Abstract
During epithelial homeostasis, stem cells divide to produce progenitor cells, which not only proliferate to generate the cell mass but also respond to cellular signaling to transition from a proliferative state to a differentiation state. Such a transition involves functional alterations of transcriptional factors, yet the underlying molecular mechanisms are poorly understood. Recent studies have implicated Kruppel-like factors (KLFs) including KLF5 in the renewal and maintenance of stem/progenitor cells. Here we demonstrate that the pro-proliferative factor KLF5 becomes anti-proliferative upon TGFβ-mediated acetylation in an in vitro model of epithelial homeostasis. In the HaCaT epidermal cell line treated with or without TGFβ, we found that KLF5 was not only essential for cell proliferation, it was also indispensable for TGFβ-induced anti-proliferation in these cells. KLF5 inhibited the expression of p15 (CDKN2B), a cell cycle inhibitor, without TGFβ, but became a coactivator in TGFβ-induced p15 expression in the same cells. Mechanistically, TGFβ recruited acetylase p300 to acetylate KLF5, and acetylation in turn altered the binding of KLF5 to p15 promoter, resulting in the reversal of KLF5 function. These studies not only demonstrate that a basic transcription factor can be both pro-proliferation and anti-proliferation in epithelial homeostasis, they also present a unique mechanism for how transcriptional regulation changes during the transition from proliferation to inhibition of proliferation. Furthermore, they establish KLF5 as an essential cofactor for TGFβ signaling.
Epithelia constitute the surface and lining of many solid tissues and have essential functions in different tissues. They are maintained through epithelial homeostasis, which involves the proliferation of stem/progenitor cells and the differentiation of their daughter cells. Epithelial homeostasis constantly occurs in vivo, and its disruption causes different diseases including cancer (1). At the molecular level, signaling from stroma compartment in a tissue such as transforming growth factor β (TGFβ)2 signaling regulates epithelial homeostasis through a transcriptional network, but the molecular details are not well understood.
The basic transcription factor KLF5 is ubiquitously expressed in many tissues including the breast, colon, intestine, lung, prostate, etc (2–5). KLF5 is highly expressed in proliferating epithelial cells such as immortal but untransformed epithelial cell lines and proliferating primary cultures of epithelial cells, which mostly represent progenitor cells (2, 3, 6, 7). In normal intestine, KLF5 is expressed at a higher level in basal rapidly proliferating cells, but at a lower level in mature and differentiated cells (8) and knock-out of one KLF5 allele significantly reduced the size of villi in mouse intestine (8). In vivo overexpression of KLF5 in epidermis causes hyperplasia of basal cells but lack of mature skin (9), further indicating a pro-proliferative role of KLF5 in epithelial homeostasis. Recently a combination of four transcription factors including KLF4, Oct4, Sox2, and c-Myc was shown to reverse differentiated cells to a pluripotent state (10–12). For the maintenance of self-renewal and pluripotency of embryonic stem cells, KLF4 was found to be dispensable but KLF5 and KLF2 appeared to be required, likely regulating key pluripotency genes (13). These studies suggest that KLF5 plays a pro-proliferative role in stem/progenitor cells including those involved in epithelial homeostasis.
In most malignant epithelial cell lines tested, however, ectopic expression of KLF5 appeared to have a contrasting function in cell proliferation (2, 3, 6). In this study, we aimed to understand whether and how KLF5 has opposite functions during different phases of epithelial homeostasis, and if yes, what mechanism is involved.
EXPERIMENTAL PROCEDURES
Cell Lines and Other Materials—The HaCaT epidermal epithelial cell line was established by Dr. Norbert E. Fusenig of the German Cancer Research Center (14), and was kindly provided to us by Dr. Robert A. Swerlick of Emory University. It was maintained following established procedures (14). The HEK293 human kidney epithelial cell line, the HepG2 hepatoma cell line, and the COS-1 cell line were purchased from American Type Culture Collection (ATCC) and propagated following ATCC instructions. The TGFβ used in this study was TGFβ1 from R&D Systems (Minneapolis, MN).
The siRNA for KLF5, as described previously (15), was used in all experiments except those shown in knockdown-reconstitution experiments, in which siRNAs targeting the UTRs of KLF5 were used (5′-AAGAGCGGAAGAAGAGTTTTGCTT-3′ and 5′-AAGGACTTAGGGTGTCGTCGTTTT-3′).
The antibodies against Smad4 or p15 were purchased from Cell Signaling Technology (Danvers, MA), and the antibody against actin was purchased from Sigma. The antibody against KLF5 was prepared as previously described (16). Polyclonal antibody specific to acetylated lysine 369 of KLF5 was produced using a synthesized peptide that had a sequence of RSNPDLEKRRIHYC (residues 362–375 of KLF5), in which the lysine was acetylated. Antisera from rabbits were affinity-purified using the same peptide, and antibodies against amino acids other than the acetylated lysine were removed by a second affinity purification using a peptide that had identical sequence but unacetylated lysine. Antibody against unacetylated lysine 369 was also produced and purified under the same manner, except that the unacetylated peptide was used as antigen for antibody production and the first affinity purification and acetylated peptide was used for removing nonspecific antibodies in the second affinity purification. Antibody production and purification were conducted by Alpha Diagnostic International (San Antonio, TX).
Cell Proliferation Assay—HaCaT cells were seeded at 2 × 104 cells/well onto 12-well tissue culture plates and grown for 48 h. Chemically synthesized siRNA were transfected into cells for 24 h. Different concentrations of TGFβ were then added and incubated with cells for 18 h, at which point [3H]thymidine (MP Biomedicals, Irvine, CA) was added at 0.5 μCi/well. Four hours later, cells were washed with phosphate-buffered saline, fixed in 5% ice-cold trichloroacetic acid for 2 h, lysed with 1 m NaOH, transferred into glass fiber filters, and dried. Radioactivity was measured using a scintillation counter.
In the knockdown-reconstitution experiments, the K369R mutant of KLF5 expression plasmid, which contained a lysine-to-arginine (AAA → CGT) mutation at amino acid residue 369 of KLF5, was produced by a PCR-based approach, with pcDNA3-FLAG-KLF5 as a template. The entire insert was sequenced to verify the mutation.
Detection of RNA or Protein Expression—Following the procedures described in our previous studies (17, 18), real-time PCR and RT-PCR were performed to analyze RNA expression, while Western blotting was conducted to examine protein expression.
Promoter-Luciferase Reporter Assay—The p15P165-Luc plasmid was kindly provided by Dr. Xiao-Fan Wang of Duke University (19). Wild-type and mutant plasmids were transfected into HaCaT cells using Lipofectamine reagent (Invitrogen) for 24 h. siRNA for KLF5 and a negative control from Dharmacon (Lafayette, CO), were then transfected. On the following day, 100 pm (2 ng/ml) TGFβ was added and incubated with cells for 20 h, before luciferase assay was carried out using the Promega luciferase assay kit as previously described (18).
Oligonucleotide Pull-down Assay with Cell Lysates—Oligonucleotides for the p15 promoter, with biotin added to their 5′-end, were synthesized by MWG-Biotech (High Point, NC). The sequences for the oligonucleotides were as follows: Inr (–12 to +11, see Li et al. (19) for nucleotide numbering), biotin-5′-GGCTGGCTCCCCACTCTGCCAGAG-3′ (wild type), and biotin-5′-GGCTGGCTCAACAATATGCCAGAG-3′ (mutant); SPS2 (–89 bp to –65), biotin-5′-CAGCGGACAGGGGGCGGAGCCTAAG-3′ (wild type), and biotin-5′-CAGCGGACAGGAAGTAGAGCCTAAG-3′ (mutant); and SBE (–443 to –385), biotin-5′-TAACTTGTATGACAGGTGCAGAGCTGTCGCTTTCAGACATCTTAAGAAAGACGGAGTTA-3′ (wild type), and biotin-5′-TAACTTGTATGACAGGTGCAGAGCTGTCGCTTTCACTCATCTTAAGAAAGACGGAGTTA-3′ (mutant). Each pair of oligos was annealed following standard protocols. HaCaT cells treated with 5 ng/ml TGFβ or control solution for 3 h under reduced serum (1%) condition were lysed in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100) containing protease and phosphatase inhibitors. After cell debris was removed by centrifugation, cell extracts were precleared with ImmunoPure streptavidin-agarose beads (20 μl/sample, Pierce) for 1 h at 4 °C. After centrifugation for 1 min at 5000 × g, the supernatant was incubated with 100 pmol of biotinylated double-strand oligonucleotides and 10 μg of poly(dI-dC)·poly(dI-dC) for 16 h at 4 °C. DNA-bound proteins were collected with 30 μl of immobilized streptavidin-agarose beads for 1 h at 4 °C, washed with lysis buffer for 4 times, separated on a 10% SDS-polyacrylamide gel, and subjected to Western blotting with different antibodies.
Co-immunoprecipitation (Co-IP) Assay—Co-IP was conducted following the standard protocol using anti-FLAG M2 affinity gel (Sigma).
Oligonucleotide Pull-down Assay with in Vitro-translated KLF5—Full-length and mutant KLF5 proteins were produced by in vitro translation in the presence of [35S]methionine (GE Healthcare, Piscataway, NJ) using the TnT® Quick-coupled Transcription/Translation Systems (Promega) as described in our previous study (18). 2 μl of in vitro-translated product were used in each oligonucleotide pull-down assay as described above.
Chromatin Immunoprecipitation (ChIP) Assay—HepG2 cells were transfected with pcDNA3-FLAG-KLF5 (16) or pcDNA3-FLAG (Invitrogen) plasmid with the Lipofectamine 2000 reagent (Invitrogen). Forty hours after transfection, cells were incubated in the presence or absence of 5 ng/ml TGFβ for 1 h. ChIP was performed according to the protocol from Upstate (Lake Placid, NY). FLAG antibody-agarose beads (Sigma) was used to protein/DNA complex. Precipitated DNA was analyzed by PCR with CDKN2B promoter-specific primers (5′-CTGGACATCCAGCGAGCAGTG-3′ and 5′-GAGCTCAAAGCCGCTCTGGCC-3′). Human β-actin gene was used as a control, with the following primers: 5′-CGGAGGG CGCCCCAACTCAG-3′ (forward) and 5′-GCGCGCGCGGCCCCAGAACA-3′ (reverse).
RESULTS
TGFβ Reverses the Function of KLF5 from Pro-proliferative to Anti-proliferative in Epithelial Homeostasis—The HaCaT epidermal epithelial cell line, treated with or without TGFβ, was chosen as a model of epithelial homeostasis for analysis because numerous previous studies have established the relevance of this model to epithelial homeostasis. For example, HaCaT cells share molecular markers and phenotypes with stem/progenitor keratinocytes (20, 21) and they also secret stem cell factor (22). In addition, TGFβ induces differentiation of HaCaT cells, and use of HaCaT cells and their TGFβ-treated counterparts has yielded great insights into epithelial homeostasis (23–25). As in other untransformed epithelial cell lines, KLF5 is highly expressed in HaCaT cells (26).
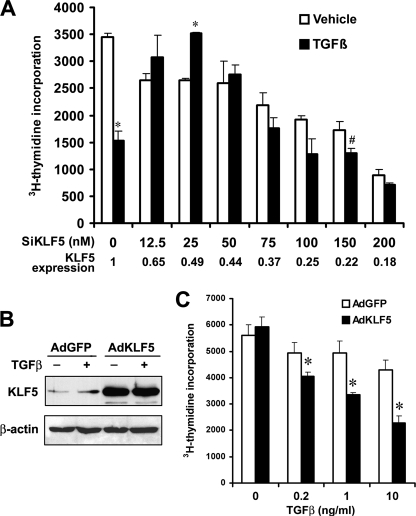
Inhibition of cell proliferation is the most significant outcome in TGFβ-mediated differentiation of HaCaT cells, in addition to other molecular and phenotypic alterations. Therefore, we examined the function of KLF5 in the proliferation of HaCaT epithelial cells. Consistent with previous findings that showed a stimulatory role of KLF5 in the proliferation of epithelial cells (6, 7) and in the renewal of embryonic stem cells (13), we found that knockdown of KLF5 expression by transfecting small interfering RNA (siRNA) significantly repressed cell proliferation (Fig. 1A). When TGFβ was added, TGFβ significantly inhibited cell proliferation without KLF5 knockdown, which is also expected (24, 25). In cells where KLF5 had been knocked down, however, TGFβ failed to inhibit cell proliferation, and even showed a trend of increasing cell proliferation to some degree with lower concentrations of siRNA (Fig. 1A). We performed similar experiments in another epithelial cell line, the PZ-HPV-7 cell line immortalized from normal human prostatic epithelial cells (27), and also found that knockdown of KLF5 significantly suppressed TGFβ's inhibitory effect on cell proliferation (supplemental Fig. S1).
FIGURE 1.
Pro-proliferative KLF5 is necessary for TGFβ-mediated inhibition of proliferation in epithelial cells. A, cell proliferation, as indicated by the incorporation rate of [3H]thymidine into DNA, in HaCaT cells treated with indicated concentrations of siRNA for KLF5 (SiKLF5) in the presence or absence of TGFβ (1 ng/ml). The negative control for siRNA treatment (the first set of bars) contained no siRNA for KLF5 but 100 nm siRNA for the luciferase gene (SiLuc). Expression of KLF5 was measured by real-time PCR. Data for each point, expressed as means (bars) ± S.E. (error bars), were from 3 wells of cells. The experiment was repeated twice and consistent results were obtained. B, KLF5 expression mediated by adenoviral infection of KLF5 (AdKLF5) in PC-3 prostate cancer cells, as detected by Western blot analysis. AdGFP serves as a control. C, increased KLF5 expression sensitized PC-3 cells to TGFβ inhibitory effect on cell proliferation. A p value of 0.05 or smaller for comparison between TGFβ-treated and untreated groups is indicated by an asterisk (*), while that between KLF5 knockdown and control in TGFβ-treated cells is indicated by the pound sign (#).
In the PC-3 prostate cancer cell line, which expresses little KLF5 protein due to excessive protein degradation (16) and showed a weak response to TGFβ growth inhibitory effect (28), we restored KLF5 expression by adenoviral infection and analyzed the function of KLF5 in cell proliferation in the context of TGFβ (Fig. 1, B and C). Infection efficiencies of adenoviruses in PC-3 cells were 86 and 81% for AdGFP and AdKLF5, respectively. Consistent with published findings, PC-3 cells infected with control viruses only showed a minimal response to the inhibitory effect of TGFβ (Fig. 1C). However, increasing KLF5 expression clearly restored the response of PC-3 cells to TGFβ in a dose-dependent manner similar to that observed in untransformed cells (Fig. 1C). These results indicate that, in some cancer cells where KLF5 expression is impaired due to different mechanisms (2, 3, 16), restoration of KLF5 expression can restore TGFβ inhibitory function.
A positive role of KLF5 in cell proliferation has been repeatedly demonstrated in different types cells including epithelial cells (2, 3, 6–8, 29). Our findings that knockdown of KLF5 reduced cell proliferation in the HaCaT epidermal epithelial cell line further support this conclusion. More interestingly, our findings in this study indicate that TGFβ, a well-known growth inhibitor in epithelial cells, converts KLF5 into an inhibitory factor in proliferating epithelial cells. Our results further suggest that a transcription factor can reverse function during the transition from proliferation to inhibition of proliferation in epithelial homeostasis.
TGFβ Also Reverses the Function of KLF5 in Gene Regulation—During TGFβ-induced epithelial differentiation, cell cycle progression is inhibited in the G1 phase through transcriptional regulation of a set of genes. The cyclin-dependent kinase inhibitor p15 (CDKN2B) is a principal effector for TGFβ-mediated growth inhibition (24, 30, 31). To address whether KLF5 functions by regulating the expression of genes including p15 in epithelial homeostasis, we determined whether KLF5 regulates p15 expression in cells treated with or without TGFβ.
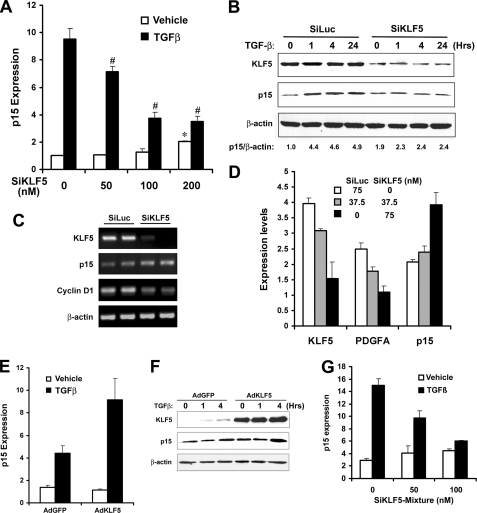
Without TGFβ signaling, knockdown of KLF5 expression increased p15 expression in HaCaT cells at both RNA and protein levels, as detected by real-time PCR, Western blot, and RT-PCR analysis (Fig. 2, A–D). Consistent with published studies (6, 15), knockdown of KLF5 also suppressed the expression of cyclin D1 and PDGFA (Fig. 2, C and D), both of which are induced by KLF5 in proliferating cells. In agreement with a positive role of KLF5 in cell proliferation without TGFβ, these results suggest that, in proliferating epithelial cells, KLF5 inhibits the expression of p15 without TGFβ. When TGFβ treatment was applied to HaCaT cells, the expression of p15 was dramatically induced at both RNA and protein levels as expected (Fig. 2, A and B), which is consistent with previous findings establishing p15 as a principal effector of TGFβ (24, 30, 31). However, p15 induction was significantly impaired by the knockdown of KLF5 expression (Fig. 2, A and B), indicating a necessary role of KLF5 in TGFβ-mediated p15 expression. These results indicate that when TGFβ is activated, the function of KLF5 in regulating p15 expression is reversed from inhibition to activation. Knockdown of KLF5 also inhibited TGFβ-induced expression of p21, another TGFβ target gene mediating its anti-proliferative effect in HaCaT cells (25) (data not shown). In the PC-3 prostate cancer cell line, restoration of KLF5 expression also enhanced TGFβ-induced p15 expression (Fig. 2, E and F).
FIGURE 2.
Inhibitory effect of KLF5 on p15 expression is reversed by TGFβ in HaCaT cells. A, knockdown of KLF5 increased p15 expression but inhibited TGFβ-induced p15 expression in HaCaT cells, as measured by real-time PCR assay. SiKLF5, siRNA for KLF5. TGFβ treatment was at 1 ng/ml for 20 h. The negative control for siRNA treatment (the first set of bars on far left) was contained no siRNA for KLF5 but 100 nm siRNA for the luciferase gene. A p value of 0.05 or smaller for comparison between KLF5 knockdown and control is indicated by asterisk (*) in cells without TGFβ treatment or by the pound sign (#) in TGFβ-treated cells. B, protein expression of p15 and KLF5, as detected by Western blot analysis, in HaCaT cells transfected with 75 nm SiKLF5 or SiLuc siRNA and treated with 1 ng/ml TGFβ for different times (hours). C, expression of KLF5, p15, and cyclin D1 in HaCaT cells treated with 100 nm SiKLF5 or SiLuc, as examined by RT-PCR assay in duplicate reactions. D, expression of p15, PDGFA, and KLF5, as detected by real-time PCR assay, in HaCaT cells transfected with different concentrations of SiKLF5 and SiLuc as indicated. E, p15 RNA expression, detected by real-time PCR assay, in PC-3 prostate cancer cells infected with adenoviruses for KLF5 (AdKLF5) or GFP control (AdGFP), with or without 1 ng/ml of TGFβ. F, protein expression of p15 and KLF5, as detected by Western blot analysis, in PC-3 cells infected with AdGFP or AdKLF5 adenoviruses and treated with 1 ng/ml TGFβ for 0, 1, and 4 h. G, transfection of a mixture of two siRNAs against sequences in the 5′- and 3′-UTR regions of KLF5 also increased p15 expression without TGFβ but impaired TGFβ-induced p15 expression, as measured by real-time PCR assay in HaCaT cells. TGFβ concentration was 1 ng/ml. β-actin served as a loading control in panels B and F.
Acetylation of KLF5 Is Responsible for TGFβ-mediated Functional Reversal of KLF5—To understand how the function of KLF5 is reversed by TGFβ, we reasoned that TGFβ-caused alterations, either directly in KLF5 or indirectly in transcription factors that interact with KLF5, should be responsible. It is known that, upon TGFβ treatment in epithelial cells, acetylase p300 is recruited to the Smad2/3/4 complex to acetylate proteins and mediate TGFβ-induced transcription (32, 33). It is also known that KLF5 can be phosphorylated and acetylated, and these modifications modulate KLF5 activity (34, 35). Taken together with the fact that the regulation of p15 by KLF5 occurs both before and after TGFβ treatment, the best possibility is that TGFβ induces the acetylation of KLF5, and KLF5 acetylation alters the binding of KLF5 to gene promoter to reverse its function in gene regulation and cell proliferation.
In testing this hypothesis, we first examined whether TGFβ alters the acetylation of KLF5. Using an antibody specific for acetylated Lys-369 residue of KLF5 (35), we found that, although acetylated KLF5 was detectable in untreated HaCaT cells, TGFβ treatment dramatically increased the acetylation of KLF5 protein (Fig. 3A). We further tested the role of TGFβ in KLF5 acetylation using COS-1 cells co-transfected with FLAG-tagged KLF5 or the acetylation-deficient K369R mutant (35), HA-tagged p300, and TGF-βRI (TβRI), a TGFβ receptor that auto-activates the TGFβ pathway (36). Whereas expression of p300 induced acetylation of KLF5 and the K369R mutation abolished KLF5 acetylation as expected (35), TGFβ signal significantly enhanced p300-induced KLF5 acetylation, (Fig. 3B).
FIGURE 3.
Acetylation of KLF5 is responsible for TGFβ-mediated functional reversal of KLF5 in gene regulation and cell proliferation. A, TGFβ increases the acetylation of endogenous KLF5 at lysine 369. HaCaT cells were incubated with 5 μm trichostatin A (TSA) for 18 h to enhance the detection of acetylation, and then treated with TGFβ (5 ng/ml) for indicated times. Cell lysates were subjected to Western blotting analysis with an antibody specifically against acetylated lysine at 369. Actin serves as a loading control. Ac-KLF5, KLF5 with acetylation at lysine 369. B, acetylase p300 induces KLF5 acetylation, which is enhanced by TGFβ, while the K369R mutation of KLF5 abolishes its acetylation. COS-1 cells were transfected with indicated plasmids, and were subjected to IP with FLAG-antibody-conjugated beads and immunoblotting with antibodies against pan-acetyl-lysines (AcK). The input was detected by using the anti-FLAG antibody. Cells were incubated with 5 μm TSA for 18 h to enhance the detection of KLF5 acetylation. C and D, acetylation of KLF5 is indispensable for TGFβ to induce p15 expression and inhibit cell proliferation. HaCaT cells were transfected with KLF5 siRNAs from UTRs along with wild type or mutant KLF5. TGFβ was at 0 or 1 ng/ml, and is indicated by – or + (D) or white or black bars (C). Protein expression was detected by Western blotting, and names of the molecules are shown at the right (D). For panels D, band intensities were determined by using the ImageJ program, and ratios of KLF5 to β-actin protein quantities are provided under the lower panel.A p value of 0.05 or smaller for comparison between TGFβ-treated and untreated groups is indicated by an asterisk (*), while that between KLF5 knockdown and control in TGFβ-treated cells is indicated by the pound sign (#).
We then determined whether acetylation of KLF5 is involved in the reversal of KLF5 function in gene regulation and cell proliferation. We developed a knockdown-reconstitution approach that enabled us to compare wildtype KLF5 and the acetylation-deficient K369R mutant of KLF5 in functional assays. We transfected siRNAs against untranslated region (UTR) sequences of KLF5, along with plasmids expressing FLAG-tagged wild type KLF5 or the acetylation-deficient K369R mutant, into HaCaT cells. These siRNAs target endogenous KLF5 but do not affect transfected KLF5, because the expression constructs do not have the UTR sequences targeted by the siRNAs. These siRNAs also efficiently knocked down KLF5 and inhibited cell proliferation as the other siRNA did (Fig. 2G), excluding the off-target effect of the KLF5 siRNAs. Interestingly, the reconstituted wild-type KLF5 still inhibited cell proliferation with TGFβ, but the reconstituted acetylation-deficient mutant lost this capability (Fig. 3C). These results not only validated the results in Fig. 1A, they also indicate that acetylation is necessary for KLF5 to reverse function and mediate TGFβ-mediated inhibition of proliferation in epithelial cells.
Using the same knockdown-reconstitution approach, we also analyzed the role of acetylation in KLF5-regulated p15 expression. Consistent with the data in cell proliferation, we found that reconstituted wild-type KLF5 still induced p15 expression with TGFβ, but the reconstituted acetylation-deficient mutant failed to do so (Fig. 3D), indicating that acetylation of KLF5 is indispensable for TGFβ to induce p15 expression.
KLF5 Regulates p15 Expression through Direct Promoter Binding in an Acetylation-responsive Manner—To further understand how KLF5 exerts opposing functions in the context of TGFβ, we conducted biochemical and molecular analyses to determine whether KLF5 directly binds to p15 promoter to regulate p15 expression, and whether acetylation of KLF5 alters such a binding.
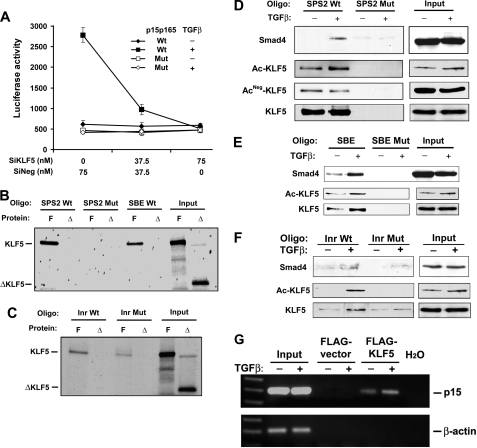
We first tested whether KLF5 binds to p15 promoter. In the promoter of p15, the transcriptional initiator (Inr) has a sequence of 5′-CCCACTCTG-3′ that belongs to the consensus KLF5 binding CA box (CCCACTC), and has been implicated in TGFβ-induced p15 expression by its binding to Miz-1, a MYC-interacting protein (24). A promoter-luciferase reporter plasmid containing this sequence, p15p165, has been established previously and shown to respond to TGFβ in expressing the luciferase reporter gene (19). We mutated this potential KLF5 binding site to 5′-AACAATATG-3′, and then determined the effect of KLF5 on TGFβ-mediated promoter activity. Upon transfection into HaCaT cells, neither wildtype nor mutant p15 promoter showed any activity when TGFβ was absent (Fig. 4A), regardless of the status of KLF5 expression, which is consistent with previous findings (19, 24). With the addition of TGFβ, significant luciferase activity was induced in cells expressing KLF5, and this activity was abolished if KLF5 expression was knocked down (Fig. 4A). The mutant promoter still did not show any activity even when TGFβ was added. These results indicate that KLF5 plays an indispensable role in TGFβ-mediated p15 induction, and that the Inr sequence could be one of the KLF5 binding sites in p15 promoter.
FIGURE 4.
KLF5 binds to p15 promoter and the binding is altered by TGFβ-mediated acetylation. ?A, knockdown of KLF5 expression diminishes TGFβ-induced p15 promoter activity, as determined by luciferase assay, in HaCaT cells. SiKLF5 and SiNeg are siRNA for KLF5 and negative control respectively. P15p165 is the p15 promoter-luciferase reporter plasmid containing both Inr and SPS2 elements but not the SBE element (19). TGFβ was at 2 ng/ml. B and C, binding of in vitro-translated full-length KLF5 (F) but not C terminus-truncated KLF5 (Δ) to different sites of p15 promoter, as detected by oligo pull-down assay. D–F, different bindings for acetylated and unacetylated KLF5 to different DNA elements (Oligo) of the p15 promoter including Inr, SPS2, and SBE, as detected by oligo pull-down assay combined with Western blot analysis in HaCaT cells. Inputs represent samples without oligo pull-down. Mutant (Mut) oligos were used as controls for wild type (Wt) oligos. TGFβ treatment was at 5 ng/ml for 1–3 h. Preparation of antibodies against acetylated and unacetylated lysine 369 is described under “Experimental Procedures.” G, in vivo binding of KLF5 to p15 promoter, as detected by the ChIP assay in HepG2 cells. Specific PCR products spanning Inr and SPS2 are seen in FLAG-tagged KLF5-bound DNA but not in controls. TGFβ treatment was at 5 ng/ml for 1 h.
In addition to the Inr sequence, two other sequences in the promoter of p15 also play important roles in its induction by TGFβ, as shown in previous studies (19, 24, 37). One is a Smad-binding element (SBE), which is located at –443/-385, and the other is the sequence harboring two Sp1 sites, especially the Sp1 site at –89/–65 (SPS2 for Sp1 site 2). They both have GC-rich sequences that are potential KLF5 binding sites (GGGCGG in SPS2 and GTCGC in SBE), and thus could also mediate KLF5 function in regulating p15 expression. We performed an oligo pull-down assay using in vitro-translated 35S-labeled full-length KLF5 protein and a truncated form of KLF5 that lacked the C-terminal DNA-binding zinc finger domains. Full-length KLF5 bound to each of the 3 sequences, but the truncated form did not (Fig. 4, B and C). Furthermore, the binding of KLF5 to SPS2 was stronger than that to SBE or Inr, and no binding was detected when mutant SPS2 oligo was used (Fig. 4B). Therefore, KLF5 can bind to p15 promoter DNA without associating with other factors.
We also performed oligo pull-down experiments for Inr, SPS2, and SBE using cell lysates from HaCaT cells treated with TGFβ, with Smad4 as a positive control because the binding of Smad4 to Inr and SBE has been reported previously (24). Biotin-labeled oligonucleotides for wild type and mutant SPS2, SBE, and Inr were incubated with lysates of HaCaT cells treated with and without TGFβ, followed by pull-down with streptavidin-agarose beads and Western blot analysis. Consistent with stronger binding of in vitro translated KLF5 to SPS2, endogenous KLF5 also had a higher affinity to the SPS2 sequence. In addition, the binding to SPS2 occurred for both acetylated and unacetylated KLF5, and TGFβ might not make a significant difference in the binding (Fig. 4D). For SBE and Inr elements, endogenous KLF5 also bound, and the binding was significantly increased by TGFβ treatment (Fig. 4, E and F). Furthermore, both the antibody against total KLF5 and that against acetylated KLF5 showed an increased binding of KLF5 after TGFβ treatment, but the antibody against unacetylated KLF5 did not detect any KLF5 binding (Fig. 4, E and F), indicating that only acetylated KLF5 binds to the SBE and Inr sequences. Mutation in each of the three sequences abolished or dramatically decreased their binding to both Smad4 and KLF5, with or without TGFβ treatment (Fig. 4, D–F). These results indicate that, without TGFβ, more KLF5 molecules are unacetylated and they only bind to the SPS2 site of p15 promoter. When TGFβ signaling is activated, a certain amount of KLF5 molecules are acetylated, and acetylated KLF5 binds to each of the three DNA elements.
To determine whether the binding of KLF5 to p15 promoter occurs in a physiological status, a ChIP assay was performed. Because our KLF5 antibodies did not work in immunoprecipitation and the transfection efficiency for HaCaT cells was rather limited, we chose to transfect FLAG-tagged KLF5 into the HepG2 carcinoma cell line, another line frequently used to study TGFβ, for the ChIP assay. As shown in Fig. 4G, binding of KLF5 to p15 promoter could be detected at least for the sequence spanning the Inr and SPS2 sites, and the binding was enhanced by TGFβ treatment.
DISCUSSION
A Transcription Factor Can Reverse Function upon Signaling-induced Modification in Epithelial Homeostasis—In this study, we first confirmed that the KLF5 transcription factor plays a positive role in the proliferation of epithelial cells (2, 3, 6, 7, 9, 13, 38) (Fig. 1). More importantly, we found that KLF5 reverses its function to mediate TGFβ-caused proliferation inhibition in epithelial homeostasis (Fig. 1). Furthermore, we found that KLF5 inhibits the expression of p15 and induces the expression of cyclin D1 but mediates TGFβ-mediated p15 induction and cyclin D1 suppression (Fig. 2). One mechanism for TGFβ-determined opposing functions of KLF5 is that KLF5 binds to gene promoters, and TGFβ-caused acetylation of KLF5 alters the binding of KLF5 to gene promoters, which in turn reverses the function of KLF5 in gene regulation (Figs. 3 and 4). Effect of acetylation on the binding of KLF5 to a promoter has been demonstrated for the PDGFA gene (35). The transition from proliferation to differentiation in epithelial homeostasis requires changes not only in the transcription of genes, but also in the function of transcription factors that regulate these genes. While conventional thinking is that, during the transition, some transcription factors are turned on while others are turned off, our findings indicate that the same transcription factor can reverse function upon signaling-induced modification. From an evolutionary point of view, it would be more advantageous to reverse the function of a factor by modification than to turn off one factor and turn on another.
We speculate that a balance between acetylated KLF5 and unacetylated KLF5 is important for maintaining a balanced proliferation and differentiation during epithelial homeostasis. Protein level of KLF5 is tightly regulated by the ubiquitin-proteasome pathway in epithelial cells (16, 18), indicating that the level of KLF5 in a cell is critical for its proper function. Insufficient KLF5 could lead to a reduced rate of cell proliferation and the production of differentiated cells, while too many unacetylated KLF5 molecules could lead to excessive proliferation and insufficient differentiation. In either case, an interruption of the balance between acetylated and unacetylated KLF5 could interrupt normal homeostasis. Two in vivo studies support this prediction (8, 9). In a knock-out study, while knock-out of both KLF5 alleles was embryonically lethal, knock-out of one KLF5 allele significantly reduced the size of villi in mouse intestine (8). We predict that the reduction in differentiated epithelium is not only due to a reduced cell proliferation rate caused by loss of KLF5, it could also be due to impaired differentiation of proliferating crypt cells. In another report, overexpression of KLF5 in epidermis caused hyperplasia of basal cells but lack of mature skin (9). Our explanation is that overexpression of KLF5 results in too much unacetylated KLF5, which not only caused hyperplasia of basal cells, but also led to a reduced ratio of acetylated KLF5 and reduced rate of differentiation.
KLF5 as an Integral Player in the TGFβ Tumor Suppressor Pathway—TGFβ signaling is an important pathway involved in multiple biological processes including epithelial homeostasis (25). Defects in the TGFβ signal transduction impair development and epithelial homeostasis, and result in uncontrolled proliferation and neoplasia in different human tissues (25, 39, 40). Understanding how TGFβ functions has been an important area of investigation, and several transcription factors mediating TGFβ function have been identified (24, 25). In this study, we found that KLF5 is necessary for TGFβ to inhibit cell proliferation and to regulate gene expression (Figs. 1 and 2), and acetylation deficiency in KLF5 interrupts TGFβ function (Fig. 3). These findings establish KLF5 as an important co-factor for TGFβ function.
In summary, we found that transcription factor KLF5 reverses its function in gene regulation and cell proliferation upon the activation of TGFβ signaling. TGFβ-mediated acetylation of KLF5 alters the binding of KLF5 to gene promoter, which in turn reverses the function of KLF5. This previously undescribed molecular process should be helpful for understanding the transition of progenitor cells from proliferation to differentiation during epithelial homeostasis.
Supplementary Material
Acknowledgments
We thank Drs. Norbert Fusenig of the German Cancer Research Centre, Dr. Robert Swerlick of Emory University, Drs. Lisa Choy and Rik Derynck of the University of California, San Francisco, Dr. Bert Vogelstein of Johns Hopkins University, Dr. Diane Simeone of the University of Michigan, Dr. Martin Eilers of the University of Marburg, Dr. Chen Yan of Indiana University, Dr. Tony Kouzarides of the University of Cambridge, and Dr. Xiao-Fan Wang of Duke University for providing various materials. We also thank Dr. Ceshi Chen for technical advice.
This work was supported, in whole or in part, by Grant R01CA87921 from the NCI, National Institutes of Health. This work was also supported by the Georgia Cancer Coalition. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: TGFβ, transforming growth factor β; KLF, Kruppel-like factor; ChIP, chromatin immunoprecipitation assay; UTR, untranslated region; siRNA, small interfering RNA; GFP, green fluorescent protein.
References
- 1.Wakefield, L. M., and Stuelten, C. (2007) Cancer Cell 12 293–295 [DOI] [PubMed] [Google Scholar]
- 2.Chen, C., Bhalala, H. V., Qiao, H., and Dong, J. T. (2002) Oncogene 21 6567–6572 [DOI] [PubMed] [Google Scholar]
- 3.Chen, C., Bhalala, H. V., Vessella, R. L., and Dong, J. T. (2003) Prostate 55 81–88 [DOI] [PubMed] [Google Scholar]
- 4.Conkright, M. D., Wani, M. A., Anderson, K. P., and Lingrel, J. B. (1999) Nucleic Acids Res. 27 1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi, H., Zhang, Z., Wang, X., Liu, S., and Teng, C. T. (1999) Nucleic Acids Res. 27 4807–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman, N. W., Tan, D., Pestell, R. G., Black, J. D., and Black, A. R. (2004) J. Biol. Chem. 279 12093–12101 [DOI] [PubMed] [Google Scholar]
- 7.Chanchevalap, S., Nandan, M. O., Merlin, D., and Yang, V. W. (2004) FEBS Lett. 578 99–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shindo, T., Manabe, I., Fukushima, Y., Tobe, K., Aizawa, K., Miyamoto, S., Kawai-Kowase, K., Moriyama, N., Imai, Y., Kawakami, H., Nishimatsu, H., Ishikawa, T., Suzuki, T., Morita, H., Maemura, K., Sata, M., Hirata, Y., Komukai, M., Kagechika, H., Kadowaki, T., Kurabayashi, M., and Nagai, R. (2002) Nat. Med. 8 856–863 [DOI] [PubMed] [Google Scholar]
- 9.Sur, I., Rozell, B., Jaks, V., Bergstrom, A., and Toftgard, R. (2006) J. Cell Sci. 119 3593–3601 [DOI] [PubMed] [Google Scholar]
- 10.Okita, K., Ichisaka, T., and Yamanaka, S. (2007) Nature 448 313–317 [DOI] [PubMed] [Google Scholar]
- 11.Wernig, M., Meissner, A., Foreman, R., Brambrink, T., Ku, M., Hochedlinger, K., Bernstein, B. E., and Jaenisch, R. (2007) Nature 448 318–324 [DOI] [PubMed] [Google Scholar]
- 12.Park, I. H., Zhao, R., West, J. A., Yabuuchi, A., Huo, H., Ince, T. A., Lerou, P. H., Lensch, M. W., and Daley, G. Q. (2008) Nature 451 141–146 [DOI] [PubMed] [Google Scholar]
- 13.Jiang, J., Chan, Y. S., Loh, Y. H., Cai, J., Tong, G. Q., Lim, C. A., Robson, P., Zhong, S., and Ng, H. H. (2008) Nat. Cell Biol. 10 353–360 [DOI] [PubMed] [Google Scholar]
- 14.Boukamp, P., Petrussevska, R. T., Breitkreutz, D., Hornung, J., Markham, A., and Fusenig, N. E. (1988) J. Cell Biol. 106 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aizawa, K., Suzuki, T., Kada, N., Ishihara, A., Kawai-Kowase, K., Matsumura, T., Sasaki, K., Munemasa, Y., Manabe, I., Kurabayashi, M., Collins, T., and Nagai, R. (2004) J. Biol. Chem. 279 70–76 [DOI] [PubMed] [Google Scholar]
- 16.Chen, C., Sun, X., Ran, Q., Wilkinson, K. D., Murphy, T. J., Simons, J. W., and Dong, J. T. (2005) Oncogene 24 3319–3327 [DOI] [PubMed] [Google Scholar]
- 17.Sun, X., Frierson, H. F., Chen, C., Li, C., Ran, Q., Otto, K. B., Cantarel, B. L., Vessella, R. L., Gao, A. C., Petros, J., Miura, Y., Simons, J. W., and Dong, J. T. (2005) Nat. Genet. 37 407–412 [DOI] [PubMed] [Google Scholar]
- 18.Chen, C., Sun, X., Guo, P., Dong, X. Y., Sethi, P., Cheng, X., Zhou, J., Ling, J., Simons, J. W., Lingrel, J. B., and Dong, J. T. (2005) J. Biol. Chem. 280 41553–41561 [DOI] [PubMed] [Google Scholar]
- 19.Li, J. M., Nichols, M. A., Chandrasekharan, S., Xiong, Y., and Wang, X. F. (1995) J. Biol. Chem. 270 26750–26753 [DOI] [PubMed] [Google Scholar]
- 20.Lemaitre, G., Gonnet, F., Vaigot, P., Gidrol, X., Martin, M. T., Tortajada, J., and Waksman, G. (2005) Proteomics 5 3637–3645 [DOI] [PubMed] [Google Scholar]
- 21.Wan, H., Yuan, M., Simpson, C., Allen, K., Gavins, F. N., Ikram, M. S., Basu, S., Baksh, N., O'Toole, E. A., and Hart, I. R. (2007) Stem. Cells 25 1286–1297 [DOI] [PubMed] [Google Scholar]
- 22.Grabbe, J., Welker, P., Rosenbach, T., Nurnberg, W., Kruger-Krasagakes, S., Artuc, M., Fiebiger, E., and Henz, B. M. (1996) J. Invest. Dermatol. 107 219–224 [DOI] [PubMed] [Google Scholar]
- 23.Fusenig, N. E., and Boukamp, P. (1998) Mol. Carcinog. 23 144–158 [DOI] [PubMed] [Google Scholar]
- 24.Seoane, J., Pouponnot, C., Staller, P., Schader, M., Eilers, M., and Massague, J. (2001) Nat. Cell Biol. 3 400–408 [DOI] [PubMed] [Google Scholar]
- 25.Siegel, P. M., and Massague, J. (2003) Nat. Rev. Cancer 3 807–821 [DOI] [PubMed] [Google Scholar]
- 26.Sur, I., Unden, A. B., and Toftgard, R. (2002) Eur. J. Cell Biol. 81 323–334 [DOI] [PubMed] [Google Scholar]
- 27.Greiff, A. H., Fischer, W. M., and Sehgal, I. (2002) Clin. Exp. Metastasis 19 727–733 [DOI] [PubMed] [Google Scholar]
- 28.Park, B. J., Park, J. I., Byun, D. S., Park, J. H., and Chi, S. G. (2000) Cancer Res. 60 3031–3038 [PubMed] [Google Scholar]
- 29.Chen, C., Benjamin, M. S., Sun, X., Otto, K. B., Guo, P., Dong, X. Y., Bao, Y., Zhou, Z., Cheng, X., Simons, J. W., and Dong, J. T. (2006) Int. J. Cancer 118 1346–1355 [DOI] [PubMed] [Google Scholar]
- 30.Hannon, G. J., and Beach, D. (1994) Nature 371 257–261 [DOI] [PubMed] [Google Scholar]
- 31.Sandhu, C., Garbe, J., Bhattacharya, N., Daksis, J., Pan, C. H., Yaswen, P., Koh, J., Slingerland, J. M., and Stampfer, M. R. (1997) Mol. Cell. Biol. 17 2458–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massague, J., Seoane, J., and Wotton, D. (2005) Genes Dev. 19 2783–2810 [DOI] [PubMed] [Google Scholar]
- 33.Simonsson, M., Kanduri, M., Gronroos, E., Heldin, C. H., and Ericsson, J. (2006) J. Biol. Chem. 281 39870–39880 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Z., and Teng, C. T. (2003) Nucleic Acids Res. 31 2196–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto, S., Suzuki, T., Muto, S., Aizawa, K., Kimura, A., Mizuno, Y., Nagino, T., Imai, Y., Adachi, N., Horikoshi, M., and Nagai, R. (2003) Mol. Cell. Biol. 23 8528–8541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagarajan, R. P., Zhang, J., Li, W., and Chen, Y. (1999) J. Biol. Chem. 274 33412–33418 [DOI] [PubMed] [Google Scholar]
- 37.Feng, X. H., Liang, Y. Y., Liang, M., Zhai, W., and Lin, X. (2002) Mol. Cell 9 133–143 [DOI] [PubMed] [Google Scholar]
- 38.Bruce, S. J., Gardiner, B. B., Burke, L. J., Gongora, M. M., Grimmond, S. M., and Perkins, A. C. (2007) BMC Genomics 8 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, A. B., and Wakefield, L. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8621–8623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts, A., and Mishra, L. (2005) Oncogene 24 5667 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.