process, activate EGFR and that this activation is coupled to downstream signaling events and cellular responses that can explain aspects of TSP1 bioactivity.
MATERIALS AND METHODS
Human Intact TSP1 Preparation—Human platelet TSP1 was purified as described (24). Briefly, fresh human platelets (Birmingham American Red Cross, Birmingham, AL) were thrombin-stimulated, and the platelet releasate was applied to a heparin-Sepharose CL-6B (Pharmacia, Piscataway, NJ) affinity column preequilibrated with Tris-buffered saline (TBS-C: 0.01 m Tris-HCl, 0.15 m NaCl, 0.1 mm CaCl2, pH 7.4). The bound TSP1 was eluted and applied to an A0.5 M gel filtration column (Bio-Rad) pre-equilibrated with TBS-C, pH 7.4.
Preparation of Recombinant TSP Proteins—Baculovirus-expressed recombinant human TSP1 domains were purified after secretion as described (38-40). These recombinant proteins (numbered from the initiating methionine of the full-length subunit) include the following: 1) the NH2-terminal heparin-binding domain + oligomerization domain + von Willebrand factor-C domain (aa 19-374) (NoC); 2) the von Willebrand factor-C domain + TSRs 1-3 (aa 312-548) (CP123); 3) TSR repeat 3 + EGF-like repeats 1-3 (aa 491-691) (P3E123); 4) EGF-like repeats 1-3 (aa 549-691) (E123); 5) EGF-like repeats 1 and 2 (aa 549-647) (E12); 6) EGF-like repeat 2 (aa 590-647) (E2); 7) EGF-like repeat 3 (aa 648-691) (E3); 8) EGF-like repeats 1-3 to the COOH terminus (aa 549-1170) (E123CaG); 9) the third EGF repeat to the COOH terminus (aa 648-1170) (E3CaG); and 10) the wire and COOH-terminal lectin-like domain (aa 692-1170) (CaG). In addition, two baculovirus-expressed TSP2 constructs, P3E123 (aa 493-693) and E123CaG (aa 551-1172), and two TSP-4 constructs, E1234 (aa 286-462) and E1234CaG, were prepared (40). Protein concentration and purity were determined by absorbance at 280 nm using an extinction coefficient based on the amino acid composition and by PAGE in SDS with and without prior reduction.
Cell Culture—Human epidermoid carcinoma A431 (American Type Culture Collection, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium (ATCC) enriched with 10% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT), 5 mm l-glutamine, nonessential amino acids, and vitamins in the presence of penicillin (50 units/ml) and streptomycin (50 μg/ml) (Sigma).
Knockdown of EGFR and MMP9 through RNA Interference—Small interfering RNA (siRNA) duplex products designed to target EGFR and MMP9, as well as an irrelevant control duplex siRNA not corresponding to any known sequence in the human genome, were introduced into A431 cells (Dharmacon, Lafayette, CO) (41). First, 5 × 105 A431 cells were centrifuged (200 × g, 10 min), after which the pellet was resuspended in 100 μl of A431 Nucleofector solution (Amaxa Biosystems) and incubated with 4.0 μg of siRNA duplexes. The A431 cell/siRNA mixture was transferred to an Amaxa-certified cuvette and subjected to programmed electroporation (program X-001) (Amaxa Biosystems). The MMP9 siRNA-transfected cells were cultured for increasing times, and the supernatants were concentrated and assayed for MMP9 in an MMP9 ELISA kit (Calbiochem). At these same time points, the EGFR siRNA-transfected cells were lysed and processed for immunoblotting with anti-EGFR antibody (BD Biosciences). To confirm equivalent protein loading and transfer, blots were stripped with 100 mm 2-mercaptoethanol, 2% SDS, 62.5 mmol/liter Tris-HCl, pH 6.7, and reprobed with 0.5 ng/ml murine anti-physarum β-tubulin IgG2b (Roche Applied Science) followed by HRP-conjugated anti-mouse IgG (BD Transduction Laboratories) and again developed with enhanced chemiluminescence (ECL).
EGFR Activation—To determine whether TSP1 activates EGFR, A431 cells were serum-starved for 6 h, after which they were exposed for 0.5 h to increasing concentrations of recombinant human EGF (R & D Systems, Inc), TSP1, or media alone, or they were exposed for increasing times to a fixed concentration of EGF (100 ng/ml or 16.7 nm), TSP1 (30 μg/ml or 214 nm), or media alone. In selected experiments, cells were pretreated for 2 h with the EGFR-selective tyrphostin, AG1478 (5 μm) (Calbiochem) (42), the MMP2/MMP9 inhibitor IV, SB-3CT (1 μm) (Calbiochem) (43), the EGFR ectodomain-blocking antibody, GR13L (Calbiochem) (44), a murine monoclonal anti-human MMP9 neutralizing antibody (Calbiochem) (45), or a species- and isotype-matched antibody control, B7-1/CD80 (R & D Systems, Inc.). In other experiments, A431 cells were transfected with EGFR or MMP9 targeting or control siRNAs. In still other experiments, A431 cells were exposed for 0.5 h to increasing concentrations of recombinant TSP1 domains or media alone or were exposed for increasing times to a fixed concentration (3.8 μg/ml or 214 nm) of TSP1 EGF-like repeats, i.e. E123 (aa 549-691) equimolar to native TSP1 at 30 μg/ml (214 nm). Finally, in one set of experiments, TSP1 (214 nm) was pre-incubated with either of two monoclonal antibodies raised against the EGF-like repeats, C6.7 and HB8432 (46, 47), or a species- and isotype-matched antibody control, B7-1/CD80. Cells were thoroughly rinsed with ice-cold HEPES buffer and solubilized with ice-cold lysis buffer containing 50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EGTA, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 μg/ml aprotinin, 1 mm Na2VO4, 1 mm NaF, 10 mm pyrophosphate, 500 μm paranitrophenol, and 1 mm phenylarsine oxide (all purchased from Sigma). The EC lysates were resolved by electrophoresis on a 4-12% SDS-polyacrylamide gel (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA). The blots were probed with murine monoclonal anti-phospho-EGFR (Tyr-1068) followed by HRP-conjugated goat anti-mouse IgG (Pierce) and developed with ECL (Amersham Biosciences). To confirm equivalent protein loading, blots were stripped and reprobed for β-tubulin. For each immunoblot, densitometric quantification of phospho-EGFR Tyr-1068 signal was normalized to the β-tubulin signal for the same lane on the same stripped and reprobed blot.
Patterns of EGFR Tyrosine Phosphorylation in Response to EGF, TSP1, and E123—A431 cells were serum-starved for 6 h after which they were exposed to media alone, EGF (10 ng/ml or 1.67 nm, 10 min), native TSP1 (30 μg/ml or 214 nm, 0.5 h), or an equimolar concentration (3.8 μg/ml or 214 nm, 1 h) of TSP1 E123 (aa 549-691). The cells were lysed and the lysates processed for immunoblotting with a series of defined, epitope-mapped, rabbit polyclonal anti-phospho-EGFR antibodies that recognize Tyr-845, Tyr-992, Tyr-1045, and Tyr-1173 (Cell Signaling Technology, Inc., Beverly, MA) and Tyr-1086 (Zymed Laboratories Inc.) all contained within the cytoplasmic domain of EGFR (27). As stated above, a murine monoclonal antibody was use to probe for phospho-EGFR Tyr-1068. The blots were stripped and reprobed with anti-β-tubulin antibodies.
Activation of PLCγ—A431 cells were cultured to ∼80% confluence in 6-well plates, serum-starved for 4 h, and incubated for increasing times with increasing concentrations of TSP1 or media alone. In selected experiments, cells were pretreated with the EGFR-selective tyrphostin, AG1478 (5 μm). In other experiments, cells were transfected with EGFR targeting or control siRNAs. In still other experiments, cells were exposed for 0.5 h to E123 (214 nm). Cells were lysed, and the lysates were resolved by 4-12% SDS-PAGE, transferred to PVDF, and blocked. The membranes were immunoblotted with rabbit anti-human phospho-PLCγ (Tyr-783) antibody (Cell Signaling Technology, Inc.) followed by goat anti-rabbit HRP-conjugated IgG (Pierce) and developed with ECL (48). To confirm equivalent protein loading and transfer, blots were stripped and reprobed for total PLCγ with rabbit anti-human PLCγ antibody (Cell Signaling Technology).
Migration Assay—A431 cells were cultured to confluence in the wells of 24-well plates (Corning Glass, Corning, NY). Using a sterile 200-μl pipette tip, a single wound was made across the diameter of each monolayer, after which cell debris was removed by washing with HEPES as described (49). The cells were then incubated for 48 h with EGF (10 ng/ml or 1.67 nm), TSP1 (30 μg/ml or 214 nm), recombinant TSP1 E123 (3.8 μg/ml or 214 nm), or media alone, each in the presence or absence of the EGFR-selective tyrphostin, AG1478 (5 μm). In selected experiments, cells were transfected with EGFR-targeting or control siRNAs. At 48 h, images of each monolayer were captured using a Nikon Eclipse TS100 microscope coupled to a Nikon Cool pix 4300 camera. Cell migration into the wound was calculated using Image J software (Rasband, WS, Image-J, National Institutes of Health, rsb.info.nih.gov). To evaluate for AG1478-induced A431 cell cytotoxicity, A431 cells cultured to confluence in 96-well plates were incubated for 48 h with AG1478 (5 μm) or media alone. At 48 h, the media were removed, and the monolayers were washed and incubated for 4 h at 37 °C with 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide dye (5 mg/ml PBS). The crystalline formazan product was solubilized for 16 h in 10% SDS, 0.01 n HCl, and A540 nm determined.
Cross-competition Binding Studies—To determine whether E123 binds to the same receptor and/or same portion of the EGFR ectodomain as do high affinity EGFR ligands, binding of fluoroprobe-labeled EGF and E123 to suspended A431 cells was studied with flow cytometry. Purified recombinant E123 was dialyzed versus borate buffer (Slide-A-Lyzer Mini Dialysis Unit, Pierce), reacted with fluorescein isothiocyanate (FITC) reconstituted in dimethylformamide, and again dialyzed to remove excess unconjugated FITC dye (EZ-Label FITC protein labeling kit, Pierce). A431 cells in fluorescence-activated cell sorter tubes (0.5-1.0 × 106 cells/tube) were incubated for 10 min at 4 °C with increasing final concentrations of either FITC-EGF (Invitrogen) or FITC-E123. To define binding over time, A431 cells were incubated at 4 °C with either FITC-EGF (50 ng/ml) or FITC-E123 (2 μg/ml) for increasing times. To establish binding specificity, FITC-EGF (3 μg/ml) was incubated for 10 min at 4 °C with increasing concentrations of unlabeled EGF (up to 500-fold relative to labeled ligand), and FITC-E123 (5 μg/ml) was incubated for 10 min at 4 °C with increasing concentrations of unlabeled E123 (up to 500-fold relative to labeled ligand). Finally, cross-competition binding studies were performed using FITC-EGF with increasing concentrations of unlabeled E123 and FITC-E123 with increasing concentrations of unlabeled EGF. The cells were washed and resuspended in PBS and analyzed by FACScan (BD Biosciences).
Chemical Cross-linking Experiments—To establish a direct receptor-ligand interaction between E123 and cell surface-expressed EGFR ectodomain, E123 (3.8 μg/ml or 214 nm) and cells were co-incubated in the presence of the H2O-soluble, cell-nonpermeable, chemical cross-linking reagent, bis(sulfos-uccinimidyl) suberate (BS3) (Mr = 572.43) (Pierce) as described (50). The two ends of this homobifunctional reagent cross-link amine groups and are separated by a flexible 11.4D spacer arm. A431 cells were serum-starved for 6 h, washed three times with ice-cold PBS (20 mm NaPO4, 0.15 m NaCl, pH 8.0, in HEPES) to remove amine-containing media, and incubated for 0.5 h at 4 °C with E123 (3.8 μg/ml or 214 nm) or media alone in the presence of freshly prepared BS3 at a final concentration of 3 mm. The reaction mixture was incubated for another 0.5 h at room temperature after which the cross-linking reaction was terminated by the addition of glycine (250 mm). The cells were washed and solubilized in the presence of glycine (250 mm). The lysates were precleared by incubation for 1 h at 4 °C with protein G cross-linked to agarose (Sigma) and pre-loaded with a species- and isotype-matched irrelevant antibody (AFAP IgG1, BD Transduction Laboratories). The lysates were then incubated overnight at 4 °C with anti-EGFR antibody (2.5 μg of antibody/500 μg of lysate) (Pharmingen) or an equivalent concentration of the irrelevant antibody control. The resultant immune complexes were immobilized by incubation with protein G cross-linked to agarose for 2 h at 4 °C. After centrifugation, the pellet was washed and boiled in sample buffer, and the eluted EGFR-binding proteins were resolved by 6% SDS-PAGE and transferred to PVDF. The blots for the EGFR immunoprecipitates were probed for either E123 with murine monoclonal C6.7 anti-human TSP1 antibody that recognizes the second EGF-like repeat (47) (Fig. 9, lanes 2-5), or EGF with rabbit polyclonal anti-human EGF antibody (Santa Cruz Biotechnology, Inc.) (lanes 6-9). Purified recombinant E123 was used as a positive control (Fig. 9, lane 1).
FIGURE 9.
Direct E123 interaction with EGFR ectodomain is not detectable. A431 cells were incubated for 10 min at 4 °C with a fixed subsaturation concentration of FITC-EGF (3 μg/ml) in the presence of increasing concentrations of either unlabeled EGF (A) or unlabeled E123 (B). In similar experiments, the cells were incubated for 10 min at 4 °C with a fixed subsaturation concentration of FITC-E123 (5 μg/ml) in the presence of increasing concentrations of either unlabeled E123 (C) or unlabeled EGF (D). A-D are representative of ≥2 independent experiments. The cells were washed, resuspended in PBS, and analyzed by FACScan. E, in other experiments, A431 cells were serum-starved, washed to remove amine-containing media, and incubated for 0.5 h at 4 °C with E123 (214 nm), EGF (16.7 nm), or media alone in the presence of the chemical cross-linking reagent BS3 (final concentration 3 mm). The cross-linking reaction was terminated by the addition of glycine (250 mm), and the cells were lysed. Lysates were immunoprecipitated with anti-EGFR antibody, and the EGFR immunoprecipitate was resolved by 6% SDS-PAGE and transferred to PVDF. The blots were probed for either E123 with murine monoclonal (C6.7) anti-human TSP1 antibody that recognizes the second EGF-like repeat (lanes 1-5) or EGF with rabbit anti-human EGF antibody (lanes 6-9). Purified recombinant E123 was used as a positive control (lane 1). The blots were stripped and reprobed for β-tubulin. IP, immunoprecipitate; IB, immunoblot; IB*, immunoblot after stripping. This blot is representative of three experiments.
Detection of MMPs—A431 cells were incubated for increasing times with TSP1 (30 μg/ml or 214 nm), E123 (3.8 μg/ml or 214 nm), EGF (100 ng/ml or 16.7 nm), or media alone. Supernatants were collected and concentrated through ultrafiltration (Centricon 30000; Millipore Corp., Bedford, MA). The concentrates were collected into Tris-glycine SDS buffer, loaded onto precast 10% zymogram gels containing 1 mg/ml gelatin polymerized within the gel (Invitrogen), and the proteins electro-phoretically resolved. The gels were incubated with renaturing buffer (0.5 h, room temperature), followed by developing buffer 2× (0.5 h at room temperature and 4 h and 37 °C), stained (Simply Blue 0.5%, 0.5 h), and destained (methanol/acetic acid/H2O, 50:10:40) according to the manufacturer's protocol. Clear bands of lysis against a dark blue background were noted, which corresponded to gel mobility of active MMPs. In other experiments, the same concentrated supernatants described above were assayed for MMP9 protein using an MMP9 ELISA kit (Calbiochem) or assayed for MMP9 catalytic activity for a fluorogenic substrate linked to a quencher molecule (Fluorokine E, R & D Systems, Minneapolis, MN). To enhance the sensitivity of the fluorogenic assay, the incubation time of samples with the fluorogenic substrate was prolonged from the prescribed 2-24 h, after which fluorescence (excitation = 355 nm, emission = 425 nm) was assayed (Fluoroskan Ascent; Thermo Scientific, Fremont, CA).
Statistical Methods—Analysis of variance was used to compare the mean responses among experimental and control groups for all experiments. The Dunnett and Scheffé F tests were used to determine significant differences between groups. A p value of <0.05 was considered significant.
RESULTS
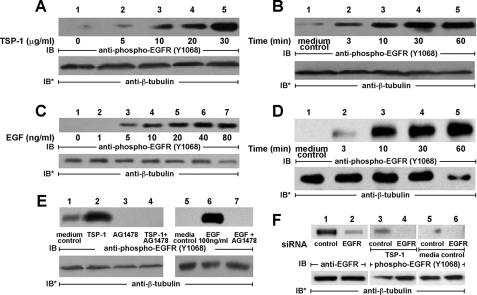
TSP1 Activates EGFR—We previously found that broad spectrum PTK inhibition protects against selected TSP1-induced cell responses (24). TSP1 contains EGF-like repeats (1) and increases ZA protein tyrosine phosphorylation (24), an activity that can be ascribed to EGFR signaling (25-27). We therefore asked whether TSP1 activates EGFR. After serum starvation, increasing concentrations of TSP1 were presented to high EGFR-expressing A431 cells (Fig. 1A). TSP1 at concentrations ≥5 μg/ml (36 nm) increased phosphorylation of EGFR on Tyr-1068. On a molar basis, the minimal TSP1 concentration required for EGFR activation (5 μg/ml, Fig. 1A) was >40-fold greater than the minimal concentration of EGF (5 ng/ml; 0.83 nm; Fig. 1C). TSP1 (214 nm) stimulated EGFR phosphorylation by 3 min with further time-dependent increases up to 60 min (Fig. 1B). These kinetics were similar to those observed with EGF (16.7 nm) treatment (Fig. 1D). Prior PTK inhibition with the EGFR-selective tyrphostin, AG1478, blocked EGFR activation in response to either EGF or TSP1 (Fig. 1E). Transfection of A431 cells with EGFR targeting siRNAs knocked down EGFR protein >90% relative to cells transfected with control siRNA (Fig. 1F, lanes 1 and 2). Prior knockdown of EGFR blocked EGFR Tyr-1068 phosphorylation in response to TSP1 (Fig. 1F, lane 4). These data indicate that TSP1 increases phosphorylation of EGFR at Tyr-1068 in a concentration- and time-dependent manner that is blocked by either an inhibitor of EGFR autophosphorylation or knockdown of EGFR through siRNA technology.
FIGURE 1.
TSP1 activates EGFR in A431 cells. A431 cells were exposed for 0.5 h to increasing concentrations of TSP1 or media alone (A) or exposed for increasing times to a fixed concentration of TSP1 (30 μg/ml or 214 nm) or media alone (B). In other experiments, A431 cells were exposed for 10 min to increasing concentrations of EGF or media alone (C) or exposed for increasing times to a fixed concentration of EGF (100 ng/ml or 16.7 nm) or media alone (D). E, cells were incubated for 0.5 h with TSP1 (30 μg/ml i.e. 214 nm), for 10 min with EGF (100 ng/ml i.e. 16.7 nm), or media alone, in the presence or absence of the EGFR-selective tyrphostin, AG1478 (5 μm). F, cells were transfected with control or EGFR-targeting siRNAs, and after 48 h were lysed and the lysates processed for EGFR immunoblotting (lanes 1 and 2). After transfection with either control or EGFR-targeting siRNAs, cells were incubated with TSP1 (lanes 3 and 4) or media alone (lanes 5 and 6). Cells were lysed and the lysates processed for immunoblotting with anti-phospho-EGFR (Tyr-1068) antibodies. To indicate protein loading and transfer, blots were stripped and reprobed with anti-β-tubulin antibody. IB, immunoblot; IB*, immunoblot after stripping. Each of these blots is representative of >2 experiments.
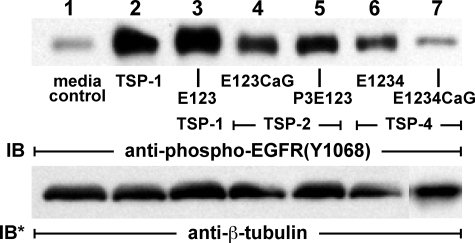
Structure-Function Studies of TSP1-induced EGFR Activation—To define which sequences within TSP1 activate EGFR, the high EGFR-expressing A431 cells were exposed to baculo-virus-encoded recombinant TSP1 constructs that correspond to overlapping sequences from the full-length protein (Fig. 2A) (38-40). Each domain, at a concentration equimolar to the monomeric subunit of TSP1 at 30 μg/ml, i.e. 214 nm, was tested for EGFR activation as measured by Tyr-1068 phosphorylation (Fig. 2B). Only the three recombinant constructs containing EGF-like repeats 1-3 were active (Fig. 2B, lanes 3-5). The E123 alone (Fig. 2B, lane 4) produced the highest level of Tyr-1068 phosphorylation. The third EGF-like repeat fused to the Ca2+-binding type 3 repeats, E3CaG, did not increase EGFR Tyr-1068 autophosphorylation (Fig. 2B, lane 6). Preincubation of TSP1 with either of two antibodies that bind to E123, C6.7, and HB8432 (46, 47) each diminished EGFR Tyr-1068 phosphorylation (Fig. 2C, lanes 4 and 5). These data indicate that the EGF-like repeats are necessary and sufficient for TSP1-induced EGFR activation.
FIGURE 2.
Structure-function analysis of TSP1-induced EGFR activation. A, schematic of baculovirus-derived recombinant TSP1 domains. B, A431 cells were exposed for 0.5 h to equimolar concentrations (214 nm) of baculovirus-derived recombinant TSP1 domains, including the following: lane 1, NH2-terminal heparin-binding domain + oligomerization + procollagen domain (NoC); lane 2, procollagen domain + properidin repeats 1-3 (CP123); lane 3, properidin repeat 3 + EGF-like repeats 1-3 (P3E123); lane 4, EGF-like repeats 1-3 (E123); lane 5, EGF-like repeats 1-3 + Ca2+-binding repeats + COOH terminus (E123-CaG); lane 6, EGF-like repeat 3 to COOH terminus (E3CaG); and lane 7, COOH terminus (CaG). C, TSP1 (214 nm) was preincubated with either of two antibodies, C6.7 and HB8432, each targeting its EGF-like repeats, or a species- and isotype-matched antibody control, B7-1, after which the TSP1 was incubated with A431 cells. D, A431 cells were exposed for 1 h to equimolar concentrations (214 nm) of E123, E12, E2. or E3 or media alone. A431 cells were exposed for 0.5 h to increasing concentrations of recombinant TSP1 EGF-like repeats (E123) or media alone (E) or exposed for increasing exposure times to a fixed concentration of recombinant TSP1 EGF-like repeats (3.8 μg/ml or 214 nm), a concentration equimolar to 30 μg/ml TSP1, or media alone (F). G, A431 cells transfected with EGFR targeting (lane 2) or control (lane 1) siRNAs were cultured for 48 h, after which they were exposed for 1 h to E123 (214 nm) or media alone. Cells were lysed and processed for immunoblotting with anti-phospho-EGFR (Tyr-1068) antibodies. The blots were stripped and reprobed with anti-β-tubulin antibody to indicate protein loading and transfer. IB, immunoblot; IB*, immunoblot after stripping; each of these blots are representative of ≥2 independent experiments. H, for each phospho-EGFR Tyr-1068 immunoblot (see Fig. 1A and Fig. 2E), densitometric quantification of each Tyr(P)-1068 signal was normalized to the β-tubulin signal for the same lane on the same stripped and reprobed blot. Vertical bars represent mean (±S.E.)-fold increase of arbitrary densitometry units of Tyr(P)-1068 signal normalized to arbitrary densitometry units of β-tubulin signal, each relative to the simultaneous control. n = 3. *, significantly increased compared with the equimolar concentration of E123 at p < 0.05.
To establish which of the three EGF-like repeats is responsible for EGFR activation, equimolar concentrations of E123, E12, E2, and E3 were compared for their abilities to stimulate EGFR Tyr-1068 phosphorylation (Fig. 2D). E123 (Fig. 2D, lane 2) stimulated the highest level of Tyr-1068 phosphorylation; E12 stimulated EGFR phosphorylation but to a lesser extent than E123 (Fig. 2D, lane 3); phosphorylation in response to E2 was still less (Fig. 2D, lane 4); and phosphorylation in response to E3 was comparable with that seen in response to E12 (Fig. 2D, lane 5). Again, E3 flanked by the COOH terminus did not increase Tyr-1068 phosphorylation (Fig. 2B, lane 6). Thus, the ability to activate EGFR appears to reside in each of the three EGF-like repeats and is enhanced when all three are assembled in tandem. Interestingly, addition of either the third TSR to form P3E123 (Fig. 2B, lane 3) or the COOH-terminal wire and lectin-like modules to form E123CaG (Fig. 2B, lane 5) attenuated the activity of E123.
When A431 cells were exposed for 1 h to increasing concentrations of purified E123, EGFR Tyr-1068 phosphorylation was increased at concentrations ≥2 μg/ml (112.6 nm) (Fig. 2E). When A431 cells were exposed for increasing times to a fixed concentration of E123 (3.8 μg/ml or 214 nm) (Fig. 2F), EGFR Tyr-1068 phosphorylation was increased after 3-10 min (lanes 2 and 3) with maximum phosphorylation at 30-60 min (lanes 4 and 5). Prior EGFR knockdown completely blocked EGFR Tyr-1068 phosphorylation in response to E123 (Fig. 2G, lane 2). To directly compare the relative potencies of native TSP1 and E123 to activate EGFR, quantitative densitometry of each phospho-EGFR Tyr-1068 signal was normalized to the β-tubulin signal for the same lane on the same stripped and reprobed blot. The mean normalized Tyr(P)-1068 value for the simultaneous media controls was assigned a value 1.0. The normalized Tyr(P)-1068 values for cells challenged with either TSP1 or E123 were expressed as fold-increase relative to the simultaneous control (Fig. 2H). When normalized Tyr(P)-1068 values in response to the TSP1 stimulus were compared with normalized values in response to equimolar concentrations of E123, at each of four tested concentrations, TSP1 was 2.1-2.8-fold more potent than was E123. Thus, the molar potency of TSP1 monomer was at least 2-fold greater than that for E123, and the stimulus-to-response lag time preceding EGFR activation for E123 (10 min) was only slightly prolonged compared with that seen for native TSP1 (3 min) (Fig. 1B).
To determine whether the EGF-like modules of other members of the TSP gene family also activate EGFR, A431 cells were incubated with equimolar concentrations (214 nm) of two recombinant TSP2 constructs, P3E123 and E123CaG, and two recombinant TSP4 constructs, E1234 and E1234 CaG (Fig. 3). Each of these four EGF motif-containing proteins increased EGFR Tyr-1068 phosphorylation. As was seen with the TSP1 constructs, addition of the wire and lectin-like modules to the EGF-like repeats of TSP4 attenuated activity (Fig. 3, lanes 6 versus 7).
FIGURE 3.
TSP-2 and TSP-4 EGF-like repeats activate EGFR. A431 cells were exposed to equimolar concentrations (214 nm) of TSP1 (lane 2), recombinant EGF-like repeats from TSP1 (lane 3), TSP-2 (lanes 4 and 5), and TSP-4 (lanes 6 and 7) or media alone (lane 1). These baculovirus-derived recombinant proteins included the following: TSP1 E123 (lane 3), TSP-2 E123CaG (lane 4), TSP-2 P3E123 (lane 5), TSP-4 E1234 (lane 6), and TSP-4 E1234CaG (lane 7). Cells were lysed and processed for immunoblotting with anti-phospho-EGFR (Tyr-1068) antibodies. The blots were stripped and reprobed with anti-β-tubulin antibody to indicate protein loading and transfer. IB, immunoblot; IB*, immunoblot after stripping. The blot is representative of ≥3 independent experiments.
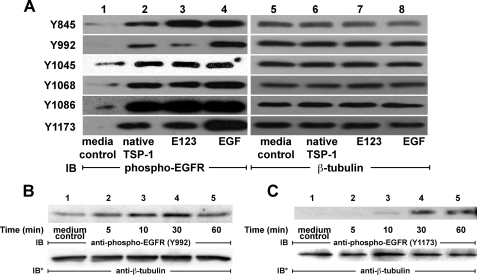
Pattern of EGFR Tyrosine Auto-phosphorylation in Response to Full-length TSP1 Versus TSP1 EGF-like Repeats—There are ≥10 tyrosine residues within the cytoplasmic domain of EGFR that have been identified as autophosphorylation sites (27). Upon phosphorylation, a number of these residues serve as docking sites for specific signaling and adaptor molecules. We asked whether TSP1 or E123 would stimulate phosphorylation of the same tyrosine residues as would be stimulated by EGF. Concentrations and incubation times for each agonist were chosen based on results presented in Fig. 1, A—D, and Fig. 2, E and F. TSP1 (214 nm, 0.5 h), E123 (214 nm, 1 h), and EGF (1.67 nm, 10 min) each increased phosphorylation of Tyr-845, Tyr-992, Tyr-1045, Tyr-1068, Tyr-1086, and Tyr-1173 (Fig. 4A, lanes 2-4). Therefore, EGFR phosphorylation in response to TSP1 and E123 was comparable with that seen in response to EGF.
FIGURE 4.
EGFR phosphotyrosine profile in response to TSP1 versus TSP1 EGF-like repeats. A, A431 cells were exposed to EGF (10 ng/ml or 1.67 nm, 10 min), native TSP1 (30 μg/ml or 214 nm, 0.5 h), an equimolar concentration of recombinant TSP1 EGF-like repeats (E123) (3.8 μg/ml or 214 nm, 1 h), or media alone. The cells were lysed and processed for immunoblotting with a series of defined epitope-mapped anti-phospho-EGFR antibodies that recognize Tyr(P)-845, Tyr(P)-992, Tyr(P)-1045, Tyr(P)-1068, Tyr(P)-1086, and Tyr(P)-1173 (lanes 1-4). In other experiments, A431 cells were exposed to a fixed concentration of TSP1 (30 μg/ml or 214 nm) for increasing times, after which the cells were lysed and the lysates processed for phospho-EGFR Tyr-992 (B) and Tyr-1173 (C) immunoblotting. To indicate protein loading and transfer, blots were stripped and reprobed with anti-β-tubulin antibodies (A, lanes 5-8). IB, immunoblot; IB*, immunoblot after stripping. These blots are representative of ≥2 independent experiments.
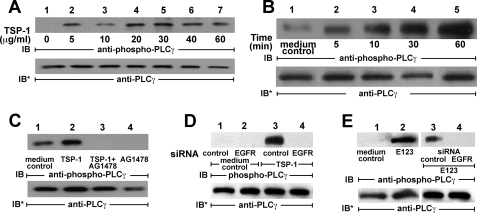
TSP1 Activates PLCγ—TSP1 and E123 activate EGFR (Figs. 1, 2, 3 and 4), stimulating autophosphorylation of EGFR Tyr-992 and Tyr-1173 (Fig. 4A). Upon EGFR activation, these two phosphotyrosines serve as docking sites for PLCγ (51). To relate these phosphorylation events to downstream signaling, phosphorylation of Tyr-992 and Tyr-1173 in response to the TSP1 stimulus was studied over time (Fig. 4, B and C). Phosphorylation of Tyr-992 was evident at ≥5 min with maximal phosphorylation at 30 min, whereas Tyr-1173 phosphorylation was not seen until ≥10 min with further increases at 30 and 60 min. We then asked whether TSP1 or E123 activates PLCγ through EGFR activation. In A431 cells, TSP1 at ≥5 μg/ml (36 nm) increased PLCγ activation; this activation plateaued at TSP1 ≥20 μg/ml (144 nm) (Fig. 5A). When A431 cells were exposed to TSP1 (214 nm) for increasing exposure times, PLCγ was activated at ≥5 min with further time-dependent increments through 60 min (Fig. 5B). The EGFR-selective PTK inhibitor, AG1478, completely blocked TSP1-induced PLCγ activation (Fig. 5C). Similarly, prior knockdown of EGFR also blocked PLCγ activation in response to TSP1 (Fig. 5D, lane 4). E123 (214 nm) also activated PLCγ (Fig. 5E, lanes 1 and 2), and again, prior EGFR knockdown prevented this activation (Fig. 5E, lanes 3 and 4). These findings indicate that the E123 domain of TSP1 activates PLCγ in a dose- and time-dependent manner and does so through EGFR.
FIGURE 5.
TSP1 activates PLCγ through EGFR activation. A431 cells were exposed for 0.5 h to increasing concentrations of TSP1 (A) or exposed to a fixed concentration of TSP1 (30 μg/ml or 214 nm) for increasing times (B) or media alone. C, TSP1 was introduced in the presence or absence of the EGFR-selective tyrphostin, AG1478 (5 μm). D, cells were transfected with either control or EGFR-targeting siRNAs, after which they were incubated with TSP1 (lanes 3 and 4) or media alone (lanes 1 and 2). E, A431 cells were exposed for 1 h to E123 (214 nm) or media alone (lanes 1 and 2). In other experiments, A431 were transfected with EGFR targeting or control siRNAs and, after 48 h, were treated for 1 h with E123 (214 nm)(lanes 3 and 4). Cells were lysed and the lysates processed for phospho-PLCγ (Tyr-783) immunoblotting. To indicate protein loading and transfer, blots were stripped and reprobed with anti-PLCγ antibodies. IB, immunoblot; IB*, immunoblot after stripping. These blots are representative of ≥2 independent experiments.
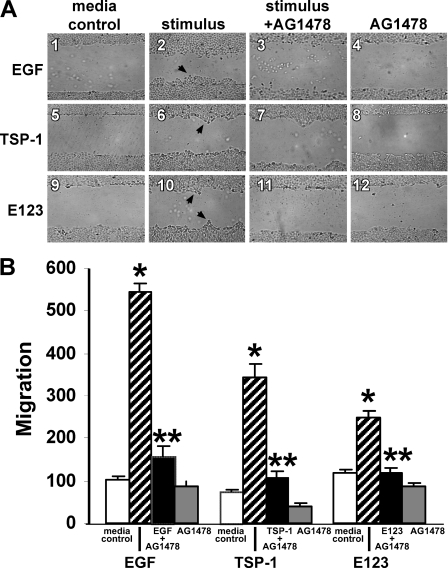
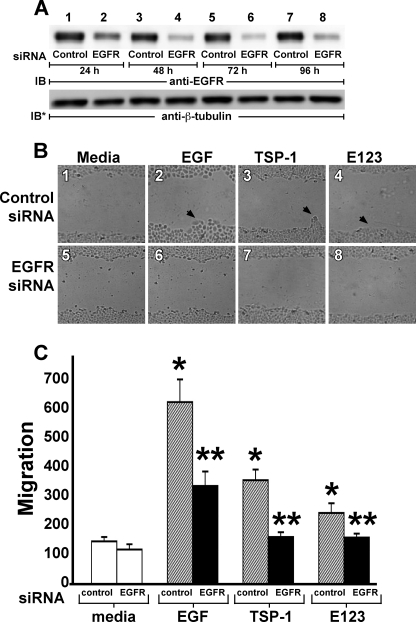
TSP1 Increases Cell Migration—Phosphorylation of EGFR Tyr-992 and Tyr-1173 (Fig. 4, B and C), and PLCγ activation (Fig. 5, A and B), are two prerequisite signaling events to EGFR-driven cell migration (19). We asked whether, like EGF, TSP1 or its EGF-like repeats might also increase cell migration through EGFR activation. The influence of these three EGFR ligands, EGF, TSP1, and E123, on A431 cell migration was analyzed in a standard wounded monolayer assay (Fig. 6) (49). At 48 h, EGF, TSP1, and E123 each increased cell migration 5.3-, 4.6-, and 2.1-fold, respectively (Fig. 6, A and B). During this same period, no increases in cell proliferation as measured by [3H]thymidine incorporation could be detected (data not shown). The increased cell migration in response to EGF, TSP1, and E123 each was diminished by 88, 88, and ∼100%, respectively, by EGFR-selective PTK inhibition with AG1478 (Fig. 6, A and B). When AG1478-exposed (5 μm, 48 h, n = 12) and media control (n = 12) A431 cells were analyzed for cytotoxicity in an 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide assay, no difference was found (1.20 ± 0.03 versus 1.22 ± 0.03; p = 0.56). Transfection of A431 cells with EGFR-targeting siRNAs knocked down EGFR protein >90% relative to cells transfected with control siRNA at 48, 72, and 96 h (Fig. 7A, lanes 4, 6, and 8). At 24 h, EGFR protein was only knocked down ∼75% (Fig. 7A, lane 2). Prior knockdown of EGFR diminished the migratory response to EGF, TSP1, and E123 by ∼60, ∼93, and ∼100%, respectively (Fig. 7, B and C). These findings indicate that the EGF-like repeats of TSP1 stimulate epithelial cell migration through EGFR activation. Because TSP1 increased migration ∼2.2-fold more than did equimolar concentrations of E123 (Figs. 6 and 7), each normalized to its simultaneous control, it is conceivable that an additional domain(s) outside the EGF-like repeats also contributes to the migratory response.
FIGURE 6.
TSP1 increases cell migration through EGFR activation. A431 cells were cultured to confluence in the wells of 24-well plates, after which they were wounded with a pipette tip, washed to remove cellular debris, and incubated for 48 h with EGF (10 ng/ml or 1.67 nm), TSP1 (30 μg/ml or 214 nm), E123 (3.8 μg/ml or 214 nm), or media alone in the presence or absence of the EGFR selective tyrphostin, AG1478 (5 μm)(n = 6). At 48 h, cellular migration into the wound was photographed in triplicate and quantified. A, representative photographs of wounded monolayers after 48 h of incubation with EGF, TSP1, E123, or media alone in the presence or absence of AG1478. Arrows in panels 2, 6, and 10 indicate increased cell migration into the wound. Magnification, ×40. B, vertical bars represent mean (±S.E.) migration into the wound at 48 h after incubation with EGF, TSP1, E123, or media alone in the presence or absence of AG1478. For each condition, n = 6. *, significantly increased compared with the media control at p < 0.05. **, significantly decreased compared with the stimulus alone at p < 0.05.
FIGURE 7.
EGFR knockdown blocks TSP1-induced cell migration. A, A431 cells were transfected with control or EGFR-targeting siRNAs and after 24, 48, 72, and 96 h were lysed, and the lysates were processed for EGFR immunoblotting (lanes 1-8). To indicate protein loading and transfer, blots were stripped and reprobed for β-tubulin. IB, immunoblot; IB*, immunoblot after stripping. This blot is representative of two independent experiments. B and C, after transfection with either control or EGFR-targeting siRNAs, A431 cells were cultured to confluence in the wells of 24-well plates, after which they were wounded with a pipette tip, washed to remove cellular debris, and incubated for 48 h with EGF (10 ng/ml or 1.67 nm), TSP1 (30 μg/ml or 214 nm), E123 (3.8 μg/ml or 214 nm), or media alone (n = 6). At 48 h, cellular migration into the wound was photographed in triplicate and quantified. B, representative photographs of wounded monolayers after 48 h of incubation with EGF, TSP1, E123, or media alone. Arrows in panels 2-4 indicate increased cell migration into the wound. Magnification, × 40. C, vertical bars represent mean (±S.E.) migration into the wound at 48 h after incubation with EGF, TSP1, E123, or media alone. *, significantly increased compared with the control siRNA-transfected media control at p < 0.05. **, significantly decreased compared with the control siRNA-transfected cells with stimulus at p < 0.05.
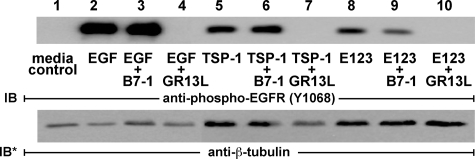
Effect of EGFR Ectodomain Blocking Antibodies on TSP1 Activation of EGFR—TSP1 or E123 could activate EGFR through the following: 1) direct binding to the ligand-binding portion of its ectodomain; 2) MMP-dependent release of cell-bound EGFR ligands; or 3) transactivation through one or more heterologous receptors (36, 52-54). To distinguish the first two possibilities from the third, A431 cells were exposed to EGF, TSP1, E123 or media alone, in the presence or absence of the specific EGFR ectodomain-blocking antibody, GR13L (44), or a species- and isotype-matched irrelevant antibody control, B7-1/CD80. Lysates of these cells were assayed for EGFR Tyr-1068 phosphorylation as an indicator for EGFR activation (Fig. 8). Inclusion of GR13L in the assay completely blocked EGFR activation in response to EGF (Fig. 8, lane 4), TSP1 (lane 7), or E123 (lane 10), whereas inclusion of the antibody control did not (lanes 3, 6, and 9). These data indicate that TSP1 and its EGF-like repeats activate EGFR through the ligand-binding portion of its ectodomain. Thus, activation appears to be due to either direct engagement of EGFR and/or through MMP-mediated release of a tethered EGFR ligand(s).
FIGURE 8.
Immunoblockade of EGFR ectodomain inhibits EGFR activation. A431 cells were exposed to EGF (10 ng/ml or 1.67 nm, 10 min), TSP1 (30 μg/ml or 214 nm, 0.5 h), E123 (3.8 μg/ml or 214 nm, 1 h), or media alone, each in the presence or absence of the EGFR ectodomain-blocking antibody GR13L or a species- and isotype-matched antibody control B7-1. The cells were lysed, and the lysates were processed for phospho-EGFR (Tyr-1068) immunoblotting. The blots were stripped and reprobed for β-tubulin to indicate protein loading and transfer. IB, immunoblot; IB*, immunoblot after stripping. This blot is representative of >2 independent experiments.
Direct Interaction of E123 with EGFR Ectodomain Is Not Detectable—To demonstrate a direct interaction between the EGF-like repeats of TSP1 and the EGFR ectodomain, purified recombinant E123 was studied vis-à-vis EGF in both cross-competition binding (Fig. 9, A-D) and chemical cross-linking (Fig. 9E) experiments. Using flow cytometry with A431 cells, FITC-E123 and FITC-EGF each displayed saturable binding that could be competitively diminished with the respective unlabeled ligand (Fig. 9, A and C). In contrast, no competitive inhibition could be demonstrated between FITC-EGF and unlabeled E123 (Fig. 9B) or between FITC-E123 and unlabeled EGF (Fig. 9D). When the high affinity EGFR ligand EGF was incubated with A431 cells in the presence of the cell-nonpermeable cross-linking reagent BS3, followed by immunoprecipitation of EGFR with immunoblotting for EGF, cross-linking of EGF to the EGFR was robust (Fig. 9E, lane 9). In contrast, when E123 was co-incubated with cells and BS3 under identical conditions, no cross-linking of E123 to the EGFR was evident (Fig. 9E, lane 5). These results argue against a direct E123-EGFR interaction at concentrations at which E123 activates EGFR.
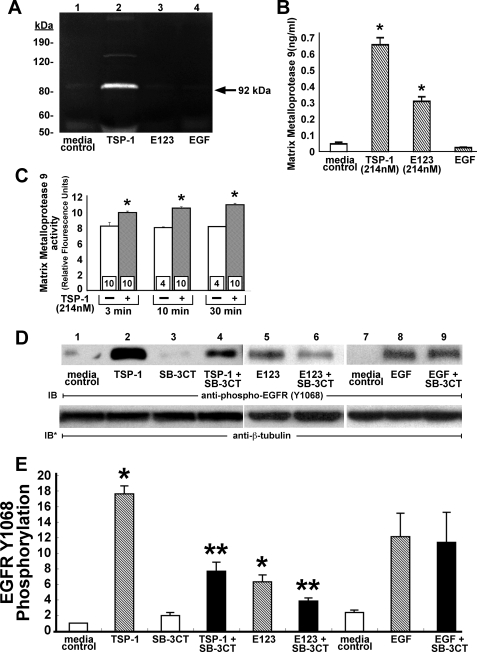
TSP1 Requires MMP Activity for EGFR Activation—Immunoblockade of the ligand-binding portion of the EGFR ectodomain blocked TSP1- and E123-induced EGFR activation (Fig. 8); however, a direct E123-EGFR interaction could not be demonstrated (Fig. 9). We therefore focused on whether TSP1 and/or E123 release EGFR ligand(s) through MMP activation as has been described (28, 54). Zymograms of supernatants of TSP1-treated A431 cells revealed gelatinase activity that migrated with an Mr of ∼92,000 (Fig. 10A, lane 2). To detect the MMP activity required, 10-fold concentrated supernatants were pooled from five 100-mm dishes. Maximal activity was apparent at 24 h (data not shown). On the basis of the apparent gel mobility of the TSP1-induced gelatinase activity, the concentrated supernatants were assayed for MMP9 protein by ELISA (Fig. 10B). After 24 h, TSP1 and E123 each increased MMP9 protein, whereas EGF did not. MMP9 catalytic activity for a fluorogenic substrate was measured at early time points following the TSP1 stimulus (Fig. 10C). TSP1 increased MMP9 activity as early as 3 min with further time-dependent increments at 10 and 30 min relative to the simultaneous media controls. Thus, the increases in MMP9 activity were temporally proximal to or coincident with EGFR activation in response to TSP1 (Fig. 1B). Although these TSP1-induced increments in MMP9 activity were no greater than 1.4-fold relative to the simultaneous controls, they were consistent and attained statistical significance. To determine whether TSP1 and/or E123 might require MMP activity for EGFR activation, EGFR phosphorylation in response to either TSP1 or E123 was studied in the presence of the MMP2/MMP9 inhibitor, SB-3CT (43). Prior MMP inhibition decreased TSP1- and E123-induced EGFR Tyr-1068 phosphorylation by ∼60% (Fig. 10, D, lanes 4 and 6, and E). In contrast, EGF-induced EGFR Tyr-1068 phosphorylation was not influenced by prior MMP inhibition (Fig. 10, D, lane 9, and E). Because TSP1 increased both MMP9 protein (Fig. 10, A and B) and activity (Fig. 10C) and prior MMP inhibition partially blocked TSP1-induced EGFR activation (Fig. 10, D and E), we asked whether MMP9 was operative. Preincubation of cells with MMP9 neutralizing antibody for immunoblockade of surface expressed MMP9 diminished TSP1-induced EGFR Tyr-1068 phosphorylation by >65% (Fig. 11A, lane 3). Transfection of A431 cells with MMP9-targeting siRNAs knocked down MMP9 protein ∼60% relative to cells transfected with control siRNA in both TSP1-treated and E123-treated cells (Fig. 11A). MMP9 levels in EGF-stimulated and media control cells were almost undetectable. Prior knockdown of MMP9 diminished EGFR Tyr-1068 phosphorylation by ∼80% relative to the control siRNA-transfected cells in response to either TSP1 or E123 (Fig. 11C, lanes 4 and 6). Similarly, prior MMP9 knockdown decreased TSP1/E123-induced PLCγ activation by >80% compared with the simultaneous controls (Fig. 11D, lanes 4 and 6). In contrast, prior knockdown of MMP9 did not alter either EGFR Tyr-1068 phosphorylation (Fig. 11C, lanes 7 and 8) or PLCγ activation (Fig. 11D, lanes 7 and 8) in response to EGF, excluding any nonspecific downstream signaling effects. These data suggest that EGFR and PLCγ activation in response to TSP1 is mediated, at least in part, through MMP activation, involving MMP9 and possibly one or more other undetected MMPs.
FIGURE 10.
TSP1 activates EGFR through MMP activation. A and B, A431 cells were treated for 24 h with TSP1 (214 nm), E123 (214 nm), EGF (1. 67 nm), or media alone, after which supernatants were concentrated 10-fold through ultrafiltration. A, concentrated supernatants were processed for zymography, after which clear bands against a dark background indicate gelatinase activity. Molecular mass is indicated at left in kDa. B, concentrated supernatants were assayed for MMP9 by ELISA (n = 6). Vertical bars represent mean (±S.E.) MMP9 levels in ng/ml. C, A431 cells were treated for increasing times (3, 10, or 30 min) with TSP1 (214 nm) or media alone, after which concentrated supernatants were incubated for 24 h with a fluorogenic substrate, and the samples were fluorometrically assayed and MMP9 catalytic activity was expressed in mean (±S.E.) relative fluorescence units. n, the number of experiments performed in each group, is indicated within each vertical bar. D and E, A431 cells were exposed for 0.5 h to TSP1 (214 nm), E123 (214 nm), EGF (1.67 nm), or mediaalone in the presence or absence of MMP-2/MMP9 inhibitor SB-3CT(1 μm) and lysed, and the lysates were processed for phospho-EGFR (Tyr(P)-1068) immunoblotting. To indicate protein loading and transfer, blots were stripped and reprobed with anti-β-tubulin antibody. IB, immunoblot; IB*, immunoblot after stripping. The blot is representative of three independent experiments. E, quantitative densitometry of immunoblots in (D)(n = 3). E, vertical bars represent mean (±S.E.) densitometry units. * indicates significantly increased compared with the simultaneous media control at p<0.05.**indicates significantly decreased compared with the TSP1/E123 control at p < 0.05.
FIGURE 11.
TSP1 increases EGFR Tyr-1068 phosphorylation through MMP9 activation. A, A431 cells were preincubated with anti-MMP9 neutralizing antibody or a species- and isotype-matched antibody control, B7-1, after which the cells were treated for 0.5 h with TSP1 (214 nm) or media alone. Cells were lysed and the lysates processed for phospho-EGFR (Tyr-1068) immunoblotting. To indicate protein loading and transfer, blots were stripped and reprobed with anti-β-tubulin antibody. B-D, A431 cells were transfected with control or MMP9-targeting siRNAs after which they were incubated with TSP1 (214 nm, 0.5 h), E123 (214 nm, 1 h), EGF (1.67 nm, 10 min), or media alone. B, supernatants were concentrated 10-fold through ultrafiltration, and the concentrate supernatants were assayed for MMP9 by ELISA. Vertical bars represent mean (±S.E.) MMP9 levels in ng/ml (n = 6). *, indicates significantly increased compared with control siRNA-transfected media control at p < 0.05; **, indicates significantly decreased compared with the control siRNA-transfected cells with stimulus at p < 0.05. C, cells were lysed and the lysates processed for phospho-EGFR (Tyr-1068) immunoblotting. To indicate protein loading and transfer blots were stripped and reprobed with anti-β-tubulin antibody. D, cells were lysed and the lysates processed for phospho-PLCγ (Tyr-783) immunoblotting. To indicate protein loading and transfer, blots were stripped and reprobed with anti-PLCγ antibodies. A, C, and D, IB, immunoblot; IB*, immunoblot after stripping. These blots are representative of ≥2 independent experiments.
DISCUSSION
In these studies, we identified a novel function for the EGF-like repeats of TSP1. Exposure of high EGFR-expressing A431 cells to human platelet-derived trimeric TSP1 or baculovirus-encoded recombinant TSP1 EGF-like repeats increased EGFR tyrosine phosphorylation. The EGF-like repeats alone were sufficient to activate EGFR, whereas no other TSP1 domains could do so. EGF-like repeats from TSP2 and TSP4 also activated EGFR. Native TSP1 and E123 each increased phosphorylation of six distinct tyrosine residues within the cytoplasmic domain of EGFR (Tyr-845, Tyr-992, Tyr-1045, Tyr-1068, Tyr-1086, and Tyr-1173). Immunoblockade of the ligand-binding portion of the EGFR ectodomain blocked EGFR activation in response to TSP1 or its EGF-like repeats. However, a direct binding interaction between E123 and EGFR could not be demonstrated. TSP1 and E123 each increased MMP9 expression and activity, and EGFR activation in response to either agonist was blocked, in part, by MMP2/MMP9-selective inhibition, immunoblockade of surface expressed MMP9, or knockdown of MMP9. These data support a scenario in which the EGF-like repeats of TSP1 stimulate MMP-mediated release of cell surface bound EGFR ligand(s), which engages the EGFR ectodomain. Finally, EGFR activation in response to TSP1 and its EGF-like repeats, including autophosphorylation of Tyr-992 and Tyr-1173, the docking sites for PLCγ, was coupled to down-stream PLCγ activation and increased cell migration.
EGFR responds to direct binding of EGF motif-containing ligands (28) but also can be transactivated through a number of heterologous receptors (36, 37, 52, 54, 55). We first asked whether the EGF-like repeats of TSP1 directly engaged the EGFR ectodomain. Using two distinct experimental approaches, direct E123 binding to EGFR could not be detected (Fig. 9). However, preincubation of A431 cells with antibody that targets the ligand-binding portion of the EGFR ectodomain completely blocked EGFR activation in response to EGF, TSP1, and E123 (Fig. 8). The requirement for an unimpeded EGFR ectodomain (Fig. 8) in the absence of a detectable E123-EGFR interaction (Fig. 9) raised the possibility that TSP1 indirectly activates EGFR through MMP-mediated release of an EGFR ligand(s) (28, 54). Of interest, TSP1 reportedly up-regulates MMP-2 and -9 expression (56, 57), and in A431 cells, we found that TSP1 and its EGF-like repeats each increases MMP9 expression (Fig. 10B) and catalytic activity (Fig. 10C). That TSP1 increased MMP9 protein expression ≥2-fold more than did an equimolar concentration of E123 (Fig. 10B) implicates participation of TSP1 domains outside of the EGF-like repeats (58). Although TSP1 is known to increase MMP9 expression (56), it also directly inhibits enzymatic conversion of pro-MMP9 to MMP9 (58, 59), up-regulates expression of TIMP1 (21) and TIMP2 (56), and protects specific substrates from proteolysis (60). These bioactivities ascribed to TSP1 may make it more difficult to detect changes in MMP9 activity in response to the same molecule. Finally, pharmacologic inhibition of MMP activity (Fig. 10D), immunoblockade of surface expressed MMP9 (Fig. 11A), or siRNA-induced knockdown of MMP9 (Fig. 11B), each substantially but incompletely reduced EGFR activation in response to either TSP1 or E123. These combined data are compatible with a mechanism in which TSP1/E123 up-regulates MMP activity, including MMP9, to proteolytically release an EGFR ligand(s), which then binds to the EGFR ectodomain and activates EGFR signaling. TSP1 reportedly regulates MMP activity but through its NH2-terminal and TSR domains (58). How E123 might up-regulate MMP expression or activity is unclear. Beyond the classical MMPs, participation of either the closely related ADAMs and/or ADAMTs (a disintegrin and metalloproteinase with TSP motifs) families has not been excluded (61, 62). ADAM-9, -10, -12, and -17 each reportedly cleave and release one or more high affinity EGFR ligands (29-32). A431 cells express mRNAs for six high affinity EGFR ligands, including EGF, transforming growth factor α, amphiregulin, heparin-binding EGF, epiregulin, and betacellulin.3 Identification of the specific EGFR ligand(s) regulated by the EGF-like repeats of TSP1 is currently under study.
Although EGFR activation in response to the EGF-like repeats of TSP1 is, in part, MMP9-mediated (Figs. 10D and 11C), other mechanisms cannot be absolutely excluded. It is conceivable that TSP1 binds to EGFR with a binding affinity so low as to preclude detection by either cross-competition binding or chemical cross-linking studies (Fig. 9, C-E). Although each of the TSP1 EGF-like repeats contain the framework of six conserved cysteine residues that form the three intramolecular 1-3, 2-4, and 5-6 disulfide bonds required for EGFR engagement, the intervening residues dramatically diverge from corresponding sequences in high affinity EGFR ligands (28). Furthermore, unlike most EGF-like repeats that contain one residue between the fourth and fifth cysteines, the EGF-like repeats of the TSPs have two, potentially altering their structural flexibility (2). Two other proteins containing divergent EGF-like repeats, tenascin-C and laminin-5, have been shown to display low affinity binding to EGFR (34, 35). Multidomain TSP1, through its ability to bind multiple heterologous receptors, may also transactivate EGFR (52, 54, 55). Of course this would not account for the activity of isolated E123. Finally, one or more TSP1 domains might also antagonize EGFR activation by the EGF-like repeats. In fact, our data indicate that the TSP1 EGF-like repeats co-expressed with either the flanking third type I repeat or the type III Ca2+-binding repeats each increased EGFR Tyr-1068 phosphorylation to lower levels than did the EGF-like repeats alone (Fig. 2B, lane 4 versus lanes 3 and 5).
EGFR tyrosine autophosphorylation is coupled to recruitment of signaling elements and cellular responses, including proliferation, differentiation, survival, and motility (27, 63). Our studies show that TSP1 and its EGF-like repeats increase phosphorylation of Tyr-992 and Tyr-1173 (Fig. 4), binding sites for PLCγ (27). PLCγ participates in EGFR-driven cell motility (63). In our studies, TSP1 and its EGF-like repeats increased PLCγ activation (Fig. 5, A, B, and E) and cell motility (Figs. 6 and 7). TSP1/E123-induced increases in PLCγ activation and cell motility were completely blocked by prior EGFR-selective PTK inhibition (Fig. 5C and Fig. 6) or siRNA-mediated EGFR depletion (Fig. 5, D and E, and Fig. 7). Although E123 also increased cell migration, it was 50% less active than native TSP1 (Fig. 6). Whether one or more domain(s) outside the EGF-like repeats is required for an optimal migratory response is unclear. Our combined data indicate that the EGF-like repeats of TSP1 can increase cell motility through EGFR and possibly through PLCγ activation.
The ability to activate EGFR, and possibly other co-expressed ErbB receptors, together with the numerous signaling elements to which they are coupled, may elucidate relatively unexplained aspects of TSP1 biology, especially apropos to the epithelial cell. In TSP1 null mice, multilineage epithelial hyperplasia with thickening and ruffling of selected epithelia was prominent (22). In human epithelium-derived cancer cells, EGF increases TSP1 expression (21). TSP1 expression is increased during tissue repair and wound healing (7, 64) where it accelerates re-epithelialization (8). In a murine model of corneal abrasion, exogenous TSP1 stimulated epithelialization, whereas anti-TSP1 antibodies inhibited it (8). In A431 cells, EGFR is co-expressed with ErbB2. TSP1 and its EGF-like repeats, through the ability to activate EGFR, transactivates its preferred heterodimerization partner ErbB2.3 Co-activation of EGFR/ErbB2 heterodimers appears to be key in epithelial tumor cell biology (65). TSP1 is known to influence tumor cell adhesion, migration, invasion, and metastasis both in vitro and in vivo (20, 65, 66). Whether TSP1 influences tumor cell behavior through EGFR/ErbB2 biology had not been tested. We now have demonstrated for the first time that increased migration of a human epidermoid carcinoma (A431) cell line, in response to TSP1, is blocked by prior EGFR-selective PTK inhibition (Fig. 6) or siRNA-induced knockdown (Fig. 7). Our findings on TSP1, TSP2, and TSP4, and the work of others on laminin-5 (34) and tenascin-C (35), collectively suggest that the phylogenetically ancient EGF-like repeat that has been conserved across a large and diverse group of proteins (33) may contribute to their bioactivity.
Acknowledgments
We thank Shirley Taylor for excellent secretarial support.
This work was supported, in whole or in part, by National Institutes of Health Grants HL084223 (to S. E. G.), HL54462 (to D. F. M.), and HL079644 (to J. M.-U.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TSP, thrombospondin; BS3, bis(sulfosuccinimidyl)suberate; E123, epidermal growth factor-like repeats 1-3; ECL, enhanced chemiluminescence; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; FITC, fluorescein isothiocyanate; HRP, horseradish peroxidase; MMP, matrix metalloprotease; PLCγ, phospholipase Cγ; PTK, protein-tyrosine kinase; PVDF, polyvinylidene difluoride; siRNA, small interfering RNA; TSR, thrombospondin type 1 repeat; aa, amino acid; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline.
A. Liu and S. E. Goldblum, unpublished observations.
References
- 1.Lawler, J., Derick, L. H., Connolly, J. E., Chen, J.-H., and Chao, F. C. (1985) J. Biol. Chem. 260 3762-3772 [PubMed] [Google Scholar]
- 2.Carlson, C. B., Lawler, J., and Mosher, D. F. (2008) Cell. Mol. Life Sci. 65 672-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson, C. B., Bernstein, D. A., Annis, D. S., Misenheimer, T. M., Hannah, B. L., Mosher, D. F., and Keck, J. L. (2005) Nat. Struct. Mol. Biol. 12 910-914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malashkevich, V. N., Kammerer, R. A., Efimov, V. P., Schulthess, T., and Engel, J. (1996) Science 274 761-765 [DOI] [PubMed] [Google Scholar]
- 5.Bornstein, P. (1995) J. Cell Biol. 130 503-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan, K., Duquette, M., Liu, J. H., Zhang, R., Joachimiak, A., Wang, J. H., and Lawler, J. (2006) Structure (Lond.) 14 33-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Streit, M., Velasco, P., Riccardi, L., Spencer, L., Brown, L. F., Janes, L., Lange-Asschenfeldt, B., Yano, K., Hawighorst, T., Iruela-Arispe, L., and Detmar, M. (2000) EMBO J. 19 3272-3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uno, K., Hayashi, H., Kuroki, M., Uchida, H., Yamauchi, Y., Kuroki, M., and Oshima, K. (2004) Biochem. Biophys. Res. Commun. 315 928-934 [DOI] [PubMed] [Google Scholar]
- 9.Orr, A. W., Pedraza, C. E., Pallero, M. A., Elzie, C. A., Goicoechea, S., Strickland, D. K., and Murphy-Ullrich, J. E. (2003) J. Cell Biol. 161 1179-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts, D. D., Haverstick, D. M., Dixit, V. M., Frazier, W. A., Santoro, S. A., and Ginsburg, V. J. (1985) J. Biol. Chem. 260 9405-9411 [PubMed] [Google Scholar]
- 11.Calzada, M. J., Sipes, J. M., Krutzsch, H. C., Yurchenco, P. D., Annis, D. S., Mosher, D. F., and Roberts, D. D. (2003) J. Biol. Chem. 278 40679-40687 [DOI] [PubMed] [Google Scholar]
- 12.Dawson, D. W., Pearce, S. F., Zhong, R., Silverstein, R. L., Frazier, W. A., and Bouck, N. P. (1997) J. Cell Biol. 138 707-717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawler, J., Weinstein, R., and Hynes, R. O. (1998) J. Cell Biol. 107 2351-2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calzada, M. J., Annis, D. S., Zeng, B., Marcinkiewicz, C., Banas, B., Lawler, J., Mosher, D. F., and Roberts, D. D. (2004) J. Biol. Chem. 279 41734-41743 [DOI] [PubMed] [Google Scholar]
- 15.Kosfeld, M. D., and Frazier, W. A. (1993) J. Biol. Chem. 268 8808-8814 [PubMed] [Google Scholar]
- 16.Gao, A.-G., Lindberg, F. P., Finn, M. B., Blystone, S. D., Brown, E. J., and Frazier, W. A. (1996) J. Biol. Chem. 271 21-24 [DOI] [PubMed] [Google Scholar]
- 17.Lahav, J. (1993) Biochim. Biophys. Acta 1182 1-14 [DOI] [PubMed] [Google Scholar]
- 18.Lawler, J. (1986) Blood 67 1197-1209 [PubMed] [Google Scholar]
- 19.O'Shea, K. S., and Dixit, V. M. (1988) J. Cell Biol. 107 2737-2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taraboletti, G., Roberts, D. D., and Liotta, L. A. (1987) J. Cell Biol. 105 2409-2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soula-Rothut, M., Coissard, C., Sartelet, H., Boudot, C., Bellon, G., Martiny, L., and Rothhut, B. (2005) Exp. Cell Res. 304 187-201 [DOI] [PubMed] [Google Scholar]
- 22.Crawford, S. E., Stellmach, V., Murphy-Ullrich, J. E., Ribeiro, S. M., Lawler, J., Hynes, R. O., Bolvin, G. P., and Bouck, N. (1998) Cell 93 1159-1170 [DOI] [PubMed] [Google Scholar]
- 23.Lawler, J., Sunday, M., Thibert, V., Duquette, M., George, E. L., Rayburn, H., and Hynes, R. O. (1998) J. Clin. Investig. 101 982-992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldblum, S. E., Young, B. A., Wang, P., and Murphy-Ullrich, J. E. (1999) Mol. Biol. Cell 10 1537-1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoschuetzky, H., Aberle, H., and Kemler, R. (1994) J. Cell Biol. 127 1375-1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mariner, D. J., Davis, M. A., and Reynolds, A. B. (2003) J. Cell Sci. 117 1339-1350 [DOI] [PubMed] [Google Scholar]
- 27.Olayioye, M. A., Neve, R. M., Lane, H. A., and Hynes, N. E. (2000) EMBO J. 19 3159-3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, R. C., Chung, E., and Coffey, R. J. (2003) Exp. Cell Res. 284 2-13 [DOI] [PubMed] [Google Scholar]
- 29.Sahin, U., Weskamp, G., Kelly, K., Zhou, H. M., Higashiyama, S., Peschon, J., Hartmann, D., Saftig, P., and Blobel, C. P. (2004) J. Cell Biol. 164 769-779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izumi, Y., Hirata, M., Hasuwa, H., Iwamoto, R., Umata, T., Miyado, K., Tamai, Y., Kurisaki, T., Sehara-Fujisawa, A., Ohno, S., and Mekada, E. (1998) EMBO J. 17 7260-7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asakura, M., Kitakaze, M., Takashima, S., Liao, Y., Ishikura, F., Yoshinaka, T., Ohmoto, H., Node, K., Yoshino, K., Ishiguro, H., Asanuma, H., Sanada, S., Matsumura, Y., Takeda, H., Beppu, S., Tada, M., Hori, M., and Higashiyama, S. (2002) Nat. Med. 8 35-40 [DOI] [PubMed] [Google Scholar]
- 32.Kurisaki, T., Masuda, A., Sudo, K., Sakagami, J., Higashiyama, S., Matsuda, Y., Nagabukuro, A., and Tsuji, A., Nabeshima, Y., Asano, M., Iwakura, Y., and Sehara-Fujisawa, A. (2003) Mol. Cell. Biol. 23 55-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Apella, E., Weber, I. T., and Blasi, F. (1988) FEB. Lett. 231 1-4 [DOI] [PubMed] [Google Scholar]
- 34.Schenk, S., Hintermann, E., Bilban, M., Koshikawa, N., Hojilla, C., Khokha, R., and Quaranta, V. (2003) J. Cell Biol. 161 197-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swindle, C. S., Tran, K. T., Johnson, T. D., Banerjee, P., Mayes, A. M., Griffith, L., and Wells, A. (2001) J. Cell Biol. 154 459-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luttrell, L. M., Daaka, Y., and Lefkowitz, R. J. (1999) Curr. Opin. Cell Biol. 11 177-183 [DOI] [PubMed] [Google Scholar]
- 37.Moro, L., Venturino, M., Bozzo, C., Silengo, L., Altruda, F., Beguinot, L., Tarone, G., and Defilippi, P. (1998) EMBO J. 17 6622-6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misenheimer, T. M., Huwiler, K. G., Annis, D. S., and Mosher, D. F. (2000) J. Biol. Chem. 275 40938-40945 [DOI] [PubMed] [Google Scholar]
- 39.Misenheimer, T. M., Hannah, B. A., Annis, D. S., and Mosher, D. F. (2003) Biochemistry 42 5125-5132 [DOI] [PubMed] [Google Scholar]
- 40.Misenheimer, T. M., and Mosher, D. F. (2005) J. Biol. Chem. 280 41229-41235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gong, P., Angelini, D. J., Yang, S., Xia, G., Cross, A. S., Mann, D., Bannerman, D. D., Vogel, S. N., and Goldblum, S. E. (2008) J. Biol. Chem. 283 13437-13449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yaish, P., Gazit, A., Gilon, C., and Levitzki, A. (1988) Science 242 933-935 [DOI] [PubMed] [Google Scholar]
- 43.Krüger, A., Arlt, M. J. E., Gerg, M., Kopitz, C., Bernardo, M. M., Chang, M., Mobashery, S., and Fridman, R. (2005) Cancer Res. 65 3523-3526 [DOI] [PubMed] [Google Scholar]
- 44.Gill, G. N., Kawamoto, T., Cochet, C., Le, A., Sato, J. D., Masui, H., McLeod, C., and Mendelsohn, J. (1984) J. Biol. Chem. 259 7755-7760 [PubMed] [Google Scholar]
- 45.O-charoenrat, P., Modjtahedi, H., Rhys-Evans, P., Court, W. J., Box, G. M., and Eccles, S. A. (2000) Cancer Res. 60 1121-1128 [PubMed] [Google Scholar]
- 46.Annis, D. S., Gunderson, K. A., and Mosher, D. F. (2007) J. Biol. Chem. 282 27067-27075 [DOI] [PubMed] [Google Scholar]
- 47.Annis, D. S., Murphy-Ullrich, J. E., and Mosher, D. F. (2006) J. Thromb. Haemostasis 4 459-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, J. W., Sim, S. S., Kim, U.-H., Nishibe, S., Wohl, M. I., Carpenter, G., and Rhee, S. G. (1990) J. Biol. Chem. 265 3940-3943 [PubMed] [Google Scholar]
- 49.Wang, W., and Passaniti, A. (1999) J. Cell. Biochem. 73 321-331 [PubMed] [Google Scholar]
- 50.Ringerike, T., Blystad, F. D., Levy, F. O., Madshus, I. H., and Stang, E. (2002) J. Cell Sci. 115 1331-1340 [DOI] [PubMed] [Google Scholar]
- 51.Chattopadhyay, A., Vecchi, M., Ji, Q.-S., Mernaugh, R., and Carpenter, G. (1999) J. Biol. Chem. 274 26091-26097 [DOI] [PubMed] [Google Scholar]
- 52.Hackel, P. O., Zwick, E., Prenzel, N., and Ullrich, A. (1999) Curr. Opin. Cell Biol. 11 184-189 [DOI] [PubMed] [Google Scholar]
- 53.Miyamoto, S., Teramoto, H., Gutkind, J. S., and Yamada, K. M. (1996) J. Cell Biol. 135 1633-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prenzel, N., Zwick, E., Daub, H., Leserer, M., Abraham, R., Wallasch, C., and Ullrich, A. (1999) Nature 402 884-888 [DOI] [PubMed] [Google Scholar]
- 55.Carpenter, G. (1999) J. Cell Biol. 146 697-702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donnini, S., Morbidelli, L., Taraboletti, G., and Ziche, M. (2004) Life Sci. 74 75-2985 [DOI] [PubMed] [Google Scholar]
- 57.Marinides, G. N., Suchard, S. J., and Mookerjee, B. K. (1994) Kidney Int. 46 350-357 [DOI] [PubMed] [Google Scholar]
- 58.Bein, K., and Simmons, M. (2000). J. Biol. Chem. 275 32167-32173 [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Manzaneque, J. C., Lane, T. F., Ortega, M. A., Hynes, R. O., Lawler, J., and Iruela-Arispe, M. L. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12485-12490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonnefoy, A., Daenens, K., Feys, H. B., De Vos, R., Vandervoort, P., Vermylen, J., Lawler, J., and Hoylaerts, M. F. (2006) Blood 107 955-964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Apte, S. S. (2004) Int. J. Biochem. Cell Biol. 36 981-985 [DOI] [PubMed] [Google Scholar]
- 62.Rocks, N., Paulissen G., El Hour, M., Quesada F., Crahay, C., Gueders, M., Foidart, J. M., Noel, A., and Cataldo, D. (2008) Biochimie (Paris) 90 369-379 [DOI] [PubMed] [Google Scholar]
- 63.Xie, H., Pallero, M. A., Gupta, K., Chang, P., Ware, M. F., Witke, W., Kwiatkowski, D. J., Lauffenburger, D. A., and Murphy-Ullrich, J. E. (1998) J. Cell Sci. 111 615-624 [DOI] [PubMed] [Google Scholar]
- 64.DiPietro, L. A., Nissen, N. N., Gamelli, R. L., Koch, A. E., Pyle, J. M., and Polverini, P. J. (1996) Am. J. Pathol. 148 1851-1860 [PMC free article] [PubMed] [Google Scholar]
- 65.Holbro, T., Civenni, G., and Hynes, N. E. (2003) Exp. Cell Res. 284 99-110 [DOI] [PubMed] [Google Scholar]
- 66.Tuszynski, G. P., Gasic, T. B., Rothman, V. L., Knudsen, K. A., and Gasic, G. J. (1987) Cancer Res. 47 4130-4133 [PubMed] [Google Scholar]